Translate this page into:
Evaluation of SSR-based genetic diversity, protein and mineral content in black gram genotypes
⁎Corresponding author. sushil254386@yahoo.com (Sushil Kumar)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Black gram [Vigna mungo (L.) Hepper] is an important food source which is rich in digestible good quality protein and many minerals. But the information on grain minerals and SSR marker based variability study is scanty. Therefore, to examine the variability for protein and minerals in black gram, 60 genotypes were screened for protein and three minerals (calcium, potassium and phosphorus) along with 11 SSR markers. The germplasm displayed ample variability for protein and minerals. The average value of protein, calcium, potassium and phosphorus content was 22.96%, 135.26 mg/100 g, 468.59 mg/100 g and, 325.96 mg/100 g. Phosphorus was significantly correlated with potassium in positive direction. A total of 66 alleles were identified from 60 black gram genotypes. The number of alleles per SSR marker was six with a range of 2–12. PIC value ranged from 0.30 (CEDG 92) to 0.90 (Vmg SSR 29 and Vmg SSR 53) with average PIC of 0.60. The major allelic frequency ranged from 0.13 to 0.78. The minimum and maximum molecular distance were found to be 0.09–1.00. The average genetic distance between 60 black gram genotypes was 0.69. The current experiment indicated that the studied genotypes exhibited ample genetic variability at biochemical and DNA level and can be used for black gram improvement during selection and hybridization program.
Keywords
Black gram
Genetic diversity
Minerals
Protein
SSR
1 Introduction
Black gram or urd bean (Vigna mungo (L.) Hepper) is an important grain legume due to it’s nutrient’s content and the aptness to cropping system. It is believed that black gram was domesticated from a wild progenitor, Vigna mungo var. silvestris in India (Kaewwongwal et al., 2015). This annual and diploid legume has 22 chromosomes. Seeds of black gram contribute 76% of carbohydrates, 25% of protein, 3–5% of fiber and 1.74% of fat. It is also a good source of lysine for vegetarians (Elangaimannan et al., 2008). Black gram flour is an important raw material for food industries which deals with snacks and cookies. In the form of soup, it works as medicines to cure dyspepsia, gastric catarrh, dysentery and diarrhoea. The foliage of black gram is also a good feed and fodder for animals and a worthy green manure crop (Anonymous, 2006).
Black gram is largely cultivated in many tropical and subtropical countries (India, Nepal, Sri Lanka, Thailand, Myanmar, etc.) of the world. In India, it is cultivated in an area of about 3.62 million hectares with production of 1.94 million tones with an average productivity of about 537 kg/ha (Anonymous, 2016). Due to a short duration legume of 70–90 days, it is mainly cultivated as fallow as well as inter-crop. Being a member of Leguminaceae family, it also fixes nitrogen (150–200 kg/ha) through symbiosis with Rhizobia and Bradyrhizobia bacteria eventually enhance/restore soil fertility (Vyas et al., 2018).
Though, black gram can persist with drought conditions, but the average productivity of this legume crop is merely 450–800 kg/ha (Kaewwongwal et al. 2015). Low genetic variability, thermal-sensitivity, deprived harvest index and vulnerability to biotic stresses especially yellow mosaic virus and wilt are some foremost limitations which hinder high yielding of black gram (Kanimozhi et al., 2009). Among pulses, black gram, a potential legume for many developing countries, is least studied crops and research is less focused as compared to peas and beans.
The information on and knowledge of genetic variability is a key criterion for crop breeding as the successful heterosis breeding rely on genetic variability (Kumar et al., 2017). Though, information on morphological variability in black gram has been reported earlier by several researchers (Ghafoor et al., 2000, Gupta et al., 2006). But little information is available on seed mineral content in urd bean. The grains of urd bean are worthy source of minerals especially potassium, calcium and phosphorus but no systematic study on seed minerals has been carried out.
Being self-pollinated crops, the genetic base of black gram is narrow, especially of released varieties. Thus, to hasten the breeding program, it is needed to measure the genetic variability available in germplasm through molecular markers. Reports are available on DNA marker-based diversity analysis in black gram, but most of the studies were conducted using Random amplified polymorphic DNA (RAPD) and intersimple sequence repeat (ISSR) (Karuppanapandian et al., 2010) which are less informative compared to co-dominant markers like simple sequence repeat (SSR) (Bharti et al., 2018). The current experiment aimed to estimate the extent of diversity for protein and mineral content in the black gram seeds and to evaluate genetic diversity with SSR markers which may provide further breeding guidelines for black gram.
2 Material and method
2.1 Plant material and field trial
A set of 60 genotypes of black gram were used in the present experiment, of which 19 were varieties released by various institutions of India. All the genotypes were grown in three replications in randomized complete block design (RCBD) in summer 2016 at Regional Research Station (RRS), Anand Agricultural University, Anand with plant × plant and row × row to distance of 10 and 45 cm, respectively, in a plot size of 4 × 0.9 m.
2.2 Estimation of protein and minerals
At the time of maturity, pods were picked from randomly tagged 5 competitive plants of every genotype for protein and mineral analysis. After de-hulling, seeds were washed with tap and distilled water to remove dust. Seeds were fine powdered in cyclone sample mill. The Kjeldahl method was used to estimate the nitrogen content in seed powder (AOAC, 1960). Protein content was acquired with the conversion factor of 6.25.
For estimation of minerals (mg/100 g), acid extract with powder was prepared by di-acid mixture (HNO3:HCLO4- 4:1). From the acid extract, potassium was determined through flame photometric method (Jackson, 1973), calcium was determined by versanate titration method (Cheng and Bray, 1951) while the vanadate-molybdate yellow colour method of Jackson (1973) was deployed for estimation of phosphorus content.
2.3 DNA extraction and SSR-PCR
DNA was isolated from young leaves of 5 plants following the protocol described by Doyle and Doyle (1990). Agarose gel (0.8%) electrophoresis was carried out to assess DNA quality followed by spectrophotometry on Nanodrop (Thermo, USA) to estimate DNA concentration. A working stock of 10 ng/μL was prepared with nuclease-free water for polymerase chain reaction (PCR) of SSR markers.
Various researchers reported that microsatellite markers from Vigna unguiculata (cowpea) and V. angularis (azuki bean) revealed a high amplification success in black gram (Kaewwongwal et al., 2015). Therefore, during PCR amplification, microsatellite primers from V. unguiculata, V. angularis and Glycine max (soybean) along with SSR markers of urdbean were used. For SSR-PCR, 15 μL reaction mixture containing 2.5 μL gDNA, 6.5 Master-mix (Takara, India), and 1.0 μL of primer (10 pM) and 5 μL distilled water was used. PCR was carried out with following thermal conditions: initial denaturation of 94 °C for 5 min, then 35 cycles of 94 °C for 45 s, T °C (specified primer) for 45 s, 72 °C for 45 s and a final extension at 72 °C for 7 min. PCR products were separated using 6% non-denaturing polyacrylamide gels (PAGE) on sequencing gel system (Bio-Rad Sequi-Gen GT Sequencing Cell) followed by silver staining of gel. The stained gels were scanned using scanner to capture gel images (Microtek, Taiwan).
2.4 Statistical analysis of biochemical and molecular data
The mean value of biochemical traits was figured out, and analysis of variance (ANOVA) was calculated as per Panse and Sukhatme (1978). For molecular diversity assessment, SSR allele calling in base pairs was carried out manually with consideration of the heterozygous or homozygous status of genotype. The number of alleles from each primer, frequency of major allele, gene-diversity, heterozygosity and polymorphism information content (PIC) was calculated by PowerMarker v.3.25 (Liu and Muse, 2005). Neighbor-Joining (NJ) algorithm in DARwin 6.0 was deployed to compute pair-wise inter-genotype genetic distance.
3 Result and discussion
3.1 Mean performance and correlation
ANOVA exposed a significant difference among the genotypes for four biochemical traits (Table 1) suggesting the presence of genetic variation in studied set of genotypes for the traits. This also gives abundant opportunity to select promising genotype from the studied material for improving protein and mineral content of black gram. The presence of ample variability might be due to a diversity and/or effect of edaphic and environmental conditions.
Character
Mean Performance
Source of variation and Mean squares (ANOVA)
Range
Mean
S.Em±
C.D (0.05)
C.V (%)
Replication (DF = 2)
Genotypes (DF = 59)
Error (DF = 118)
Protein (%)
20.28–26.41
22.96
0.22
0.63
1.70
1.21
7.00**
0.15
Potassium (mg/100 g)
337.96–548.08
468.59
4.83
13.54
1.79
179.74
3758.15**
70.07
Phosphorus (mg/100 g)
237.00–398.07
325.96
7.41
20.76
3.94
386.17
3913.92**
164.89
Calcium (mg/100 g)
117.17–148.33
135.26
0.62
1.73
0.79
0.95
132.65**
1.15
Pulses are inexpensive source of protein in comparison to animal proteins in developing countries (Singh and Jambunathan, 1991). In general, legumes contain 18–25% protein, about double that of the cereals. Moreover, the protein content of black gram is considerably higher than chickpea and pigeonpea (Singh and Singh, 1992). Being a pulse, high protein is a necessary trait in black gram. The average value of protein content was 22.96%. Among the genotypes studied, VUG-14 (26.41%) had the highest protein content and was at par with VUG-10 (26.26%), VUG-55 (26.12%) and VUG-7 (26.12%). Lowest protein was recorded in genotype PU-30 (20.28%). Significant difference for this trait has also reported by Konda et al. (2009) and Patidar et al. (2018).
In human cell, calcium and phosphorus play important roles in countless biochemical reactions. They are also the main components of the skeleton. Moreover, phosphorus is an important constituent of soft tissue and has function in the formation of bones and teeth. The deficiency of dietary calcium is widespread, especially in developing countries (Pettifor, 2014). Black gram contains a high amount of calcium (Patwardhan 1962). In the current study, the mean for calcium was 135.26 mg/100 g in seed with a range from 117.17 (VUG-56) to 148.33 mg/100 g (LBG-20). Kavitha et al. (2013) also detected a wide range of variation for calcium content in black gram. The level of calcium in black gram was up to twice as many legumes like red gram, green gram and peanut (Singh and Singh, 1992; Bukya, 2014). Though, the calcium in black gram was significantly lower than soybean (Singh and Singh, 1992). Thus, regular consumption of black gram can help in maintaining bone health.
The population mean for seed phosphorus content was 325.96 mg/100 g. Among all genotypes, highest phosphorus content (398.07 mg/100 g) was detected in VUG-22, and it was statistically at par with VUG-5 (394.60 mg/100 g), VUG-67 (388.56 mg/100 g), VUG-39 (377.43 mg/100 g). The genotype PANT U 40 (237.00 mg/100 g) exhibited the lowest phosphorus content. Significant difference for this trait was recorded by Kavitha et al. (2013) in black gram.
Similar to calcium, black gram is also a rich source of potassium (K). Potassium is linked with movement of H2O, nutrients and sugars in plant cell. It is a key mineral for activation of many enzymes which affects protein, starch and adenosine triphosphate production (ATP) in the plant. The results revealed that the mean potassium content was 468.59 mg/100 g in seeds of black gram though it ranged from 337.96 (VUG-63) to 548.08 mg/100 g (VUG-66) which indicated sufficient variation. After VUG-66, VUG-22 also exhibited high K (539.01 mg/100 g) in the current study. Earlier, Gopalan et al. (1989) reported high potassium (800 mg/100 mg) in black gram.
The genotypic and phenotypic correlation coefficient among characters is presented in Table 2. In the present study, phosphorus was significantly correlated with potassium at both levels in a positive direction. Vandemark et al. (2017) recorded a similar result for phosphorus and potassium content in chickpea and lentil.
Character
Protein
Potassium
Phosphorus
Potassium
rg
0.026
1.000
rp
0.020
1.000
Phosphorus
rg
0.105
0.337**
1.000
rp
0.091
0.326**
1.000
Calcium
rg
−0.083
−0.057
−0.127
rp
−0.077
−0.047
−0.133
3.2 Molecular diversity analysis
PCR dependent co-dominant marker based diversity estimation has proven useful due to high polymorphism level loci and easy analysis. Molecular markers can overcome the limitations especially of genotype × environment interaction associated with morphological/biochemical markers (Kumar et al., 2016). Hence, molecular markers are very powerful for genetic diversity estimate. In the current study, a total of 42 markers were used for amplification and polymorphism survey in sixty genotypes. Out of 42 primers, 27 (66.66%) were amplified successfully. However, out of 27, 16 markers (59.25%) were found to be monomorphic while remaining 11 markers (40.74%) were recorded polymorphic (Table 3). The no amplification and high rate of monomorphism of SSR primers have been reported by Tondonba et al. (2018).
Locus Name
Amplicon size (bp)
Number of Alleles
Major Allele Frequency
He
Hl
PIC
CEDG 68*
120–128
5
0.41
0.69
0.05
0.64
CEDG 08*
110–118
6
0.35
0.77
0.40
0.74
CEDG 92*
148–158
3
0.78
0.35
0.00
0.30
CEDG 86*
124–134
4
0.46
0.60
0.00
0.52
CEDG 44*
166–198
8
0.38
0.77
0.00
0.74
Vmg SSR 29^
352–378
12
0.13
0.91
0.00
0.90
Vmg SSR 82^
315–328
5
0.78
0.37
0.00
0.35
Vmg SSR 53^
336–386
12
0.15
0.91
0.00
0.90
Vmg SSR 31^
310–328
7
0.26
0.81
0.00
0.78
AG 81#
144–146
2
0.62
0.47
0.00
0.36
Bmd 12@
174–177
2
0.58
0.49
0.00
0.37
Range
110–386
2–12
0.15–0.78
0.35–91
0.0–0.40
0.35–0.90
Average
–
6
0.44
0.65
0.04
0.60
The molecular weight of the amplified DNA ranged from 110 (CEDG 08) to 386 bp (Vmg SSR 53) reflecting a significant difference in the repeat motifs between the different alleles. A total of 66 alleles were identified from 60 black gram genotypes. The average number of allele per locus was 6 with a range of 2 (AG 81, Bmd 12) to 12 (Vmg SSR 53 and Vmg SSR 29). The average number alleles per locus in the current investigation was lesser than the earlier reported by Kaewwongwal et al. (2015) where mean alleles per locus was 9.05 and higher than Gupta and Gopalakrishna (2009) where mean alleles per locus were 4.1 (Table 3). The major allelic frequency is ranging from 0.13 (Vmg SSR 29) to 0.78 (CEDG 92 and Vmg SSR 82).
The maximum gene diversity (0.91) was linked for Vmg SSR 29 and Vmg SSR 53 while, it was minimum (0.35) for CEDG 92. Though, mean gene diversity was 0.65. Kaewwongwal et al. (2015) reported mean gene diversity 0.75 in wild black gram species which was higher than present investigation, while 0.59 mean diversity reported in cultivated black gram species which was lower than present investigation.
Heterozygosity varied widely from 0.00 to 0.40 (CEDG 08) with an average of 0.04. In overall, low heterozygosity is due to self-pollinating nature of legumes which prevent inter-population gene flow eventually precludes the blending of the different gene pools and thereby, decreasing the genetic variation and increasing the homozygosity (Gediya et al., 2019).
The PIC value is a discriminating power of a particular marker in a defined germplasm (Smith et al., 2000). The PIC value of a marker dependents on the number of alleles and their relative frequencies in studied germplasm. In the present study, PIC values for SSR markers ranged from 0.30 (CEDG 92) to 0.90 (Vmg SSR 29 and Vmg SSR 53) with average 0.60 which was close agreement with PIC value of SSR marker (0.60) reported by Kaewwongwal et al. (2015). The PIC was higher than Gupta and Gopalakrishna (2009) where mean PIC was 0.49. However, except four, seven markers were found to be more informative as displayed PIC >0.5.
Sixty black gram genotypes were grouped into three main clusters A, B and C with 28, 28 and 4 genotypes, respectively (Fig. 1). Gupta and Gopalakrishna (2009) and Tondonba et al. (2018) also reported three groups with SSR markers. In the current study, main cluster A was further divided into two sub clusters A1 (18 genotypes) and A2 (10 genotypes). Main cluster B was divided into two sub-clusters B1 (22 genotypes) and B2 (6 genotypes). The pair wise comparison value of Nei’s (1973) genetic distance ranged from 0.09 to 1.00. A total of 31 pairs depicted extreme genetic distance (1.00) indicated that these pairs of genotypes are highly dissimilar at the genomic level and can be used in heterosis breeding to widen the genetic background and to create mapping populations to ignite molecular breeding program. The lowest genetic distance (0.09) was observed between VUG-31/VUG-41 and T-9/GU-1. The mean genetic distance in current study was 0.69. Pyngrope et al. (2015) recorded 0.50 average distance was in 30 black gram genotypes which were lower than the present study.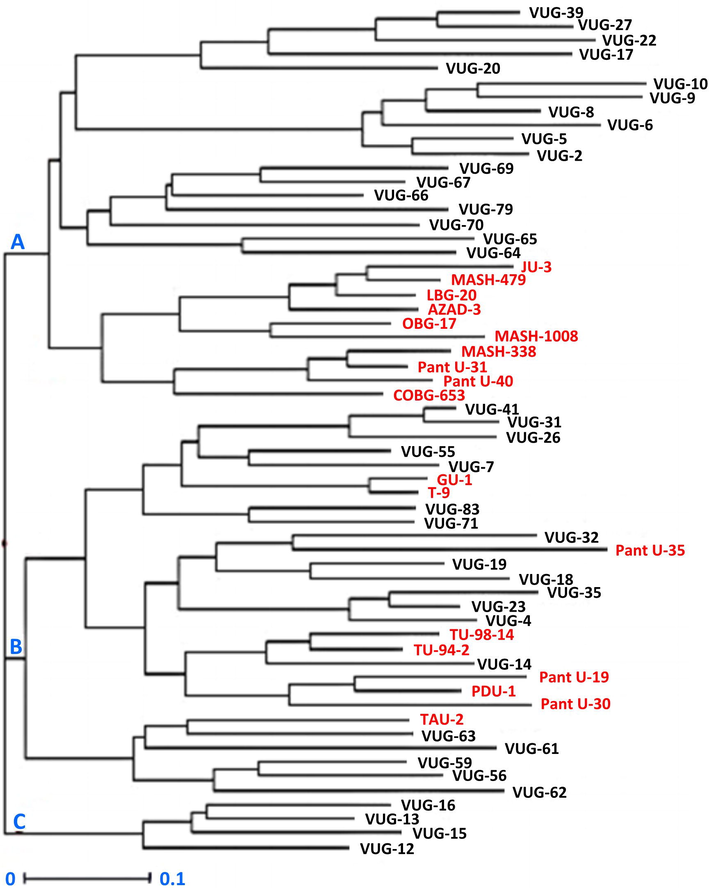
Dendrogram showing clustering of sixty black gram genotypes constructed using un-weighted Neighbour-Joining analysis (Genotypes in red font are released varieties).
4 Conclusion
The findings of present investigation lead to the conclusion studied genotypes are having sufficient diversity for protein, calcium, phosphorus and potassium. Phosphorus exhibited high significant and positive correlation with potassium. On the basis of genetic diversity study through SSR markers, it could be concluded that the genotypes with same genetic make-up were harbored in same cluster, and this would helpful during breeding program as breeder can perform hybridization between genotypes harbored in different clusters.
Acknowledgements
Authors acknowledge Anand Agricultural University, India for providing all necessary facilities.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
References
- Anonymous (2006). Post Harvest Profile of Black Gram. Government of India, Ministry of Agriculture, Directorate of Marketing and Inspection, Nagpur, India, pp 5.
- Anonymous (2016). Agricultural Statistics at a Glance. Directorate of Economics and Statistics, Ministry of Agri., Govt. of India (Website http://www.dacnet.nic.in/ean), pp 13.
- Development of genomic simple sequence repeat (gSSR) markers in cumin and their application in diversity analyses and cross-transferability. Ind. Crops Prod.. 2018;111:158-164.
- [Google Scholar]
- Development of a genome wide anchored microsatellite map for common bean [Phaseolus vulgaris L.] Theor. Appl. Genet.. 2003;107:1362-1374.
- [Google Scholar]
- Bukya, A. (2014). A Handbook on Legumes in Indian Agriculture and Health Benefits International. E-Publication www.isca.me, www.isca.co.in Indore-452005 (MP) India.
- Determination of calcium and magnesium in soil and plant material. Soil Sci.. 1951;72(6):449-458.
- [Google Scholar]
- Genetic diversity in black gram [Vigna mungo (L.) Hepper] Legumes Res.. 2008;31(1):57-59.
- [Google Scholar]
- Phenotypic variability, path analysis and molecular diversity analysis in chickpea (Cicer arietinum L.) Vegetos. 2019;32(2):167-180.
- [Google Scholar]
- Cluster analysis and correlation in black gram germplasm. Pak. J. Biol. Sci.. 2000;3(5):836-839.
- [Google Scholar]
- Nutritive Value of Indian Foods. Hyderabad, India: National Institute of Nutrition; 1989.
- Genetic variability, heritability and genetic advance for some traits in black gram [Vigna mungo (L.) Hepper] Prog. Agric.. 2006;6(2):164-166.
- [Google Scholar]
- Genetic diversity analysis in black gram [Vigna mungo (L.) Hepper] using AFLP and transferable microsatellite markers from azuki bean. Genome. 2009;52:120-128.
- [Google Scholar]
- Soil Chemical Analysis. New Delhi: Prentice Hall of India Pvt. Ltd.; 1973. p. :38-56.
- Genetic diversity of black gram [Vigna mungo (L.) Hepper] gene revealed by SSR markers. Breed. Sci.. 2015;65(2):127-137.
- [Google Scholar]
- Genetic diversity as assessed by ISSR markers in black gram [Vigna mungo (L.) Hepper] Electron. J. Plant Breed. 2009;1:12-17.
- [Google Scholar]
- Efficiency of RAPD and ISSR markers in assessing genetic diversity and relationships in black gram (Vigna mungo L. Hepper) varieties. Can. J. Plant Sci.. 2010;90:443-452.
- [Google Scholar]
- Physicochemical, functional, pasting properties and nutritional composition of selected black gram (Phaseolus mungo L.) varieties. Indian J. Sci. Technol.. 2013;6:5386-5394.
- [Google Scholar]
- Genetic variability studies for Productivity and its component in black gram [Vigna mungo (L.) Hepper] Legumes Res.. 2009;32(1):59-61.
- [Google Scholar]
- Development and validation of EST-derived SSR markers and diversity analysis in cluster bean (Cyamopsis tetragonoloba) J. Plant Biochem. Biotechnol.. 2016;25(3):263-269.
- [Google Scholar]
- Assessment of genetic diversity among okra genotypes using SSR markers. J. Plant Biochem. Biotechnol.. 2017;26(7):172-178.
- [Google Scholar]
- PowerMarker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21:2128-2129.
- [Google Scholar]
- Statistical Method for Agricultural Workers. New Delhi, India: Indian Council of Agricultural Research; 1978.
- Genetic variability studies in black gram (Vigna mungo (L.) Hepper) Int. J. Chem. Stud.. 2018;6(2):1501-1503.
- [Google Scholar]
- Cross-Species amplification of soybean (Glycine max) simple sequence repeats within the genus and other legumes genera: implication for the transferability of SSR in plants. Mol. Biol. Evol.. 1998;15(10):1275-1287.
- [Google Scholar]
- Calcium and vitamin D metabolism in children in developing countries. Ann. Nutr. Metab.. 2014;64(suppl. 2):15-22.
- [Google Scholar]
- Genetic diversity analysis of black gram [Vigna mungo (L.) Hepper] using morphological and molecular markers. Int. J. Appl. Pure Sci. Agri.. 2015;1(8):104-112.
- [Google Scholar]
- Pulses as a substitute for animal protein. In: The Proceedings of Advances in Pulses Research in Bangladesh, National Workshop on Pulses. Bangladesh: Bangladesh Agricultural Research Institute (BARI); 1991. p. :191-197.
- [Google Scholar]
- Genetic diversity among elite sorghum inbred lines assessed with simple sequence repeats. Crop Sci.. 2000;40:226-232.
- [Google Scholar]
- De novo assembly, characterization of immature seed transcriptome and development of genic-SSR markers in black gram [Vigna mungo (L.) Hepper] PloS One. 2015;10(6):1-15.
- [Google Scholar]
- Crossability studies and genetic diversity analysis in black gram (Vigna mungo L. Hepper) using molecular markers. Agrotechnology. 2018;7(2):179.
- [Google Scholar]
- Mineral concentration of chickpea and lentil cultivars and breeding lines grown in the U.S. pacific northwest. The Crop J. 2017;6(3):253-262.
- [Google Scholar]
- Assessment of genetic diversity in black gram [Vigna mungo (L.) Hepper] genotypes based on ISSR. Legume Res.. 2018;41(2):175-181.
- [Google Scholar]
- The development of SSR markers by a new method in plants and their application to gene flow studies in azuki bean [Vigna angularis (Willd.) Ohwi & Ohashi] Theor. Appl. Genet.. 2004;109:352-360.
- [Google Scholar]







