Translate this page into:
Epigenetic variation as a new plant breeding tool: A review
⁎Corresponding author. aalshayeed@kku.edu.sa (Ahlam Khalofah)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Epigenetic variations are inherited or uninherited effects that occur beyond the DNA sequence of an individual. However, DNA sequence has a critical role in shaping epigenetic variation. The great diversity of epigenetic markers confers an advantage of various uses without interrupting its highly environmental independence. The epigenetic effects are highlighted by many vital events, especially the regulation of gene expression in hybrid vigor and inbreeding depression, even in the absence of genetic diversity. However, various stress genes can include many repeats that undergo alternately methylation and demethylation states to regulate gene expression positively or negatively. After all the arguments raised on the genetic basis of hybrid vigor in its both traditional and molecular aspects, the term “epigenome” strongly emerged as one of the main causes of performance deviation among offspring. These include both histone and DNA biochemical modifications, which play a key role during successive stages of development and differentiation in addition to the regulation of gene expression in response to biotic and abiotic stresses. Evidence has shown a correlation between unique DNA methylation and heterosis in many plant species as well as between inbreeding and the sharp decline in fitness of most naturally cross-pollinated species. Although detailed molecular mechanisms laying behind many of these plant breeding aspects remain little understood, epigenetics has provided some explanations.
Keywords
Acetylation
Chromatin remodeling
DNA methylation
ncRNA
1 Introduction
Traditional plant breeding is focused on capturing and gathering many variable and desirable alleles for possible improvement of traits of interest, in addition to plant efficiency in using limited natural resources (Postnote, 2017; Kaiser et al. 2020). The development of modern molecular tools facilitates achieving breeders’ goals (Sadder et al. 2014). Plant breeding goals can be categorized into two major aspects; namely the improvement of agronomic traits (including yield and quality components) and stress mitigation including various abiotic and biotic factors (Hamany Djande et al. 2020).
Until recently, genetic variation that commonly points to the heritable variation of genetic information of individuals and populations was thought to be the only responsible factor for revealing phenotypic traits (Goulet et al., 2017). However, researchers discovered yet another source of variation in view of various traits among individuals within the same species, which has no direct correlation with DNA polymorphisms. These newly discovered variations which coined the term “epigenetics” has received considerable attention to better understand their stability through successive generations (Cavalli and Heard, 2019; Springer and Schmitz, 2017; Zenda et al. 2021).
The heritable or reversible changes in gene expression happen at a level higher than that of the nucleotide sequence. In other words, it is not attributed to alterations in the type and/or sequences of DNA nucleotides (Alvarez-Venegas et al. 2003; McKeown and Spillane, 2014; Liu et al. 2017b).
Genetic and environmental variations and their interactions naturally induce phenotypic variations. Altered phenotypic traits may be resulting from an identical genetic structure and such alterations result from identical alleles acting in different ways in response to biotic or abiotic stress (Fortes and Gallusci, 2017; Zhi and Chang, 2021). Naturally characterized epialleles are relatively few, consequently, the role of epigenetic variation in revealing large phenotypic alterations is still vague (Springer and Schmitz, 2017). Epigenetic changes usually participate with a variety of chromatin marks, such as cytosine methylation, modifications of histone tail, chromatin remodeling and non-coding RNAs (Saleh et al. 2008; Rajewsky et al. 2017).
The objective of this communication is to briefly introduce epigenetic variations asa breeding tool that may support traditional genetic variation in widening the genotypic gap, hence improving the opportunity for plant breeders to achieve hybrid vigor in the desired attributes.
2 Histone acetylation
Nucleosome is a 147 nucleotide (nt) of DNA wrapped tightly around a core of eight polymer histones considered the basic unit of chromatin. The described octamer consists of two of each of the following molecules: H2A, H2B, H3 and H4. Histones are highly positively charged proteins, including 24 amino acids of lysine and arginine. As DNA can be methylated, histones can be subjected to numerous posttranslational modifications, e.g., acetylation, methylation, and phosphorylation (Demetriadou et al., 2020; Grabsztunowicz et al., 2017).
Histone acetylation is being widely studied as a major form of epigenetic modifications in addition to DNA methylation. It is obvious that the core histones are acetylated in a reversible manner (Liu et al. 2017b), service in posttranslational modifications, like acetylation, methylation, ubiquitination, phosphorylation and ribosylation of adenosine diphosphate (ADP) . Surprisingly, many of the epigenetic mechanisms operate cooperatively via organizing the work of each other to regulate gene expression during cell differentiation. This “fine-tuning” mode of action would guarantee a precise operating system, such as in histone deacetylation which helps in the maintenance of DNA methylation (Blevins et al., 2014; Lee et al. 2017).
In the last two decades, DNA methylation was proved to be a major component of the plant epigenome; likewise, histone acetylation was recently investigated as another major player in epigenetics (Shi et al. 2019). These claims created compelling arguments for lysine’s acetylation in histone tails, which is often correlated with an increased expression of accompanied genes (Xiao et al. 2017). It should be noted here that DNA acetylation involves the addition of acetyl (CH3COO–) group to the NH3+ group of lysine amino acid, whereas histone deacetylation eradicates the acetyl groups (Boycheva et al., 2014).
Advancement in epigenetic research enables understanding of the function and regulation of histone acetylation in plants, which delivers more accurate assessments as compared to inhibitors. In maize, several copies of histone acetyltransferases and histone de-acetyltransferases have been biochemically characterized (Zhou et al., 2017; Kopytko et al., 2020). However, it is not clear how chromatin structure is modulated by different histone acetylation states, the most common scenario is deduced from the crystal structure of nucleosomes in which non-acetylated tails of histone are available to interact with nearby nucleosome beads and moderate higher-order chromatin wrapping (De Ioannes et al. 2019). Therefore, this suggests the norm of action of histone deacetylases; on the contrary, histone acetyltransferases would moderate chromatin relaxation (Boycheva et al., 2014; Peng et al., 2017).
An important histone modification, attracting great attention, is the acetylation of protected lysine α-amino acid, particularly in amino-terminal tails. The competitive effect of either HAT or HAC histone acetyltransferase enzymes against histone deacetylase (HDA) enzymes determines histone acetylation levels (Ma et al. 2013; Wang et al. 2014). Dozens of HATs, HACs and HDAs have been characterized in plants, which have critical roles as biotic and abiotic stimulants and as functional regulators in normal developmental processes (Peng et al., 2017; Zhao and Zhou, 2012).
In plants, like most eukaryotes, HAT-A and HAT-B are the two major types of histone acetyltransferase enzymes (Liu et al. 2017b). The HAT-A enzymes have special importance since occupying the nucleus and operate to acetylate core histones which have been integrated into the chromatin; therefore, they are involved in controlling gene expression (Boycheva et al., 2014; Giaimo et al., 2019).
Certain changes in histone acetylation were found to be related to DNA replication at the cytological level rather than to their transcriptional activity (Vergara and Gutierrez, 2017). Such changes have been identified by using acetylated histone antibody isoforms (Li et al., 2017). During the cell life cycle, there are various oscillations with histone acetylation which are controlled by HAT-B enzymes (Class 2 of histone acetyltransferases), (Yongfeng et al. 2019). Free cytoplasmic histone (H4 or H3) would initially be acetylated by HAT-B enzymes, enter the nucleus to be deposited into recently replicated chromatin (Yang et al., 2011a,b). According to single nucleotide homology, there are three groups of plant HDAs: the first named reduced potassium dependency 3 (RPD3)/HDA1, while the second is histone deacetylase 2 (HD2) and silent information regulator 2 (SIR2) is the third type (Ma et al. 2013; Wang et al. 2014).
3 Non-coding RNA
The tremendous revolution in the epigenomic era has provided researchers with invaluable findings prompting a continuous revision of plant genetics. In general, the eukaryotic genome is not simple, and over time, accumulated evidence has endorsed such complexity (Jin et al. 2017). Absolute scientific facts were brought back to the discussion table. One of these abolished statements restricts transcripts to be exclusively derived from protein-coding domains (Berretta and Morillon, 2009). Surprisingly, the encoding proteins come from a tiny portion (2–25 %) of the total genome space (Liu et al. 2015).
Although non-coding RNAs (ncRNAs) do not end up with manufactured proteins, as usual, their regulatory role for various biomechanics cannot be overlooked (Liu et al. 2017a). Several ncRNAs have been suggested as effective tools in directing cell division and differentiation, as well as regulating plant response to environmental stresses at transcriptional and posttranscriptional levels (Cavalli and Heard, 2019; Matsui et al., 2013; Sunkar et al., 2012).
Non-coding RNAs can be organized into two classes, the well-studied small ncRNAs (sncRNAs), which consist of less than 200 nt and long ncRNAs (lncRNAs), which consist of more than 200 nt, the latter being less studied (Zhang and Chen, 2017).
The sncRNAs have very small molecular weights, and they have been studied extensively for decades in both plants and animals (Bhatia et al. 2017). There are two main groups of plant sncRNAs, short interfering RNAs (siRNAs) and microRNAs (miRNAs). The two groups differ in their genetic origin and their final consequence of regulation (Rajewsky et al. 2017).
Typically, siRNA refers to exogenous double-stranded RNA (dsRNA) that is brought from outside of the cell, whereas the endogenous stem-loop non-coding RNA produces the single-stranded miRNA (Guleria et al. 2011). The RNase Dicer-Like will be responsible for processing each of the three non-coding RNAs, miRNA and siRNA, to be transported in the next step out of the nucleus to bind with Argonaute (AGO) proteins into the cytoplasm and integrate them to shape the RNA-induced silencing complex (RISC) (Prathiba et al. 2017; Setten et al. 2019), which in many occasions regulates the expression of the target gene at a post-transcriptional level (Carthew and Sontheimer, 2009; Layton et al., 2020; Vaucheret et al., 2006). The mobility of sncRNA molecules inside the organism may serve to ease gene silencing in different plant cells and tissues (Sarkies and Miska, 2014). In addition, sncRNAs have been found to play a critical role in regulating DNA methylation, histone modifications and gene silencing, consequently, controlling the transcriptional system in living organisms (Holoch and Moazed 2015).
During the past decades, plant miRNAs have been intensively studied. Although they have a small molecular size (21–24 nt), miRNAs roles are particularly important in the regulation of various biological processes through targeted mRNA repression, either by degradation or translation inhibition (Wu, 2013). To fulfill their function ideally, miRNAs have a complementary sequence, which closely matches their respective mRNAs targets. The binding of miRNAs to their complementary sequences confers their ability to regulate gene expression, which is more advanced in plants as compared with their counterparts in animals, where just limited regions of miRNA are available (Narjala et al. 2020); therefore, they have a restricted complementary action (Xu et al. 2017). Recent biochemical and genetic evidence indicates that many miRNAs could regulate their own targets during translation (Bordersen et al. 2008; Prathiba et al. 2017). They have key functions in controlling plant differentiation, a transition from vegetative to reproductive phase, morphogenesis of reproductive organs, phytohormone stimulation, stress-response regulation and they may even control the pathway of their biogenesis (Zhao et al. 2017).
Interestingly, miRNAs can move from one cell to another (cell-to-cell movement), probably via plasmodesmata (Skopelitis et al. 2018). Such a property allows miRNA to play the regulatory role in the differentiation of various cells and organs where they have been synthesized (Kitagawa and Jackson, 2017).
In many flowering plants, the molecular basis for the formation and periodic maintenance of epiallelic states is provided by the RNA-directed DNA methylation pathway (Law and Jacobsen, 2010). Gene expression will be repressed through the described pathway by setting a reverse loop between siRNAs and DNA methylation, enabling the broadcasting of the epialleles through the entire genome (Kawakatsu and Ecker, 2019).
In addition, sequence-specific guides can be provided by siRNAs, and the former is simplifying silencing at farther loci on the same or different chromosomes (Chow and Ng, 2017). Therefore, sncRNAs have a significant role in the expressed hybrid vigor by guiding DNA methylation via RNA-directed DNA methylation pathway. Recently, patterns of small RNAs showed differential expression in hybrids compared with their respective parents in different crops (He et al. 2010).
4 Chromatin remodeling
Chromatin is the most condensed and complicated form of DNA. This compactness ensures the fitness of DNA packaged into the nucleus, protects the DNA structure, organizes gene expression, and controls DNA replication (Rajewsky et al. 2017). Chromatin shows two different coiling intensities, euchromatin which are the less compacted shape and heterochromatin representing the more coiled and compacted form (Santos et al. 2017; Pecinka et al. 2020). During the plant cell cycle (mitosis and meiosis), chromatin pass-through various dynamic structural changes termed “Chromatin Remodeling”. These structural changes serve to mediate the attachment between transcription factors and their respective DNA sequence targets (Arya et al., 2010; Bhadouriya et al., 2021; Weaver et al., 2017).
On many occasions, chromatin remodeling exposes its action by facilitating the passage of transcriptional factors to the nucleosome octamer core; this will eventually permit an aberrant pattern of gene expression (Secco et al. 2017). Modulation of chromatin depends on the disruption of the nucleosome macromolecules-DNA package which represents the basic structural unit of DNA (Goldstein et al. 2013). Several findings demonstrate that epigenetic changes of chromatin remodeling could be transmitted via successive generations, and this mode of action operates like a cell memory that provokes organisms to acclimatize and overcome stress conditions (Lämke and Bäurle, 2017; Santos et al., 2017).
RNA and DNA polymerases are key players in the chromatin re-modulation process facilitated with the aid of SWI2/SNF2 proteins family (SWItch/Sucrose Non-Fermentable). The two polymerases form a complex of SWI2/SNF2 and two copies from each of the H3 and H4 dimer histones. The incorporation process requires energy in the form of ATP (Clapier et al. 2017; Bhadouriya et al. 2021). The required energy will guarantee smooth access of the transcriptional factors to the DNA nucleosome. Although the DNA is still wrapped around the histone octamer, this procedure will make the chromatin less compact and more flexible in a norm of action similar to acetylation (Rajewsky et al. 2017). The SWI2/SNF2 has been demonstrated by various regulation functions in the biosystem. Based on its function and phylogenetic pathway, the SWI2/SNF2 family could be further divided into several subfamilies. The most important member is BRAHMA (BRM) which plays a pivotal role in several vital biological events especially post-embryonic stage via regulation of gene activation and suppression (Zhang et al. 2017). Furthermore, the importance of BRM goes beyond that as it may activate the upstream regulation of the transition to the reproductive phase (Yang et al. 2015).
5 DNA methylation
From a biochemical point of view, DNA methylation is a chemical change that includes a covalent bonding between the methyl group (CH3) and the cytosine residue in the DNA helix (Villicaña and Bell 2021). This chemical modification is the most studied example of epigenetic variations and could inheritably control the dynamics of chromatin structure through what has become known as “Cell Memory” (Alvarez-Venegas et al. 2014). Thus, the combination DNA-methyl group can significantly alter the biological activity of the DNA by silencing and re-activation of transposable elements and eventually regulating the gene expression.
Developmental abnormalities may result from alterations in DNA methylation, and such defects could be induced by a classical genetic mutation. In other words, it is like a combination of sudden genetic modification accumulated in backgrounds of hypomethylated DNA resulting in heritable epigenetic mutations “Epimutations” (Ashapkin et al., 2020; Fortes and Gallusci, 2017). Variation in DNA methylation is evident between individuals that belong to wild or domestic plant species. Therefore, it is suggested that the plant phenotypic variation caused by epialleles may be wider than what has been foreseen. The level of cytosine methylation is highly different from one taxon to another in the plant kingdom and it may reach 30 % of the total cytosine. Furthermore, there is a higher level of DNA methylation in monocots as compared to dicots (Rajewsky et al. 2017).
In plants, DNA methylation happens at cytosine residue in different sequences, asymmetric CHH and both CHG and CG symmetric status (H is any nucleotide except G). Through meiosis and mitosis, patterns of DNA methylation are propagated through different pathways by distinct DNA methyltransferase enzymes (Akhter et al., 2021; Catania et al., 2017). Methyltransferase 1 and the unique plant chromomethyltransferase maintain the CHG and CG methylation via DNA replication, whereas methyltransferase 2 establishes DNA methylation in all sequence status by a siRNA-directed DNA methylation pathway (Law and Jacobsen, 2010). There is a great similarity between DNA mutations and heritable DNA methylation, as both can take place in a state of induced or natural manner. The creation of novel epialleles affects gene expression that in turn could lead to aberrant final products, and consequently, de novo phenotypic traits will emerge (Becker and Weigel, 2012; Rajewsky et al., 2017; Schmitz and Ecker, 2012).
The genetic architecture of many individuals is the main cause of most variations in DNA methylation that were observed among them. For example, the polymorphism among individuals such as repeated sequences or transposons will determine the epigenetic status of epialleles which are haplotype specific (Eichten et al. 2013).
Different studies have focused on comparative global genome analysis of DNA methylation in plants with limited genetic diversity (Rodrigues and Zilberman, 2015). These studies concluded that the epigenetic differentiation of individuals was caused by spontaneous variation of DNA methylation within a short time (Zhang et al. 2020). Several examples showed a natural variation in cytosine methylation, which is caused by an alteration in cytosine methylation levels over time indicating a potential semi-stable heritability of epigenetic information (Gutzat and Scheid, 2012). On the contrary, the outcome of another study performed on some inbred lines and their hybrids (Lauria et al., 2014), indicated that the variation of DNA methylation was due to alterations in CHG or CG/CHG methylation at the same time. They found that novel methylation of the unmethylated alleles revealed all changes in DNA methylation, and 88 % of these changes in DNA methylation were inherited.
6 DNA methylation effects on hybrid vigor
Hybridization remains the most important tool in traditional breeding programs despite the tremendous evolution of molecular markers (Goulet et al., 2017). Over the long run of evolution, plant breeders can guide the local adaption through the transgressive segregation and creation of novel alleles and/or epialleles, which in turn will result in the formation of new hybrid genotypes (Bradshaw, 2017). The three proposed models; dominant, over-dominant and epistasis have achieved remarkable success; however, they did not offer a distinct understanding of heterosis since each model has certain weaknesses (Khotyleva et al. 2017). Moreover, these models cannot cover all aspects of the heterosis and on various occasions have failed to give a full explanation for this phenomenon (Groszmann et al. 2013). In addition to genetic effects, heterosis could be influenced by “epigenetic” effects, which represent a part of the non-Mendelian inheritance, cell fate and regulation of gene expression (McKeown and Spillane, 2014).
Different modifications could prompt epigenetic effects; consequently, the same genotype may result in different phenotypes (Alvarez-Venegas et al., 2014; Almelhami, 2017). It has been proven through different studies that the epigenetic effects of cytosine methylation can contribute to the development of the heterosis phenomenon. Such studies have exposed key variations in cytosine patterns of DNA methylation of superior maize hybrids against their inbred parents (Zhao et al. 2007).
Molecular analysis of the distributed methyl groups in the genomes of 11 maize inbred lines and their respective half-diallel hybrids has been conducted (Yang et al., 2011a,b). The analyzed data of methylation-sensitive amplified polymorphism (MSAP) technique revealed a negative relationship between methylation level and hybrid vigor for the number of kernel rows trait, but the relationship goes in the opposite direction as being positive for kernel numbers trait. However, the trait of 300 kernel weight did not correlate with methylation level in hybrid genomes.
The same technique was followed by Eichten et al. (2013) as they studied the differentially methylated regions (DMRs) in the genomes of 20 maize parental lines. They succeeded in detecting 1966 common DMRs and 1754 rare domains. Most of the detected DMRs were inherited, and approximately-one-half of the total DMRs were found to be tightly linked to single nucleotide polymorphism within DMRs domains or next to them. There was not only a highly significant relationship between DNA methylation and gene expression pattern, but also, but the latter was also affected by the DMRs even in relatively distant regions. The epigenetic variations may represent a new genetically independent source for phenotypic variations (Lauria et al., 2014). The authors attempted to track the variation in the DNA methylation pattern across eight generations of maize inbred lines by using MSAP. The authors reached an interesting conclusion that 12 % of DNA methylation has been memorized by the maize plant's genome and transmitted through six generations, while the cytosine residue was differentially methylated from one individual to another at a ratio of 7.4 %.
Two hybrids with their inbred parents were selected to monitor the possible alterations in the DNA methylation level at different organs and growing stages (Liu et al. 2014). The DNA methylation was analyzed in embryos and endosperm 15 days after fertilization, and in leaves and primary roots at the germination stage. Methylation levels in all hybrid organs were reduced as compared with their counterpart inbreds, and de-methylation was at its highest level in the hybrids, which indicates its tendency to permit higher levels of gene expression and de-activation of gene suppressor. The total of polymorphic DNA fragments reached 63, and 11 of these were found to encode well-known functional proteins in maize.
The level of DNA methylation had an understandable relationship with the phenotypic performance of eight CMS sunflower parental lines and their half-diallel hybrids (Kanoosh et al. 2021). The MSAP results pointed to significant alterations in the DNA methylation pattern due to hybridization, and this has a key role in hybrid vigor shown by the resulted half-diallel hybrids (Tables 1 and 2). Estimation of DMRs revealed a general decline in DNA methylation level in hybrid populations compared with their parental lines. The percentages of unmethylated (HPA+/MSP + ) and hemimethylated (HPA+/MSP-) regions were 13.1 % and 9.9 % in hybrids and 9.5 % and 6.6 % in parents, respectively. The percentage of internal cytosine methylation (HPA-/MSP + ) in the parental population was 20.7 % compared with a similar percentage in the hybrid’s population (19.5 %). Parents showed a higher percentage of full methylation (HPA-/MSP-) or absence of target (63.2 %) against this percentage in their half-diallel hybrids which was a bit lower (57.6 %).
Methylation Status %
Populations
Unmethylated
Hemimethylated
Internal cytosine methylation
Full methylation or absence of target
HPA+/MSP+
HPA+/MSP-
HPA-/MSP+
HPA-/MSP-
A-Lines
0.23
0.1
0.1
0.57
R-Lines
0.31
0.08
0.14
0.46
F1′s
0.22
0.13
0.13
0.5
Comparisons
Methylation Status %
Unmethylated
Hemimethylated
Internal cytosine methylation
Full methylation or absence of target
HPA+/MSP+
HPA+/MSP-
HPA-/MSP+
HPA-/MSP-
Self-poll. Lines vs Original Lines
Self-poll. Lines
22.7
27.3
26
24
Original Lines
22.7
28
22.7
22.7
Self-poll. Lines vs their Hyb.
Self-poll. Lines
26
27.3
25.3
21.3
Self-poll. Hyb.
26
26.7
24
23.3
Original Lines vs their Hyb.
Original Lines
20
31.1
24.4
24.4
Original Hyb.
20.7
30.4
24.4
24.4
7 Inbreeding and DNA methylation
Within the relentless endeavors of traditional genetics, feasible methods have been suggested to estimate the best vigor, unfortunately, the mechanisms that lie beneath this multifaceted phenotype are still beyond sensible understanding (Chodavarapu et al. 2012).
The understanding of the heterosis mechanism is based on an accurate understanding of inbreeding depression. Classically, the definition of inbreeding depression is of two parts, the first is the mating between two individuals being identical in their genetic composition which represents the cause that will result in the second part which is the depression of the offspring's overall performance (Labroo et al. 2021). On the other hand, in cross-pollinated crops, self-pollination leads to the production of inbred lines that represent the keystone in the production of elite hybrids through the crossing process (Shuro, 2017).
Despite the obvious effect of repeated self-pollination and its combination of deleterious alleles, the adoption of this idea did not help so much to set a convincing explanation of the hybrid vigor shown by genotypes propagated by self-pollination (Hartfield et al., 2017).
Considering modern epigenetic, there are still many questions that need to be addressed. For instance, will the inbreeding process lead to the gathering of harmful “epialleles” as it gathers harmful alleles? Epialleles are genetically identical alleles, while they vary in the way they are epigenetically modified (Lauss, 2017).
The inbreeding process is accompanied by many genetic as well as epigenetic alterations including DNA methylation, which is considered the most prominent epigenetic change. Many studies proved that variation in DNA methylation between inbred parents and F1 hybrids indicates the importance of inbreeding in the development of epigenetic differences (Vergeer et al. 2012; Lauss et al. 2018; Pang et al. 2019).
Song et al. (2010) proposed that the greater part of gene expression alterations showed by F1 hybrids are not associated with shifting of cis-acting DNA methylation, alternatively, the transcriptional differences revealed by offspring could be mediated by the different levels of trans effects that existed in the inbred parents. Furthermore, intergenerational epimutations could be an alternative model resulting in a subset modification in gene expression within hybrid genomes.
In a previous comparative study (Hussein, 2018), although, significant differences were detected in the differentially methylated regions (DMRs) within the studied maize, the practiced two rounds of selfing did not affect the unmethylated state (HPA+/MSP + ) of the genomic DNA, where both original and self-pollinated inbreds had the same number of unmethylated loci (22.7 %). The MSAP results revealed that the majority of the detected loci (30 loci) were Methylation Sensitive (MSL), against only 9 loci described as Non-Methylated Loci (NML). According to the 16 polymorphic loci, the polymorphism percentage reached 53 %.
The hemimethylated state (HPA+/MSP-) was at a higher rate in the original populations (28.0 %) than in the self-pollinated counterparts (27.3 %). The level of internal cytosine methylation (HPA-/MSP + ) was significantly magnified due to selfing to be 26.0 %, meanwhile, it was 22.7 % in the original inbreds. Moreover, the fully methylated loci (HPA-/MSP-) in the original inbreds was less than what existed in the self-pollinated populations (22.7 % and 24 %, respectively). The Principal Coordinate Analysis (PCoA) of MSL data (Figs. 1 & 2) assured that the compared populations were more varied at the first coordinate (C1), while the second coordinate (C2) was in less variation (33.3 % and 25 %, respectively). The cluster analysis indicated that the inheritance of MSL has differed from one inbred to another. The self-pollination process has a significant impact on the methylation state of some descended population's genomes, hence some self-pollinated inbreds separated from their original parents, and consequently occupied different clusters. Hybrid vigor was directly affected by the apparent variation of the epigenetic content. The joint work of genetic and epigenetic variations can create a larger gap that may, in turn, improve the opportunities to get a hybrid vigor in the desired direction.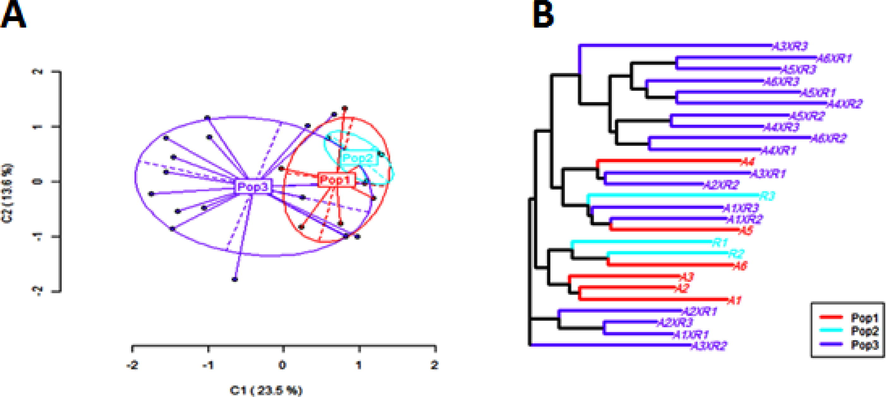
(A) Principal Coordinates Analysis (PCoA); (B) hierarchical clustering using the nearest neighbor method for the Methylated Sensitive Loci (MSL) in the A-lines (Pop.1), R-lines (Pop.2) and F1 hybrids (Pop.3) of CMS sunflower populations (Kanoosh et al., 2021).
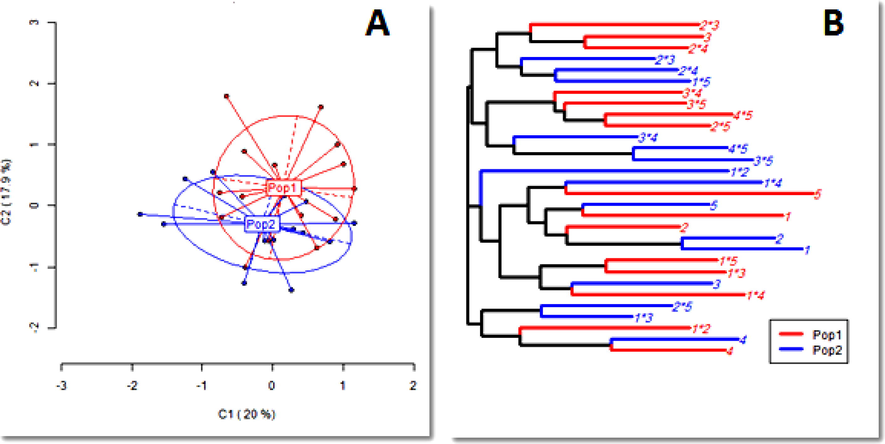
(A) Principal Coordinates Analysis (PCoA); (B) hierarchical clustering using the nearest neighbor method for the Methylated Sensitive Loci (MSL) in the self-pollinated (Pop.1) and original (Pop.2) maize populations (Hussein, 2018) .
Other efforts have focused on the role of DNA methylation in updating the “epigenetic memory” either during plant development (mitotically) or among individuals belonging to the same species but at different genetic backgrounds (meiotically), where the patterns of DNA methylation can vary (Eichten et al. 2011). A study performed on maize inbred lines which were developed by at least four generations of self-pollination followed by three back-crosses was subjected to DMRs analysis, which reflects a relatively stable inheritance of DNA methylation pattern. In another study, cytosine methylation was found to increase in hybrids as compared to their parents (Shen et al. 2012). This proved the impact of hybridization in the modification of DNA methylation levels.
The long-term inheritance of DMFs was investigated by selecting two individuals from the second generation to be advanced to the sixth generation via self-pollination (Lauria et al., 2014). Authors identified 15 out of 102 methylated regions to be DMFs in different members of the first generation. Part of the original DMFs has been transmitted to the next generation in a ratio ranging from 50 to 72.5 %. MSAP analysis assured that 82 % (14 out of 17) of the studied DMFs were still measurable in the sixth progeny. These results indicated that at least 12 % (11 out of 87 at leaf 2 stage; and 11 out of 93 at leaf 9) of the total DMFs originally recognized in the base population generation were meiotically transmitted to the six subsequent generations. Feng et al. (2015) could not find a link between the heterozygosity of the DMRs and the levels of DNA methylation. However, they reported that about 85 % of the assays showed an expected level of DNA methylation, whereas just 5 out of 150 assays revealed exclusive shifting in the methylation status and another (17 out of 150) assays showed a partial increase or decrease in DNA methylation.
8 Modulation of stress through epigenetic regulation
Although different stress forms affecting plants would have some overlaps in their responsive genes, they still show unique stress-specific gene expression profiles (Sadder et al. 2014). Based on several demonstrated evidence, it is believed that such unique responses are modulated through specified epigenetic changes, where chromatin remodeling is an indispensable step to organize the whole genome and regulating gene expression (Wang and Chang, 2018). To adapt to environmental changes, the expression of stress-responsive genes is synchronized with the methylated DNA and the amendment of histone modification (Kim et al. 2015).
Arabidopsis histone deacetylase complex 1 Swi-Indipendent3 was found to be upregulated under salt stress (Perrella et al. 2013). Under salinity stress, maize roots were reported to show enhanced expression of cell wall-related genes, which were correlated with elevated histone H3 (H3K9) acetylation (Li et al. 2014). On the other hand, H3K27me3 islands were found to undergo a shortening process under salinity stress as compared to the intact H3K4me2, H3K4me3, and H3K9me2 islands, which would imply a release of gene repression in these islands (Sani et al. 2013).
Drought-stressed Arabidopsis showed H3K4me3 modifications through activation of NCED3 by trithorax homolog ATX1 (Ding et al., 2011). Meanwhile, drought stress in rice was found to induce HAT genes and elevated acetylation of H3K9, H3K18, H3K27, and H4K5 (Fang et al. 2014). On the other hand, the severity of a specific stress condition like drought will determine the final enrichments of histone modifications in a specific gene. For instance, H3K4me3 and H3K9ac on drought stress-upregulated genes, such as RD20 and RD29A, were found to be highly enriched compared to fewer drought conditions (Kim et al. 2012). Moreover, threonine 3 of histone H3 was found to be phosphorylated in pericentromeric regions upon the onset of osmotic stress in Arabidopsis (Wang et al., 2015). Several reports support the importance of the histone variant H2A.Z in response to heat stress, e.g., in Arabidopsis (Coleman-Derr and Zilberman, 2012) and in Brachypodium (Boden et al. 2013). Unique acetylation such as H3K56 was enhanced upon implementation of heat stress (Weng et al. 2014).
9 Stress mitigation via epigenetic memory
The histone H3K4 methyltransferase of trithorax (ATX1) homolog in Arabidopsis was found to be an epigenetic regulator (Alvarez-Venegas et al. 2003). ATX1 expression was shown to depend on environmental and developmental stimuli in roots through the lipid messenger, phosphatidylinositol 5-phosphate (Alvarez-Venegas et al. 2006).
Furthermore, signaling of dehydration stress was found to be effectively regulated by ATX1 in abscisic acid (ABA)-dependent and/or independent pathways (Ding et al., 2011; Ruschhaupt et al., 2019). Several tri thorax homologs were isolated and characterized in other plant species, e.g. tomato (Sadder et al., 2011), barley (Papaefthimiou and Tsaftaris, 2012), maize (Qian et al. 2014) and rice (Choi et al. 2014),
Application of chromatin immune precipitation followed with sequencing (ChIP-Seq) in Arabidopsis thaliana reveals a polymorphism among histone H3 methylation patterns under dehydration stress (van Dijk et al. 2010). H3K4me3 positioning the 5′-ends of differently transcribed genes, however genes persuadable by drought stress revealed broader H3K4me3 distribution profiles than what existed before and/or after stress conditions (Murray et al., 2019).
Such a change in chromatin marks was correlated to short memory in plants exposed to multiple stresses. Stress memory refers to the primary, sub-lethal stress that may shift a plant's general responses to the same subsequent stresses (Ding et al., 2013; Virlouvet et al., 2018). Evidence from previous studies has been documented in Arabidopsis thaliana (Ding et al., 2012), maize (Ding et al. 2014), wheat (Allouzei, 2016), and rice (Li et al. 2019).
Endogenous factors such as abscisic acid (as phytohormone) and DNA methylation are found to effectively participate in short-term rehydration memory in rice plants, probably serving as memory factors that trigger transcripts of dehydration-related memory in photosynthesis and proline biosynthesis pathways, to mediate flexible response to the successive stresses (Li et al. 2019).
After removal of the drought stress, [+/+] and [+/-] states of transcription memory of revised response memory genes persisted for 5–7 days (Ding et al., 2012; Liu et al., 2016). In the same context, at the [+/+] memory genes, both Ser5P Pol II and H3K4me3 levels also lasted for 5–7 days, indicating epigenetic modifications of their transcriptional memory. As a short-term memory, dehydration stress memory may function as a mechanism for transcriptional responses during seasonal attacks of drought and is doubtful to be meiotically inherited through successive generations (Ding et al., 2012; Li et al., 2019). The temporary alteration of specific gene expression may increase the survival opportunity of individual plants under frequent drought stress (Avramova, 2018). As some plants subjected to frequent drought stress, alternating with full-watered recovery, demonstrated transcriptional and/or physiological memory responses by reducing water loss, during the next drought stress, in contrast to plants subjected to first-time drought stress (Virlouvet et al., 2018). Thus, the plasticity of the generationally transmitted stress memory can have a great impact on breeding programs, especially as it functions principally in the female gametes (Wibowo et al. 2016).
Nonetheless, certain contradictions were found in the short-term memory responses to stress. Memory genes of jasmonic acid-dependent stress [+/-] provide developed different transcriptional responses to drought stress memory that are opposite to those of secondary herbivore stress (Avramova, 2017; Liu et al., 2016; Vos et al., 2013).
10 Genetic and epigenetic aspects of Somaclonal variations
Tissue culture is a widely applied technique serving in the micropropagation and production of new plant genotypes in a relatively short time (De Schepper et al., 2003). Tissue culture techniques are expected to produce clone-identical individuals, however, on many occasions, this technique proved to create variants at a relatively high rate (Ghosh et al., 2021).
Like in tissue culture, plant cells that are propagated in-vitro are exposed to a wide range of environment-derived stresses, which leads to genetic and epigenetic instability, especially at the initial stages of cell differentiation in cultured tissues.
The stressful in-vitro conditions may induce specific cell organelles like cell membranes to resist the harsh surrounding environment by triggering an extensive range of signals that in turn essentially contribute to the accumulation of free radicals, like ROS (Bednarek and Orłowska, 2020b; Bednarek and Orłowska, 2020a). Depending on the nature, the severity of the stress and the plant species, the propagated plants respond to these stressful conditions at varying levels. In general, most of these are reflected in different genotypes with different qualitative and quantitative phenotypic characteristics (Kaeppler et al, 2000).
Chromosomal abnormalities and altered DNA sequence are the two common examples of genetic modifications, whereas DNA methylation, transposable elements (TEs), and chromatin remodeling are considered the main epigenetic reasons laying behind phenotypic variations revealed by the regenerated plants compared to their original clones (Srikant and Drost, 2021). Somaclonal variations could be adopted to improve ornamental plants which continuously will agronomically produce economically valuable genotypes (Jain et al., 1998). Therefore, Nhut et al. (2013) exploited this type of variation to induce flower color in Torenia fournieri lind via invitroing selected Somaclonal lines and subsequently micropropagated. Invitroingly, somaclonal variation is very important in the improvement of date palm because of the growth anomaly that results from micropropagation (Jain et al., 2011).
11 Transposable elements and epigenetic effect
Since Ac/Ds system (activator/dissociation) was firstly coined by Barbara McClintock during 1940 s describing the regulatory role of transposable elements TEs (Pappalardo et al., 2021), TEs evolutionary importance was evidenced in several eukaryotic genomes (Ali et al., 2021; Chuong et al., 2017; Wells and Feschotte, 2020). TEs fall into two groups, retrotransposons, and DNA transposons, both represent a valuable part of plant genomes that efficiently participate in rearranging and modifying DNA recombinations (Almojil et al, 2021). The two TEs groups act via two different mechanisms, a cut-paste mechanism that results in changing TE loci and a copy-paste mechanism that increases TE copies.
Being deleterious, TEs can be epigenetically repressed by different mechanisms evolved by hosts which is an indirect way to regulate nearby gene expression (Qin et al, 2015; Kelleher et al, 2020).
Several studies pointed to a unique relationship between transposable elements mobilization and DNA methylation pattern as the most prominent form of genetic influence through which genetic material can be reshuffled (Baduel and Colot, 2021), thus gene expression is redrawn (Srikant and Drost, 2021). Remarkably, these TEs are subjected to waves of silencing and reactivation according to the shifting pattern of DNA methylation, and this highlights the importance of integrating the effect of the two epigenetic mechanisms in reshaping genomes. However, this kind of epigenetic crosstalk may not only suppress the invaded TEs but also silence one or more vital genes (Muyle et al, 2020). Additionally, functional RNA-mediated mechanisms may involve efficiently in the regulation of TEs activity (Hsu et al, 2021).
The role of the TEs may extend beyond protein-coding sequences to the establishment of entirely novel genes. Notably, the genomic distribution, abundance and diversity are key factors that determine the norm of action of TEs that may affect important bioprocesses like fitness (Eyre-Walker and Keightley, 2007).
12 In vitro culture influence on epigenetic
Tissue culture effectively contributes to the precise production of new plant variants to cope with the magnifying need in a relatively short time. Yet, the evolution of new rearrangements has been detected frequently, where not all the developed plants are identical at the morphological level. The tissue culture-derived rearrangements can be originated from genetic and/or epigenetic modifications (Orłowska et al., 2016; Orłowska and Bednarek, 2020).
In general, the epigenetic effect is magnified in response to the increasing number of regenerative cycles which verify the transgenerational heritability of epigenetic variations across successive generations (Rosato et al, 2016). Conversely, some other studies reported no relation between the long-term cell culture and the increased level of epigenetic variations (Bobadilla Landey et al., 2015; Franzen et al., 2021).
Epigenetic changes exposed by tissue culture-derived plants can be categorized into DNA and histone methylation, chromatin remodeling, and small RNAs (Orłowska, 2021; Yu et al., 2020; Zhang et al., 2021; Jiang et al., 2011; Miguel and Marum, 2011; Miyao, 2011; Neelakandan and Wang, 2012; Sabot et al., 2011). Nevertheless, several reports indicated that the most common form of epigenetic modification was DNA methylation, which may or may not be associated with the activation of transposons (Bednarek and Orłowska, 2020b; Machczyńska et al., 2014; Gimenez et al., 2016; Machczyńska et al., 2015; Orłowska et al, 2016). The DNA methylation was found to be plant species-dependent, although the methylation level was at a higher rate in the regenerated plants compared to donor plants (Bednarek and Orłowska, 2020a) .
The rapid development of molecular tools proved that epigenetic changes which occur during the initial stages of cell differentiation can effectively participate in the production of new plant regenerates with new phenotypic aspects that their mother donors did not possess (Miguel and Marum, 2011). Some elements can take part in epigenetic cases under an in-vitro environment when supplied in media such as molybdenum (Rihan et al., 2015), who confirmed that this element possessed a large positive effect on cold tolerance of cauliflower micro shoots.
13 Regulation of reproductive plant development by epigenetics
Plants in their growing environment are surrounded by biotic and abiotic factors that regularly interact. In view of recent findings, plants use epigenetic modifications to control early floral initiation (Yaish et al., 2011).
Epigenetic modifications may serve direct (activation/deactivation) or indirect regulation (hormone production) of somatic embryogenesis (SE) via fine-tuning of gene expression. Considering indirect regulation, epigenetic factors like microRNA (miRNA) usually involve post-transcriptional alterations. In view of recent findings, miRNA contributes to regulating the specific type of transcription factors that control auxin metabolism and ultimately SE development (D’Ario et al., 2017; Song et al., 2019).
Plants have two main schemes to reproduce, sexual and asexual. The nature of epigenetic influences on the development of sexual reproductive organs, particularly seeds, remains poorly understood. In flowering plants, the dynamic process of epigenetic modifications such as DNA methylation exchange between gametes and their companion cells. However, the DNA methylation level was found to be magnified in the mature embryos, then decreased significantly as the seed germinate (Han et al, 2019).
Additionally, polycomb group (PcG) proteins is another epigenetic mechanism that has a pivotal role in the regulation of normal seed development (Salaün et al, 2021). PcG contributes significantly to controlling cell proliferation through silencing specific genes (Simonini et al, 2021).
14 Reversal of flower color in transgenic petunia under field conditions
Besides producing more and superior quality yields, genetic engineering is an alternative approach used to combine attractive traits in ornamental plants, like unique flower colors. The undeclared genetically modified petunia (Petunia hybrida) was first diagnosed in the middle of the last decade. Petunia was genetically engineered by introducing the maize gene responsible for encoding dihydroflavonol 4-reductase (DFR, A1) to develop orange pelargonidin in the innovative flowers (Bashandy and Teeri, 2017; Haselmair-Gosch et al., 2018).
The CHS-A gene encoding chalcone synthase was identified in transgenic petunia, in which the developed flowers have unpigmented sectors, or it will be completely white. Surprisingly, the unpigmented petal tissues can be noticed even in nontransgenic petunia, in the same context recent findings strongly recommended the responsibility of post-transcriptional modifications due to short interfering RNAs (siRNAs) and/or altered DNA methylation patterns (Kasai et al, 2013).
15 Active compounds as epigenetic modulators
In many parts of the world where conventional medicine is practiced, people have exploited natural products such as extracts, powders, teas, juices, or mixtures. As epigenetic factors induced variations in traditional crops, they also affect medicinal plants and their constituents. Thus, they could be exploited in medicinal plant improvements (Shahrajabian et al., 2019). Methylated DNA may assist in obtaining new chemical compounds. Different epigenetic mechanisms act inside medicinal plant cells such as cytosine methylation, histone modification and SRNA (miRNA and siRNA) led to uncovering the epigenomics complications that are influenced by environments (Hao and Xiao, 2018). from these chemicals, melatonin that modulates ATP and glucosinolates led to an increase in some fungi resistance infested Brassica rapa ssp. Pekinensis resulted in proteomic expression (Teng et al., 2021). These phytochemicals could modulate epigenetic efficacies thereby assisting to extract new bioindicators that will be used as cancer preventers (Carlos-Reyes et al., 2019) like flavonoids (Busch et al., 2015). For instance, nutritional phenolic constituents could be regulated by DNA methylation, histone alteration and miRNA making these molecules as a target for preventing many diseases that are epigenetically controlled via gene expression (Ciz et al., 2020). Furthermore, epigenetic markers triggered bioactive nucleosides and bisabolene biosynthesis in Aspergillus versicolor (Wu et al., 2020). This technique could be economically used to produce active compounds.
16 Conclusions and recommendations
Various epigenetic mechanisms are triggered in response to different stresses e.g., temperature, dehydration, and salinity. Notably, both double-stranded RNA (dsRNA) and posttranscriptional gene silencing (PTGS) are the key transgenerational epigenetic regulators of variable stresses, hence a potential breeding tool. Hybridization is the most widespread and influential phenomenon in the genetic and morphologic formation of plant populations. This phenomenon is still playing a crucial role in reducing the level of DNA methylation and contributes efficiently to releasing the gene expression, thus, the outstanding performance of the different crosses compared with their ancestral pure parents. Noticeably, hybrid vigor tends to be more positive along with the advanced selfing generations even for a few generations, hence it can result in a considerable alteration in the genetic and epigenetic recital of plant population. The traditional direct relationship between selfing and inbreeding depression has been supported by increasing the methylation level in line with self-pollinated generations.
Finally, plant breeders must pay more attention to the propagation method of inbred lines because the continuous practice of selfing may result in a significant depression in the performance of these lines. There is a need to adopt new tools in the epigenetic assessment of the new genotypes that significantly reflects on the actual evaluation. Furthermore, epigenetic variations and their relationship with the management practices should be studied for a deeper understanding of the environmental-epigenetic interference.
17 Competing interests statement
Authors declare that they have no significant competing financial, professional, or personal interests that might have influenced the performance or presentation of the work described in this manuscript.
Acknowledgment
This work is ostensibly supported by the University of Anbar through program of adopting sober scientific research. We'd like to thank Dr. Andrés Pérez-Figueroa, University of Vigo, Spain for his sage advice and helping in analyzing MSAP data.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- In response to abiotic stress, DNA methylation confers epigenetic changes in plants. Plants. 2021;10:1096.
- [CrossRef] [Google Scholar]
- Role of Transposable Elements in Gene Regulation in the Human Genome. Life. 2021;11:118.
- [CrossRef] [Google Scholar]
- Allouzei, M. Evaluation of durum wheat (Triticum turgidum L. var. durum) accessions for yield, agronomic traits and drought responsive genes. 2016, [MSc. Thesis.] Amman, University of Jordan.
- Almelhami, A.S. Molecular relationship between DNA methylation pattern and hybrid vigor in maize (Zea mays L.) using MSAP. 2017, [MSc. Thesis.] Anbar, University of Anbar.
- The Structural, Functional and Evolutionary Impact of Transposable Elements in Eukaryotes. Genes. 2021;12:918.
- [CrossRef] [Google Scholar]
- ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Current Biology. 2003;13(8):627-637.
- [Google Scholar]
- The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proceedings of the National Academy of Sciences. 2006;103(15):6049-6054.
- [Google Scholar]
- Epigenetics in Plants of Agronomic Importance: Fundamentals and Applications. Switzerland, Springer International Publishing 2014:152.
- [CrossRef] [Google Scholar]
- A structural perspective on the where, how, why, and what of nucleosome positioning. Journal of Biomolecular Structure and Dynamics. 2010;27:803-820.
- [Google Scholar]
- Epigenetic mechanisms of plant adaptation to biotic and abiotic stresses. International Journal of Molecular Sciences. 2020;21(20):7457.
- [CrossRef] [Google Scholar]
- The jasmonic acid-signalling and abscisic acid-signalling pathways cross talk during one, but not repeated, dehydration stress: a non-specific ‘panicky’ or a meaningful response? Plant, Cell and Environment. 2017;40(9):1704-1710.
- [Google Scholar]
- Defense-related priming and responses to recurring drought: Two manifestations of plant transcriptional memory mediated by the ABA and JA signalling pathways. Plant, Cell and Environment. 2018;42(3):983-997.
- [Google Scholar]
- The epiallelic potential of transposable elements and its evolutionary significance in plants. Phil. Trans. R. Soc. B. 2021;376:20200123.
- [CrossRef] [Google Scholar]
- Genetically engineered orange petunias on the market. Planta. 2017;246:277-280.
- [CrossRef] [Google Scholar]
- Epigenetic variation: Origin and transgenerational inheritance. Current Opinion in Plant Biology. 2012;15:562-567.
- [CrossRef] [Google Scholar]
- Bednarek, P.T.; Orłowska, R. Plant tissue culture environment as a switch-key of (epi)genetic changes. Plant Cell Tissue Organ Culture (PCTOC). 2020, https ://doi.org/10.1007/s1124 0-019-01724 -1.
- Demethylation Leads to Sequence Mutations in an Anther Culture of Barley Due to the Presence of Cu, Ag Ions in the Medium and Culture Time. Int J Mol Sci.. 2020;21(12):4401. PMID: 32575771; PMCID: PMC7353013
- [CrossRef] [Google Scholar]
- Pervasive transcription constitutes a new level of eukaryotic genome regulation. EMBO Reports. 2009;10:973-982.
- [Google Scholar]
- Role of chromatin architecture in plant stress responses: an update. Frontiers in Plant Science. 2021;11:603380
- [CrossRef] [Google Scholar]
- Present scenario of long non-coding RNAs in plants. Non-coding RNA. 2017;3:16.
- [CrossRef] [Google Scholar]
- A two-step process for epigenetic inheritance in Arabidopsis. Molecular Cell. 2014;54:30-42.
- [CrossRef] [Google Scholar]
- Assessment of genetic and epigenetic changes during cell culture ageing and relations with somaclonal variation in Coffea arabica. Plant Cell Tiss Organ Cult. 2015;122:517-531.
- [CrossRef] [Google Scholar]
- Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A.Z-nucleosomes. Genome Biology. 2013;14:R65.
- [Google Scholar]
- Widespread translational inhibitory plant miRNAs and siRNAs. Science. 2008;320:1185-1190.
- [CrossRef] [Google Scholar]
- Histone acetyltransferases in plant development and plasticity. Current Genomics. 2014;15(1):28-37.
- [CrossRef] [Google Scholar]
- Epigenetic activites of flavonoids in the prevention and treatment of cancer. Clinical epigenetics.. 2015;7:64.
- [CrossRef] [Google Scholar]
- Dietary compounds as epigenetic modulating agents in cancer. Frontiers in Genetics.. 2019;10:79.
- [CrossRef] [Google Scholar]
- Epigenetic maintenance of DNA methylation after evolutionary loss of the de novo methyltransferase. Cell 2017
- [CrossRef] [Google Scholar]
- Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489-499.
- [CrossRef] [Google Scholar]
- Transcriptome and methylome interactions in rice hybrids. Proceedings of the National Academy of Sciences.. 2012;109(30):12040-12045.
- [CrossRef] [Google Scholar]
- Trithorax group protein Oryza sativa Trithorax1 controls flowering time in rice via interaction with early heading date. Plant physiology. 2014;164(3):1326-1337.
- [Google Scholar]
- Regulation of miR163 and its targets in defense against Pseudomonas syringae in Arabidopsis thaliana. Scientific Reports. 2017;7:46433.
- [CrossRef] [Google Scholar]
- Regulatory activities of transposable elements: From conflicts to benefits. Nat. Rev. Genet.. 2017;18:71-86.
- [Google Scholar]
- The role of dietary phenolic compounds in epigenetic modulation involved in inflammatory processes. Antioxidants.. 2020;9:691.
- [CrossRef] [Google Scholar]
- Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nature Reviews Molecular Cell Biology. 2017;18:407-422.
- [CrossRef] [Google Scholar]
- Deposition of histone variant H2A.Z within gene bodiles regulates responsive genes. PLoS Genetics. 2012;8:e1002988.
- [Google Scholar]
- Structure and function of the Orc1 BAH-nucleosome complex. Nature Communications. 2019;10(1):2894.
- [CrossRef] [Google Scholar]
- Genetic and epigenetic aspects of somaclonal variation: flower colour bud sports in azalea, a case study. South African Journal of Botany. 2003;69(2):117-128.
- [Google Scholar]
- Histone N-alpha terminal modifications: genome regulation at the tip of the tail. Epigenetics and Chromatin. 2020;13:29.
- [CrossRef] [Google Scholar]
- The Arabidopsis trithorax-like factor ATX1 functions in dehydration stress responses via ABA-dependent and ABA-independent pathways. The Plant Journal. 2011;66(5):735-744.
- [Google Scholar]
- Multiple exposures to drought 'train' transcriptional responses in Arabidopsis. Nature Communications. 2012;3:740.
- [Google Scholar]
- Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biology. 2013;13:229.
- [Google Scholar]
- Dehydration stress memory genes of Zea mays; comparison with Arabidopsis thaliana. BMC Plant Biology. 2014;14:141.
- [Google Scholar]
- Epigenetic and genetic influences on DNA methylation variation in maize populations. The Plant Cell. 2013;25:2783-2797.
- [Google Scholar]
- S.R. Eichten R.A. Swanson-Wagner J.C. Schnable A.J. Waters P.J. Hermanson S. Liu C.T. Yeh Y. Jia K. Gendler M. Freeling P.S. Schnable M.W. Vaughn N.M. Springer Heritable epigenetic variation among maize inbreds PLoS Genetics 7 2011 10.1371/journal.pgen.1002372 e1002372.
- The distribution of fitness effects of new mutations. Nat. Rev. Genet.. 2007;8:610-618.
- [Google Scholar]
- Expression analysis of histone acetyltransferases in rice under drought stress. Biochemical and Biophysical Research Communications. 2014;443:400-405.
- [Google Scholar]
- Recent advances in understanding plant heterosis. Agricultural Sciences. 2015;6:1033-1038.
- [CrossRef] [Google Scholar]
- Plant stress responses and phenotypic plasticity in the epigenomics era: Perspectives on the grapevine scenario, a model for perennial crop plants. Frontiers in Plant Science. 2017;8:82.
- [CrossRef] [Google Scholar]
- DNA methylation changes during long-term in vitro cell culture are caused by epigenetic drift. Commun Biol. 2021;4:598.
- [CrossRef] [Google Scholar]
- Tissue culture-induced DNA methylation in crop plants: a review. Mol Biol Rep. 2021;48:823-841.
- [CrossRef] [Google Scholar]
- The histone variant H2A.Z in gene regulation. Epigenetics and Chromatin. 2019;12:37.
- [CrossRef] [Google Scholar]
- Gimenez, M.D.; Yañez-Santos, A.M.; Paz, R.C.; Quiroga, M.P.; Marfil, C.F.; Conci, V.C.; García-Lampasona, S.C. Assessment of genetic and epigenetic changes in virus-free garlic (Allium sativum L.) plants obtained by meristem culture followed by Plant Molecular Biology Plant Cell Rep 2016, 35, 129–141. https ://doi. org/10.1007/s0029 9-015-1874-x.
- Nucleolin mediates nucleosome disruption critical for DNA double-strand break repair. Proceedings of the National Academy of Sciences. 2013;110(42):16874-16879.
- [CrossRef] [Google Scholar]
- Hybridization in plants: Old ideas, new techniques. Plant Physiology. 2017;173:65-78.
- [CrossRef] [Google Scholar]
- Post-translational modifications in regulation of chloroplast function: Recent advances. Frontiers in Plant Science. 2017;8:240.
- [CrossRef] [Google Scholar]
- The role of epigenetics in hybrid vigour. Trends in Genetics. 2013;29(12):684-690.
- [CrossRef] [Google Scholar]
- Plant small RNAs: Biogenesis, mode of action and their roles in abiotic stresses. Genomics, Proteomics and Bioinformatics. 2011;9(6):183-199.
- [CrossRef] [Google Scholar]
- Epigenetic responses to stress: Triple defense? Current Opinion in Plant Biology. 2012;15:568-573.
- [CrossRef] [Google Scholar]
- Metabolomics: A tool for cultivar phenotyping and investigation of grain crops. Agronomy. 2020;10:831.
- [CrossRef] [Google Scholar]
- Epigenetics Regulates Reproductive Development in Plants. Plants (Basel).. 2019;8(12):564. PMID: 31810261; PMCID: PMC6963493
- [CrossRef] [Google Scholar]
- Deep in shadow: epigenetic and epigenomic regulation of medicinal plants. Chinese herbal medicines. 2018
- [CrossRef] [Google Scholar]
- The evolutionary interplay between adaptation and self-fertilization. Trends in Genetics. 2017;33(6):420-431.
- [CrossRef] [Google Scholar]
- Great Cause-Small Effect: Undeclared Genetically Engineered Orange Petunias Harbor an Inefficient Dihydroflavonol 4-Reductase. Frontiers in Plant Science. 2018;9:149.
- [CrossRef] [Google Scholar]
- Global epigenetic and transcriptional trends among two rice subspecies and their reciprocal hybrids. The Plant Cell. 2010;22:17-33.
- [CrossRef] [Google Scholar]
- More than causing (epi)genomic instability: emerging physiological implications of transposable element modulation. J Biomed Sci. 2021;28:58.
- [CrossRef] [Google Scholar]
- Hussein, Z.T. The molecular effect of selfing in the genetic and epigenetic diversity and their relationship with hybrid vigor in maize. 2018, [MSc. Thesis.] Anbar, University of Anbar.
- Somaclonal Variation and Induced Mutation in crop improvement. Dordrecht: Kluwer academic publisher; 1998. p. :81-104.
- S.M. Jain J.M. Al-Khayri D.V. Johnson Date Palm Biotechnology” SPRINGER NATURE. P762 (2011)“.
- Jiang, C.; Mithani, A.; Gan, X.; Belfield, E. J.; Klingler, J. P.; Zhu, J.K.; Ragoussis, J.; Mott, R.; Harberd, N P. Regenerant Arabidopsis lineages display a distinct genome-wide spectrum of mutations conferring variant phenotypes. Curr Biol, 2011, 21, 1385–1390. https ://doi.org/10.1016/j. cub.2011.07.002.
- Transcriptome analysis reveals the complexity of alternative splicing regulation in the fungus Verticillium dahliae. BMC Genomics. 2017;18:130.
- [CrossRef] [Google Scholar]
- Epigenetic aspects of Somaclonal variation in plants. Plant Mol Biol. 2000;43:179-188.
- [CrossRef] [Google Scholar]
- The role of conventional plant breeding in ensuring safe levels of naturally occurring toxins in food crops. Trends in Food Science and Technology. 2020;100:51-66.
- [CrossRef] [Google Scholar]
- Epigenetic role of DNA methylation in hybrid vigor of cytoplasmic male sterility in sunflower. Pakistan Journal of Botany. 2021;53(1):191-196.
- [CrossRef] [Google Scholar]
- Deep sequencing uncovers commonality in small RNA profiles between transgene-induced and naturally occurring RNA silencing of chalcone synthase-A gene in petunia. BMC Genomics. 2013;14:63.
- [CrossRef] [Google Scholar]
- Diversity and dynamics of DNA methylation: epigenomic resources and tools for crop breeding. Breeding Science. 2019;69(2):191-204.
- [CrossRef] [Google Scholar]
- Taming the TurmoilWithin: New Insights on the Containment of Transposable Elements. Trends Genet.. 2020;36:474-489.
- [Google Scholar]
- Theoretical aspects of heterosis. Russian Journal of Genetics: Applied Research. 2017;7(4):428-439.
- [CrossRef] [Google Scholar]
- Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiology. 2012;53:847-856.
- [Google Scholar]
- Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Frontiers in Plant Science. 2015;2(6):114.
- [CrossRef] [Google Scholar]
- Plasmodesmata-mediated cell-to-cell communication in the shoot apical meristem: How stem cells talk. Plants. 2017;6:12.
- [CrossRef] [Google Scholar]
- Garcinol—A natural histone acetyltransferase inhibitor and new anti-cancer epigenetic drug. Int. J. Mol. Sci.. 2020;22:2828.
- [CrossRef] [Google Scholar]
- Heterosis and Hybrid Crop Breeding: A multidisciplinary review. Front. Genet. 2021
- [CrossRef] [Google Scholar]
- Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biology. 2017;18:124.
- [CrossRef] [Google Scholar]
- Lauria, M.; Piccinini, S.; Pirona, R.; Lund, G.; Viotti, A.; Motto, M. Epigenetic variation, inheritance, and parent-of-origin effects of cytosine methylation in maize (Zea mays). Genetics, 4, 196, 653–666.
- Phenotypic variation in plants: Roles for epigenetics [PhD. Thesis.]. Amsterdam: (FNWI), University of Amsterdam; 2017.
- K. Lauss R. Wardenaar ,; Oka, R., Hulten, M.H.A., Guryev V., Keurentjes, J.J.B., Stam, M., Johannes, F. Parental DNA Methylation states are associated with heterosis in epigenetic hybrids Plant Physiology 176 2 2018 1627 1645 10.1104/pp.17.01054.
- Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature Reviews Genetics. 2010;11(3):204220
- [CrossRef] [Google Scholar]
- Regulatory RNAs: A universal language for inter-domain communication. International Journal of Molecular Sciences. 2020;21(23):8919.
- [CrossRef] [Google Scholar]
- Fine-tuning of gene expression dynamics by the Set2-Rpd3S pathway. BMB Reports. 2017;50(4):162-163.
- [CrossRef] [Google Scholar]
- Histone acetylation modifications affect tissue-dependent expression of poplar homologs of C4 photosynthetic enzyme genes. Frontiers. Plant Science. 2017;8(8):950.
- [CrossRef] [Google Scholar]
- P. Li H. Yang W.L. Liu H., Huo, H., Zhang, C., Liu, A., Zhu, A., Hu, J., Lin, Y., Liu, L. Physiological and transcriptome analyses reveal short-term responses and formation of memory under drought stress in rice Frontiers in Genetics 8 10 2019 55 10.3389/fgene.2019.00055.
- Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biology. 2014;23(14):105.
- [CrossRef] [Google Scholar]
- New technologies accelerate the exploration of non-coding RNAs in horticultural plants. Horticulture Research. 2017;4:17031.
- [CrossRef] [Google Scholar]
- Memory responses of jasmonic acid-associated Arabidopsis genes to a repeated dehydration stress. Plant, Cell and Environment. 2016;39(11):2515-2529.
- [Google Scholar]
- Analysis of DNA methylation patterns and levels in maize hybrids and their parents. Genetics and Molecular Research. 2014;13(4):8458-8468.
- [Google Scholar]
- Long noncoding RNA transcriptome of plants. Plant Biotechnology Journal. 2015;13:319-328.
- [Google Scholar]
- Histone acetylation and plant development. Enzymes. 2017;40:173-199.
- [CrossRef]
- Histone deacetylases and their functions in plants. Plant Cell Report. 2013;32:465-478.
- [CrossRef] [Google Scholar]
- Machczyńska, J.; Zimny, J.; Bednarek, P. Tissue culture-induced genetic and epigenetic variation in triticale (× Triticosecale spp. Wittmack ex A. Camus 1927) regenerants. Plant Mol Biol, 2015, 89, 279–292. https ://doi.org/10.1007/s1110 3-015-0368-0.
- Extended metAFLP approach in studies of the tissue culture induced variation (TCIV) in case of triticale. Mol Breed. 2014;34(845–854)
- [CrossRef] [Google Scholar]
- Arabidopsis non-coding RNA regulation in abiotic stress responses. International Journal of Molecular Sciences. 2013;14:22642-22654.
- [Google Scholar]
- Landscaping plant epigenetics. Methods in Molecular Biology. 2014;1112:1-24.
- [CrossRef] [Google Scholar]
- C. Miguel L. Marum An epigenetic view of plant cells cultured in vitro: somaclonal variation and beyond J Exp Bot. 62 11 2011 3713 3725 10.1093/jxb/err155 Epub 2011 May 26 PMID: 21617249.
- Miyao, A.; Nakagome, M;. Ohnuma, T.; Yamagata, H.; Kanamori, H.; Katayose, Y.; Takahashi, A.; Matsumoto, T.; Hirochika, H. Molecular spectrum of Somaclonal variation in regenerated rice revealed by whole-genome sequencing. Plant Cell Physiol, 2011, 53, 256–264. https ://doi.org/10.1093/pcp/pcr17 2.
- Murray; S.C., Lorenz, P., Howe; F.S., Wouters; M., Brown; T., Xi, S., Fischl, H., Khushaim, W., Rayappu; J.R., Angel, A., Mellor, J. H3K4me3 is neither instructive for, nor informed by transcription bioRxiv 709014 2019 doi:org/10.1101/709014 (Preprint).
- Gene capture by transposable elements leads to epigenetic conflict in maize. Mol. Plant. 2020
- [CrossRef] [Google Scholar]
- A conserved sequence signature is essential for robust plant miRNA biogenesis. Nucleic Acids Research. 2020;48(6):3103-3118.
- [CrossRef] [Google Scholar]
- Neelakandan, A.K.; Wang, K. Recent progress in the of tissue culture-induced genome level changes in plants and potential applications. Plant Cell Rep, 2012, 31,597–620. https ://doi. org/10.1007/s0029 9-011-1202-z.
- Nhut, D.T.; Hai, N.T.; Thu, P.T.M.; Thi, N.N.; Hien, T.T.D.; Tuan, T.T.; Nam, N.B.; Huy, N.P.; Chien, H.X.; Jain, S.M. (2013): Protocol for inducing flower color Somaclonal variation in Torenia (Torenia fournieri Lind). Methods in Molecular Biology. 2013, 11013, 455-62.V994, DOI: 10.1007/978-1-62703-074-8_34.
- (2021): somatic embryogenesis-an attempt to modify variation induced in tissue culture. J of Biol Res-Thessaloniki. 2021;28:9.
- [CrossRef] [Google Scholar]
- Orłowska, R.; Bednarek, P.T. Precise evaluation of tissue culture-induced variation during optimisation of in vitro regeneration regime in barley. Plant Mol Biol. 2020, 103, 1-2, 33-50. doi: 10.1007/s11103-020-00973-5. Epub 2020 Feb 11. PMID: 32048207; PMCID: PMC7170832.
- DNA methylation changes and TE activity induced in tissue cultures of barley (Hordeum vulgare L.) J Biol Res. 2016;23(19–19)
- [CrossRef] [Google Scholar]
- Y.-Y. Pang R.-H. Lu P.-Y. Chen Behavioral epigenetics: perspectives based on experience-dependent epigenetic inheritance Epigenomes 3 3 2019 18 10.3390/ epigenomes3030018.
- Characterization of a drought inducible trithorax-like H3K4 methyltransferase from barley. Biologia Plantarum. 2012;56(4):683-692.
- [Google Scholar]
- Transposable Elements and Stress in Vertebrates: An Overview. Int. J. Mol. Sci.. 1970;2021:22.
- [CrossRef] [Google Scholar]
- Pradillo, MChromatin dynamics during interphase and cell division: similarities and differences between model and crop plants. Journal of Experimental Botany. 2020;71(17):5205-5222.
- [CrossRef] [Google Scholar]
- Genome-Wide identification of histone modifiers and their expression patterns during fruit abscission in Litchi. Frontiers in Plant Science. 2017;8(639):1-16.
- [CrossRef] [Google Scholar]
- Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. Plant Cell. 2013;25:3491-3505.
- [CrossRef] [Google Scholar]
- New Plant Breeding Techniques. The Parliamentary Office of Science and Technology, Westminster, London-UK. 2017;548:1-4.
- [Google Scholar]
- Computational identification of miRNAs and their targets from Niger (Guizotia abyssinica) Journal of Applied Biology and Biotechnology. 2017;5(2):53-58.
- [CrossRef] [Google Scholar]
- Identification and characterization of the SET domain gene family in maize. Molecular Biology Reports. 2014;41(3):1341-1354.
- [CrossRef] [Google Scholar]
- S. Qin P. Jin X. Zhou L. Chen F. Ma The Role of Transposable Elements in the Origin and Evolution of MicroRNAs in Human PLoSOne. 10 6 2015 10.1371/journal.pone.0131365 e0131365.
- Plant Epigenetics. 2017;p:536.
- Plant abiotic stress tolerance analysis in cauliflower using curd micropropagation system. Acta Hort.. 2015;1083:43-52.
- [CrossRef] [Google Scholar]
- Evolution and function of genomic imprinting in plants. Genes and Development. 2015;29:2517-2531.
- [CrossRef] [Google Scholar]
- Nuclear rDNA instability in in vitro-generated plants is amplified after sexual reproduction with conspecific wild individuals. Bot J Linn Soc. 2016;181(127–137)
- [CrossRef] [Google Scholar]
- Rebuilding core abscisic acid signaling pathways of Arabidopsis in yeast. EMBO Reports. 2019;2, 38, 17:e101859.
- [Google Scholar]
- F. Sabot N. Picault M. El-Baidouri C. Llauro C. Chaparro B. Piegu A. Roulin E. Guiderdoni M. Delabastide R. McCombie O. Panaud Transpositional landscape of the rice genome revealed by paired-end mapping of high-throughput resequencing data Plant J 66 2011 241 246 10.1111/ j.1365-313X.2011.04492.x.
- Transcriptomic analysis of tomato lines reveals putative stress-specific biomarkers. Turkish Journal of Agriculture and Forestry. 2014;38(5):700-715.
- [Google Scholar]
- Sadder, M.T.; Alsadon, A.; Al-Thamra, M.; Zakri, A.M.; Al-Doss, A. Phylogenetic analysis of SET domain in trithorax SlTX1 of 'Solanum lycopersicum'. Plant Omics, 2011,4, 2, 95- 99. 025 Plant Omics J 2011 SlTX1-SET-.pdf.
- Genetic and Molecular Control of Somatic Embryogenesis. Plants. 2021;10(7):1467.
- [CrossRef] [Google Scholar]
- The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. The Plant Cell. 2008;20(3):568-579.
- [Google Scholar]
- Hyperosmotic priming of Arabidopsisseedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome. Genome Biology. 2013;14:R59.
- [CrossRef] [Google Scholar]
- Concerted flexibility of chromatin structure, methylome, and histone modifications along with plant stress responses. Biology (Basel). 2017;6(1):3.
- [CrossRef] [Google Scholar]
- Small RNAs break out: the molecular cell biology of mobile small RNAs. Nature Reviews Molecular Cell Biology. 2014;15(8):525-535.
- [CrossRef] [Google Scholar]
- Epigenetic and epigenomic variation in Arabidopsis thaliana. Trends in Plant Science. 2012;17:149-154.
- [CrossRef] [Google Scholar]
- Nutrient stress-induced chromatin changes in plants. Current Opinion in Plant Biology. 2017;39:1-7.
- [CrossRef] [Google Scholar]
- The current state and future directions of RNAi-based therapeutics. Nature Reviews Drug Discovery. 2019;18:421-446.
- [CrossRef] [Google Scholar]
- DNA methylation as the most important content epigenetics in traditional Chinese herbal medicine. Journal of Medicinal Plants Research.. 2019;13(16):357-369.
- [CrossRef] [Google Scholar]
- Genome-wide analysis of DNA methylation and gene expression changes in two Arabidopsis ecotypes and their reciprocal hybrids. The Plant Cell. 2012;24:875-892.
- [CrossRef] [Google Scholar]
- Transient stability of epigenetic population differentiation in a clonal invader. Frontiers in Plant Science. 2019;1(9):1851.
- [CrossRef] [Google Scholar]
- Review paper on approaches in developing inbred lines in cross-pollinated crops. Biochemistry and Molecular Biology. 2017;2(4):40-45.
- [CrossRef] [Google Scholar]
- The Polycomb group protein MEDEA controls cell proliferation and embryonic patterning in Arabidopsis. Developmental Cell.. 2021;56(13):1945-1960.e7.
- [CrossRef] [Google Scholar]
- Gating of miRNA movement at defined cell-cell interfaces governs their impact as positional signals. Nature communications. 2018;9(1):1-10.
- [Google Scholar]
- MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol.. 2019;70:489-525.
- [CrossRef] [Google Scholar]
- Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Molecular Plant. 2010;3:1012-1025.
- [CrossRef] [Google Scholar]
- Exploiting induced and natural epigenetic variation for crop improvement. Nature Reviews Genetics. 2017;18:563-575.
- [CrossRef] [Google Scholar]
- Drost, H-G How Stress Facilitates Phenotypic Innovation Through Epigenetic Diversity. Front. Plant Sci.. 2021;11:606800
- [CrossRef] [Google Scholar]
- Functions of microRNAs in plant stress responses. Trends in Plant Science. 2012;17:196-203.
- [CrossRef] [Google Scholar]
- Z. Teng Y. Yu Z. Zhu S.B. Hong B. Yang Y. Zang Melatonin elevated Sclerotinia sclerotium resistance via modulation of ATP and glucosinolates biosynthesis in Brassica rapa ssp Pekinensis. Journal of proteomics. (2021): 243:doi:10.1016/j.jprot.2021.104264.
- Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biology. 2010;10:238.
- [CrossRef] [Google Scholar]
- AGO1 homeostasis entails co expression of MIR168 and AGO1 and preferential stabilization of miR168 by AGO1. Molecular Cell. 2006;22:129-136.
- [CrossRef] [Google Scholar]
- Emerging roles of chromatin in the maintenance of genome organization and function in plants. Genome Biology. 2017;18:96.
- [CrossRef] [Google Scholar]
- Evidence for an epigenetic role in inbreeding depression. Biology Letters. 2012;8:798-801.
- [CrossRef] [Google Scholar]
- Dehydration stress memory: Gene networks linked to physiological responses during repeated stresses of Zea mays. Frontiers in Plant Science. 2018;9:9.
- [CrossRef] [Google Scholar]
- Costs and benefits of hormone-regulated plant defences. Plant Pathol.. 2013;62:43-55.
- [CrossRef] [Google Scholar]
- The roles of histone acetylation in seed performance and plant development. Plant Physiology and Biochemistry. 2014;84:125-133.
- [CrossRef] [Google Scholar]
- Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proceedings of the National Academy of Sciences. 2015;112(27):8487-8492.
- [Google Scholar]
- Epigenomics—technologies and applications. Circulation Research. 2018;122(9):1191-1199.
- [CrossRef] [Google Scholar]
- Stress and the emerging roles of chromatin remodeling in signal integration and stable transmission of reversible phenotypes. Frontiers in Behavioral Neuroscience. 2017;11:41.
- [CrossRef] [Google Scholar]
- A field guide to eukaryotic transposable elements. Annu. Rev. Genet.. 2020;54:539-561.
- [CrossRef] [Google Scholar]
- Histone chaperone ASF1 is involved in gene transcription activation in response to heat stress in Arabidopsis thaliana. Plant Cell Environment. 2014;37:2128-2138.
- [Google Scholar]
- A. Wibowo C. Becker G. Marconi J. Durr J. Price J. Hagmann Hyperosmotic stress memory in Arabidopsis is mediated by distinct epigenetically labile sites in the genome and is restricted in the male germline by DNA glycosylase activity Elife 5 2016 10.7554/eLife.13546 e13546.
- Plant microRNAs and development. Journal of Genetics and Genomics. 2013;40(5):217-230.
- [Google Scholar]
- Epigenetic agents trigger the production of bioactive nucleoside derivatives and bisabolanes sesquiterpenes from the marine-derived fungus Aspergillus versicolor. Frontiers in microbiology.. 2020;11:85.
- [CrossRef] [Google Scholar]
- Developmental transitions: integrating environmental cues with hormonal signaling in the chromatin landscape in plants. Genome Biology. 2017;18:88.
- [CrossRef] [Google Scholar]
- Altered expression profiles of microRNA families during de-etiolation of maize and rice leaves. BMC Research Notes. 2017;10:108.
- [CrossRef] [Google Scholar]
- The role of epigenetic processes in controlling flowering time in plants exposed to stress. Journal of Experimental Botany. 2011;62(11):3727-3735.
- [CrossRef] [Google Scholar]
- The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell. 2015;27:1670-1680.
- [CrossRef] [Google Scholar]
- Parental epigenetic difference in DNA methylation-level may play contrasting roles for different agronomic traits related to yield heterosis in maize. African Journal of Biotechnology. 2011;10(46):9253-9263.
- [Google Scholar]
- HAT4, a Golgi apparatus-anchored B-type histone acetyltransferase, acetylates free histone H4 and facilitates chromatin assembly. Molecular cell. 2011;44(1):39-50.
- [CrossRef] [Google Scholar]
- Histone acetylation dynamics integrates metabolic activity to regulate plant response to stress. Frontiers in Plant Science. 2019;10:1236.
- [CrossRef] [Google Scholar]
- Effects of Hormones and Epigenetic Regulation on the Callus and Adventitious Bud Induction of Fraxinus mandshurica Rupr. Forests. 2020;11:590.
- [CrossRef] [Google Scholar]
- Advances in cereal crop genomics for resilience under climate change. Life. 2021;11:502.
- [CrossRef] [Google Scholar]
- Epigenetic Regulation by Noncoding RNAs in Plant Development. In: Rajewsky N., Jurga S., Barciszewski J., eds. Plant Epigenetics. Cham: Springer International Publishing:; 2017. p. :183-198.
- [CrossRef] [Google Scholar]
- The SWI2/SNF2 chromatin-remodeling ATPase BRAHMA regulates chlorophyll biosynthesis in Arabidopsis. Molecular Plant. 2017;10:155-167.
- [CrossRef] [Google Scholar]
- Assessment of Epigenetic and Phenotypic Variation in Populus nigra Regenerated via Sequential Regeneration. Front. Plant Sci.. 2021;12:632088
- [CrossRef] [Google Scholar]
- Natural variation in DNA methylation homeostasis and the emergence of epialleles. Proceedings of the National Academy of Sciences. 2020;117(9):4874-4884.
- [CrossRef] [Google Scholar]
- Epigenetic inheritance and variation of DNA methylation level and pattern in maize intra-specific hybrids. Plant Science. 2007;172:930-938.
- [Google Scholar]
- Editorial: Molecular and cellular plant reproduction. Frontiers in Plant Science. 2017;8:199.
- [CrossRef] [Google Scholar]
- Epigenomic modification and epigenetic regulation in rice. Journal of Genetics and Genomics. 2012;39:307-315.
- [CrossRef] [Google Scholar]
- Exploiting variations for crop disease resistance improvement. Front. Plant Sci.. 2021;04
- [CrossRef] [Google Scholar]
- Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. The Plant Cell. 2017;29:1088-1104.
- [CrossRef] [Google Scholar]
Further reading
- The complex language of chromatin regulation during transcription. Nature. 2007;447:407-412.
- [CrossRef] [Google Scholar]







