Translate this page into:
Enhanced protection of tomato against Fusarium wilt through biopriming with Trichoderma harzianum
⁎Corresponding author. mukeshmeenamlsu@gmail.com (Mukesh Meena) mukeshmeenabhu@gmail.com (Mukesh Meena)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objective
Microbial priming represents an adaptive strategy to enhance the plant defense against subsequent challenges incited by pathogenic microbes. The aim of the study was to investigate the effect of priming with Trichoderma harzianum (Th) on the induced resistance potential of tomato after challenged with Fusarium oxysporum f. sp. lycopersici (Fol) pathogen.
Methods
This work demonstrated antioxidative and defense related enzyme activities and qRT-PCR to study the resistance mechanisms of tomato plants bioprimed with T. harzianum against Fol pathogen.
Result
Microbial biopriming with T. harzianum resulted into enhanced expression of tomato defense-related genes and was accompanied by increased antioxidative enzymic activities. The study reported that the T. harzianum primed plants showed 2.71-fold higher SOD than control and 1.34-fold (Fol + Th) higher SOD activity compared to Fol challenged plants. In contrast, Fol + Th treated showed 5.87-fold and 1.34-fold higher CAT enzyme activity as compared to control and pathogen exposed plants. T. harzianum bioprimed plants noted 1.47- and 11.47-fold enhanced PPO activity as compared to Fol challenged and controls, respectively. PAL and PO activities were also found higher in T. harzianum primed plants. The qRT-PCR revealed that expression of defense related gene showed higher up-regulation in T. harzianum primed plants as compared to pathogen challenged plants. As compared to control, Fol + Th treated plants also showed higher up-regulation of all the studied genes.
Conclusion
The study concluded T. harzianum priming aggravates the plant defense system against the Fol challenged condition and accompanied by higher expression of defense related genes and increased antioxidative activities against subsequent Fol attack.
Keywords
Microbial biopriming
Trichoderma
Plant defense
Pathogen
Defense genes
- BCAs
-
Biological control agents
- PAMPs
-
Pathogen-associated molecular patterns
- MAMPs
-
Microbe-associated molecular patterns
- SA
-
Salicylic acid
- PRRs
-
Pattern recognition receptors
- SAR
-
Systemic acquired resistance
- PR genes
-
Pathogenesis-related genes
- PCR
-
Polymerase chain reaction
- NCBI-BLAST
-
National center for biotechnology information-Basic local alignment search tool
- ROS
-
Reactive oxygen species
- PDA
-
Potato dextrose agar
- NaOCl
-
Sodium hypochlorite
- SOD
-
Superoxide dismutase
- CAT
-
Catalase
- PO
-
Peroxidase
- PAL
-
Phenylalanine ammonia-lyase
- PPO
-
Polyphenol oxidase
- CT
-
Cycle threshold
- ANOVA
-
Analysis of variance
- ISR
-
Induce systemic resistance
- PTI
-
Pathogen-triggered immunity
- ETI
-
Effector-triggered immunity
- JA
-
Jasmonic acid
- ET
-
Ethylene
Abbreviations
1 Introduction
Tomato (Solanum lycopersicum Mill.) plant is one of the most important vegetable crops grown for high nutritional aspects as well as its medicinal values (Alimohammadi et al., 2017). Fusarium wilt known for the causing serious diseases of tomato (Zhu et al., 2019) which could lead to 30–40% damages to the tomato crops and under the more favourable conditions, it might go upto 80% losses. Classical methods to control the disease are now being questioned, and the use of beneficial microbes, to control plant diseases has been found to be an environmentally friendly, economically useful and therefore, enhancing agricultural productivity in a sustainable manner (Abdelrahman et al., 2016). Biological control agents (BCAs) directly suppress the growth of phytopathogens and microbial priming could results into pre-activation of immune response against the challenging pathogens (Walters et al., 2013; Singh et al., 2020).
Trichoderma species helps to promote growth of the plant by following various direct and indirect mechanisms and induce systemic resistance against subsequent pathogen attack (Al-Ani, 2018; Meena et al., 2017a). Trichoderma interaction with plants could be responsible to express defense-related genes in plant which induces immune system of the plants against the various pathogen challenge and affect the growth and development (Galletti et al., 2020; Pimentel et al., 2020). Mostly plants known to recognize microbial elicitors or surface derived molecules (PAMPs; pathogen-associated molecular patterns or MAMPs; microbe-associated molecular patterns) of microbes through their immune receptors. However, the endogenous salicylic acid (SA) during pattern recognition receptors (PRRs) activation results into transcriptional re-programming of defense-related genes developing systemic acquired resistance (SAR). Infact, PRRs activation results into activation of a broad spectrum type basal immunity that leads into transcription of pathogenesis-related (PR) genes triggering production of the reactive oxygen species (ROS) molecules, biosynthesis of defense-related proteins and secondary metabolites and programmed cell death (Husaini et al., 2018).
In many studies, Trichoerma spp. mediated priming against phytopathogens and its effect on key defense players have been demonstrated (Boccardo et al., 2019). However, srtudies on T. harzianum mediated defense programming against the Fusarium wilt of tomato is limited. Therefore, the present study was undertaken to evaluate the expression profile changes of key defense genes, and to measure the specific activities of defense enzymes acts in tomato leaves, pre-inoculated with T. harzianum, following the infection of the Fol pathogen to gain a better insights into possible relationship in between the induction of defense enzymes and induced disease resistance.
2 Material and methods
2.1 Pathogen isolation
Pathogen was isolated from the infected tomato plants on the Petri-plates containing potato dextrose agar (PDA) media (Zehra et al., 2017a). After inoculation, the plates were incubated at 25–27 °C for 5–7 days.
2.2 Isolation of Trichoderma spp.
For isolating Trichoderma spp., soil samples were collected from tomato rhizosphere located at different places of the agricultural field of Banaras Hindu University campus, Varanasi (25°20′N latitude and 83°01′E longitude), Uttar Pradesh, India and were kept at 4 °C in the laboratory. Serial dilution method was adopted for isolating different Trichoderma spp. (Zehra et al., 2017b).
2.3 Fungal DNA extraction and ITS sequencing
Genomic DNA was extracted from the 1 week old from the fungal isolate following the method of Doyle and Doyle (1987). The universal primers ITS1/ITS4 were used to amplify ITS-rDNA using polymerase chain reaction (PCR) (Meena et al., 2017b). Finally, the purified products were transferred for both end sequencing and analysis. Using, National center for biotechnology information-Basic local alignment search tool (NCBI-BLAST), the sequences were matched in GenBank (https://www.ncbi.nlm.nih.gov/).
2.4 Plant material
Tomato seeds (Lycopersicon esculentum cv. Punjab Chhuara) were collected from Indian Institute of Vegetable Research (IIVR), Varanasi, Uttar Pradesh and further surface of the seeds were sterilized in 4% sodium hypochlorite (NaOCl) solution having Tween-20 (0.02% v/v), rinsed systematically using disinfected water, and then allowed for germination (Meena et al., 2016).
2.5 Experimental design, biological treatments and growth condition
The experiment was performed in plastic pots (30 cm diameter; surface sterilized with 0.1% mercuric chloride). The treatments were arranged as (1) uninoculated control, (2) pathogen (Fol) challenged, and (3) plants primed with T. harzianum LMMT spores and challenge inoculated with pathogen (Fol). The individual seedlings were transferred to the pot filled with a sterile soil mixed with sand (autoclaved with 121 °C and 15 psi pressure for 15 min):soil (4:1) mixture along with Trichoderma inoculums 106 conidia/g of soil. Experiments was performed in three biological replicates (five seedlings per replicate) for each treatment. The same amount of sand:soil mix but free from T. harzianum LMMT were added to the uninoculated control plants. All the plants were kept and grown in a green house that was maintained on a temperature 25–29 °C with 70% humidity, and 14 h light and 10 h dark cycle. After 3 weeks of seedling transplantation, plants were made infected with the Fol pathogen. For this the Fol spore suspension was inoculated in the same soil sand mixtures at a concentration of 106 conidia/g in all the Fol challenged and T. harzianum LMMT supplemented plants, whereas the uninoculted control plants were kept as such. The leaves from all the three treatments were plucked at five different stages at different time intervals 0, 24, 48, 72 and 96 h post inoculation (hpi) with the pathogen and used for biochemical analysis. The expression profile changes of the defense associated genes encoding defense-related antioxidative enzymes like SOD, CAT, PO and other proteins like PR proteins, PAL and PPO were also measured. The transcript abundance and relative gene expression were measured in all the treatments during the different interval of time varying from 0 to 96 h.
2.6 Superoxide dismutase (SOD), catalase (CAT) and peroxidase (PO) activity assay
The activity of SOD (EC 1.15.1.1) was measured using a method proposed by Fridovich (1974). The estimation of CAT (EC 1.11.1.6) activity was done spectrophotometrically using the previous described method exemplified by McKersie et al. (1993). The approach given by Hammerschmidt et al. (1982) was used to calculate the activity of PO (EC 1.11.1.7).
2.7 Phenylalanine Ammonia-Lyase (PAL) and polyphenol oxidase (PPO) activity assay
PAL (EC 4.1.3.5) activity was assayed according to the procedure as described by El-Shora (2002) with minor modifications. PPO (EC 1.14.18.1) activity was performed using the method of Gauillard et al. (1993).
2.8 PR-2 protein (β-1,3-glucanase) and PR-3 protein (Chitinase) assay
The activity of β-1,3-glucanase and chitinase (EC 3.2.1.14) was measured using Pan et al. (1991) and Giri and Aggarwal's (1998) technique, respectively with some minor changes.
2.9 qRT-PCR analysis
The expression patterns of defense-related genes in RNA samples of all the treatments of tomato leaves were verified by real time-PCR. Total RNA were isolated from the control and treated tomato leaf tissues using Chromous RNA isolation Kit. First strand of cDNA was synthesized using 1 μg of RNA sample with random primers with the help of cDNA synthesis kit according to the manufacture’s instructions. Primer was designed by Primer 3 software. Primers for qRT-PCR analysis were synthesized according to the following references (Song et al., 2015; Jayanna and Umesha, 2017; Aazami et al., 2021). The RT-PCR was performed by following the method of Cheng et al. (2017). All reactions were performed in triplicates on ABI Step-one RT-PCR machine. Primers with master mix was used as a negative control in all RT-PCR runs. Real time software was used to analyze the output of RT-PCR runs for the determination of relative expression used by cycle threshold (CT) values. To normalize each reaction rubisco transcript was used. All three independent biological replicates followed the same trends of expression change, of which one such value has been represented in this study.
2.10 Statistical analysis
SPSS Version 16.0 was used to perform the statistical analysis of three independent biological replicates of all the experiments. All the data were used to analyze one-way analysis of variance (ANOVA) and Duncan’s multiple range test to determine the significance of differences at P ≤ 0.05 between the treatments.
3 Results
3.1 Pathogen identification
The pathogen recovered from wilted tomato plants when grown on PDA plates showed pink-white color of the fungal hyphae with cottony surface. The hyphae when observed in the microscope showed asexual structures including microconidia and macroconidia. Koch’s postulates followed for pathogenicity test of all the isolates of Fol. A highly virulent based on percentage occurrence of disease development was further selected for experiments.
3.2 Identification of Trichoderma
The species of isolated Trichoderma was identified as Trichoderma harzianum based on the morphological and conidial characteristics using the key as suggested by Rifai (1969) and Nagamani et al. (2002). The raw data obtained from the sequencing results were submitted to NCBI after annotation. The annotated ITS sequence was perform for BLASTn analysis using the standard database nucleotide collection (nr/nt). The BLASTn results showed sequence similarity with T. harzianum.The accession number provided by NCBI Genbank for T. harzianum BHU LMMT (Th) was KX 091167.
3.3 Disease incidence and Biocontrol efficacy
The T. harzianum pre-treated induced resistance in tomato seedlings were recorded (14 days after challenging the pathogen) through the decrease in severity of wilt disease infected with Fol. Observations noted maximum disease percentage for pathogen infected plants and it was 84.16%. It was noted that Th primed tomato plants exhibited significant reduction in disease severity (10.18%) while compared with Fol challenged plants. Biocontrol efficacy of Th against Fol was found to be 87.90%.
3.4 SOD, CAT and PO activity
After inoculation both non-inoculated pathogen control and the Th inoculated plants enzyme activity enhances with time. As shown in Fig. 1A, Th primed plants showed elevation in SOD generation at 48 hpi in compare to the pathogen challenged and control plants. SOD activity level rises at 48 hpi and then decreased thereafter. Th primed plants showed 2.71- and 1.34-fold higher SOD enzyme activity in compare to pathogen challenged and control plants. Natural SOD activity was regularly present in control during the assays. Activity of enzyme in non-inoculated pathogen control and the Th inoculated plants rises with time after inoculation. CAT activity in tomato primed with Th decreased after pathogen inoculation. An increase in CAT activity was observed gradually after 24 h of pathogen challenge. Maximum activity was observed at 96 hpi in Fol + Th treated plants. Fol + Th treated showed 5.87- and 1.34-fold higher CAT enzyme activity in compare to control infected plants (Fig. 1B). Elevation in activity of PO was noted in both treatments compare to the control. At 72 h, PO activity recorded maximum in all treatment, after that it gets reduce. Th prepared plants noted 1.26- and 10.94-fold increase activity of PO in compare to pathogen-infected and without treated plants, individually (Fig. 1C). A little sum of PO action was reliably recorded in sound control plants.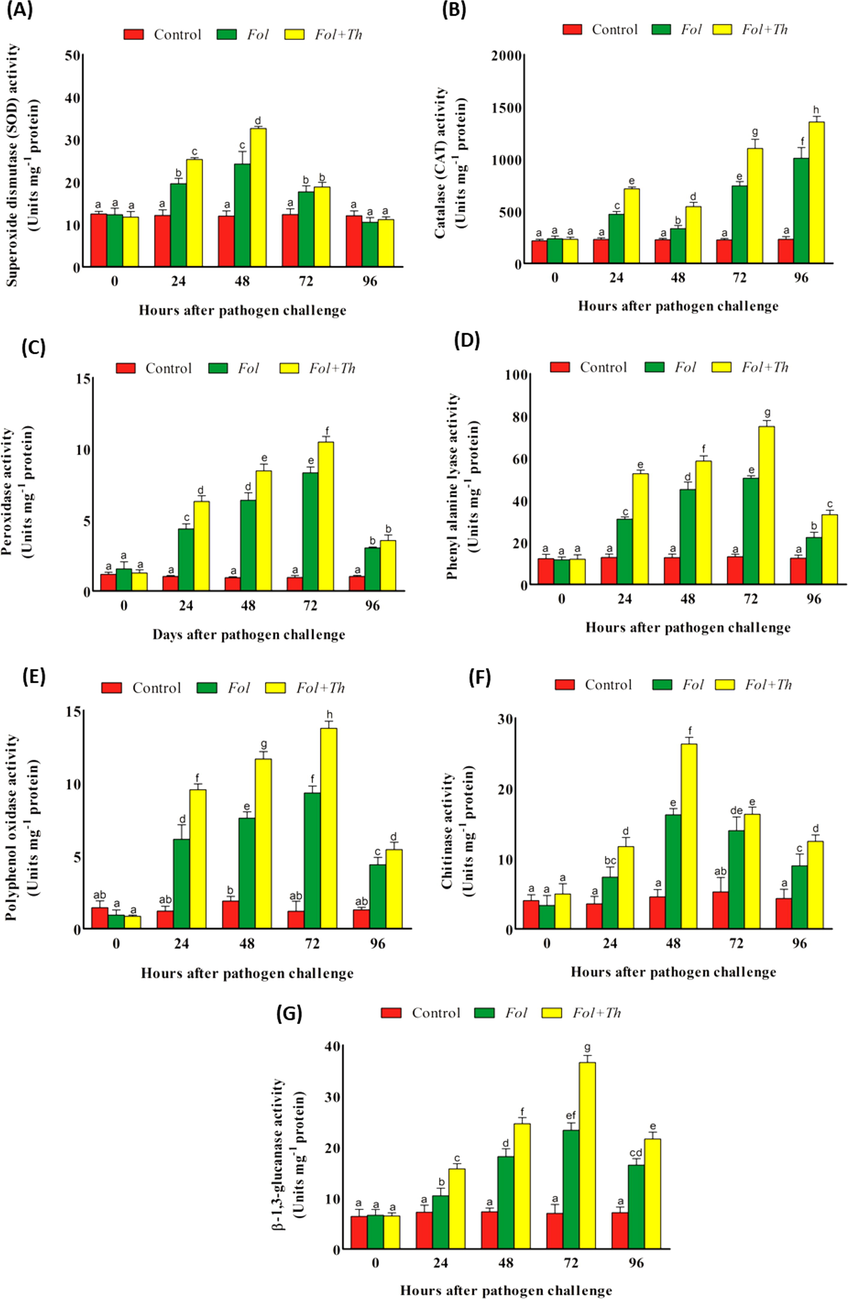
Estimation of changes in the activities of (A) SOD, (B) CAT, (C) PO, (D) PAL, (E) PPO, (F) Chitinase and (G) β-1,3-Glucanase in the leaves of tomato plants pre-treated with T. harzianum, after challenge inoculation with F. oxysporum f. sp. lycopersici (Fol).
3.5 PAL and PPO activity
PAL increases significantly in both treatments during the first 72 h, then falls. In comparison to control and pathogen-challenged plants, Th primed plants showed 1.48- and 5.70-fold higher PAL activity after 72 h (Fig. 1D). PPO activity increases in both treatments when compared to control plants. PPO activity was at its peak at 72 h in all of the treatments, however it gradually decreased. When compared to pathogen-inoculated and untreated controls, Th primed plants had 1.47- and 11.47-fold greater PPO activity, respectively (Fig. 1E). In healthy control plants, a small level of PPO activity was routinely recorded.
3.6 Chitinase and β-1,3-Glucanase activity
In comparison to control plants, chitinase activity increased after pathogen exposure. Fol + Th treated plants had a 1.62- and 5.78-fold increase in chitinase activity at 72 h as compared to pathogen challenged and control plants. Following that, all of the treatments showed a decrease in chitinase activity (Fig. 2F). In comparison to control plants, activity of β-1,3-glucanase was reported to increase after pathogen challenge. At 72 h, Fol + Th treated plants had a 1.96- and 16.08-fold increase in β-1,3-glucanase activity, respectively, when compared to pathogen challenged and control plants (Fig. 2G). Following that, all of the treatments showed a decreasing trend in β-1,3-glucanase activity.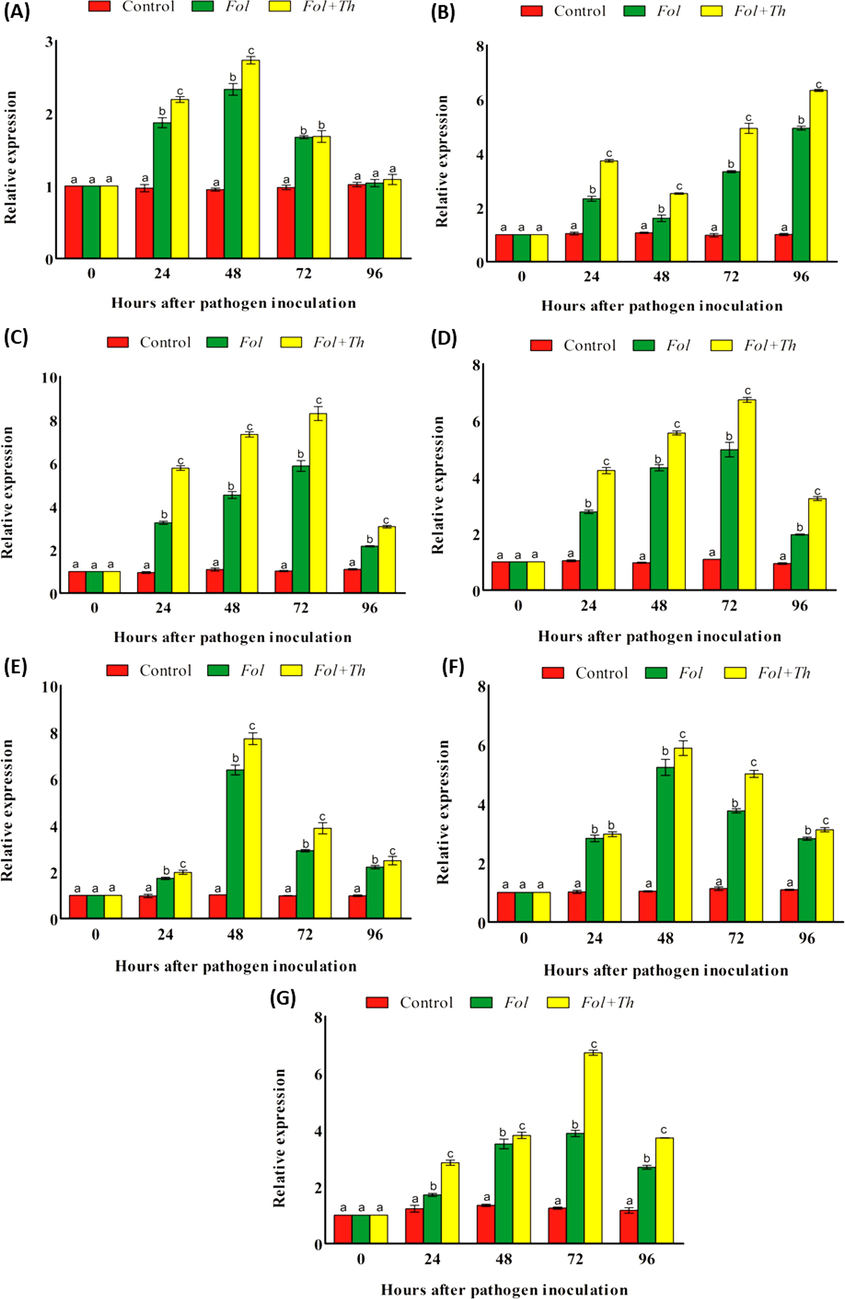
Relative expression of different antioxidant enzyme genes (A) SOD, (B) CAT, (C) PO, (D) PAL, (E) PPO, (F) PR-3 and (G) PR-2 in the leaves of tomato plants pre-treated with T. harzianum, after challenge inoculation with F. oxysporum f. sp. lycopersici (Fol).
3.7 Regulation of defense related genes
In control plants of 24, 48 and 72 h duration leaf samples the SOD gene expression reduces non-significantly compared to the 0 h leaf samples (Fig. 2A). Although, a non-significant rise compared to control noted in 96 h leaf sample. However, in Fol and Fol + Th treated plants the expression of SOD increases compared to the control plants. In 24 and 48 h, Fol and Fol + Th treated plants it was of 1.87, 2.33 and 2.19, 2.73-fold, respectively. Although, under similar treatment after 72 and 96 h, it was 1.67, 1.04 and 1.68, 1.09-fold, respectively (Fig. 2A).
Similar to enzyme activity catalase gene expression also recorded, in non-treated plants of 24, 48, 72 and 96 h duration a non-significant rise in catalase gene expression noted upto 48 h, then it decreases non-significantly upto 72 h, then again it increases under 96 h. Compared to control plants respective values of fold change compared to control plants in Fol treated plants was 2.33, 1.61, 3.33 and 4.94-fold, respectively under 24, 48, 72, and 96 h. Under similar condition in Fol + Th treated plants the values compared to control plants was 3.73, 2.52, 4.93 and 6.33-fold (Fig. 2B).
Similar to other enzyme gene expression the expression of peroxidase showed varied expression pattern, in control plants leaf samples of different hour sampling, it gets reduced in 24 h leaf sample while it increases significantly in 48 h leaf samples and non-significantly increases in 72 and 96 h leaf samples compared to control 0 h leaf samples. However, increased expression of same gene recorded when the leaf samples of 24, 48, 72 and 96 h is exposed to Fol and Fol + Th. In Fol treated 24, 48, 72 and 96 h leaf samples the fold change compared to 0 h leaf samples was 3.25, 4.53, 5.87 and 2.17-fold, under similar condition Fol + Th treated plant leaf sample’s gene expression fold change was 5.77, 7.33, 8.29 and 3.07-fold, respectively (Fig. 2C).
Gene expression of PAL was not uniform, in control plants compared to control in 24 h and 72 h leaves its expression increases, while it decreases in the leaf samples of 48 h and 96 h. Although in Fol and Fol + Th treated, all leaf samples it increases compared to control. It was maximum 4.97 fold compared to control in Fol treated plants after 72 h, while it was minimum 1.97-fold compared to control after 96 h under similar treatment. Similarly in Fol + Th treated plants, it was maximum 6.73-fold after 72 h and minimum 3.24-fold compared to control after 96 h (Fig. 2D). PPO gene expression was non-significantly decreased in 24, 72 and 96 h leaf samples under control condition, while it get increased non-significantly in 48 h leaf samples under control condition. However, it increases in all leaf samples compared to control plants when treated with Fol and Fol + Th. In Fol treated plants, it was maximum in 48 h leaf samples while minimum in 24 h leaf samples, however it was still higher compared to control plant. Similarly in Fol + Th treated plants, it was maximum in 48 h leaf sample and minimum in 24 h leaf sample (Fig. 2E).
Expression level of chitinase (PR-3) genes were recorded under control condition. A very slight changes noted even in case of 24, 48, 72 and 96 h of leaf samples, initially it increases upto 72 h, then decreases in 96 h leaf samples. In case of Fol and Fol + Th treatment, a significant increase noted, however, it rises sharply upto 48 h in both case and the fold change values compare to control was 2.83, 5.23 and 2.97 and 5.88-fold, respectively for Fol treatment and Fol + Th treatment. In case of Fol treatment after 72 h, a slight reduction noted, however, it was high reduction after in 96 h exposed plants. While in Fol + Th treated plants, a sharp reduction observed after 72 and 96 h of Fol + Th exposure (Fig. 2F). β-1,3 Glucanase (PR-2) gene expression under control, Fol and Fol + Th, treatment increases, however, in control plants it increases upto 48 h of compared to plants, then decreases after 72 and 96 h, however, it was still higher compared to the control plants of 0 h duration. Under Fol and Fol + Th treatment, this gene expression compared to 0 h exposed plants under 24, 48 and 72 h of Fol and Fol + Th exposure, although after 96 h of both exposure, it decreases (Fig. 2G).
4 Discussion
Microbial biopriming speaks to a versatile methodology to improve the defense system of the plants that outcomes in enhanced resistance/stress resilience, or potentially an increased defense mechanisms against the future biotic and abiotic stress (Zehra et al., 2017a). In this study, priming of tomato plants with T. harzianum BHU LMMT (Th) was tested to enhance the defense response of the plants against the Fol wilt disease.The percentage of disease incidence were found reduced by the Th priming. This study was also supported by the findings of Saravanakumar et al. (2017) where T. harzianum strain induce systemic resistance (ISR) in maize plants against Fusarium graminearum, and therefore, could be considered as potential BCA.
It has been reported that transcriptomic studies done on ISR mediated by Trichoderma-root colonization which results upregulation of PR proteins encoding genes (Siddaiah et al., 2017). The transcriptomic studies also stated that some vital defense enzymes encoded by upregulated vital genes and involved in plant defense against phytopathogens (Siddaiah et al., 2017; Zehra et al., 2021). However, the molecular mechanism behind the SA mediated resistance induced by Trichoderma spp. is very complex and involve many signaling components, proteins, transcription factors and is regulated by phytohormonal crosstalk (Fig. 3). Pathogen effectors or microbe associated molecular patterns are molecular signatures employed by pathogens that are identified by the host via. R gene products and specific pathogen recognition receptors. Pathogen-triggered immunity (PTI) is formed in response to MAMPs, while effector-triggered immunity (ETI) is generated in response to effectors. The activation of the plethora of signal transduction pathways mediated by phytohormones such as salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) is the result of this recognition. The detection of pathogenic elicitors or components is accompanied by a shift in membrane potential, which leads to the activation of the NADPH complex, which is implicated in the generation of ROS molecules, and thus the activation of the ROS scavenging mechanism. Ca+2 signaling may be activated as a result of the shift in ionic flux.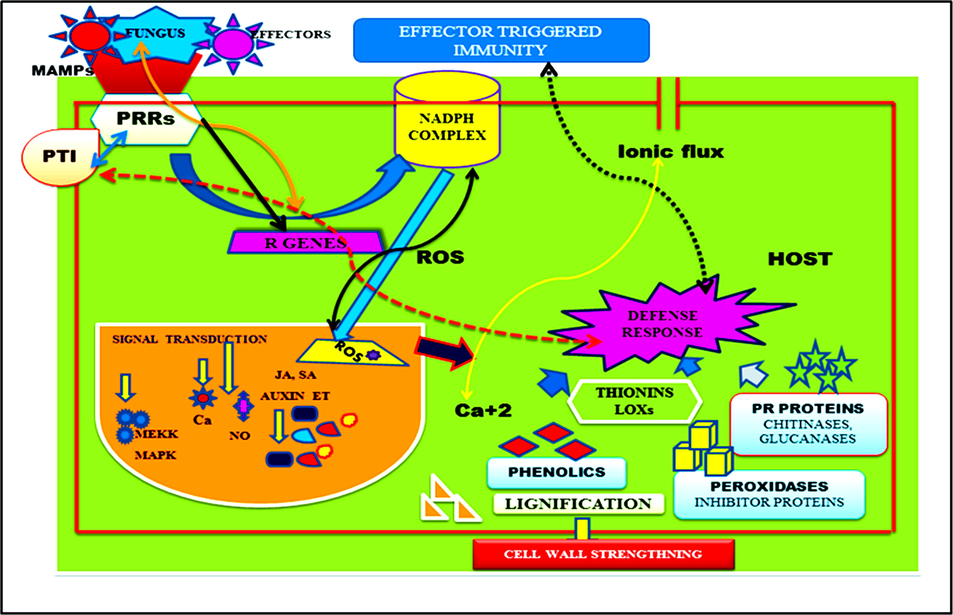
The recognition of a pathogen and the production of a host defence response. Pathogen effectors or microbe associated molecular patterns (MAMPs) are molecular signatures employed by pathogens that are identified by the host via. R gene products and specific pathogen recognition receptors (PRRs). Pathogen-triggered immunity (PTI) is formed in response to MAMPs, while effector-triggered immunity (ETI) is generated in response to effectors. The activation of the plethora of signal transduction pathways mediated by phytohormones such as SA, JA, and ET is the result of this recognition. The detection of pathogenic elicitors or components is accompanied by a shift in membrane potential, which leads to the activation of the NADPH complex, which is implicated in the generation of ROS molecules, and thus the activation of the ROS scavenging mechanism. Ca+2 signalling may be activated as a result of the shift in ionic flux.
In our results, we found increased expression of PR-genes (chitinases and β-1,3-glucanases) in tomato plants primed with Th and challenged with Fol. Furthermore, we reported that Fol + Th treated tissues were found to have higher expression of antioxidants as well as PAL and PPO enzymes. PAL an inducible enzyme plays a crucial role in plant defense against various abiotic and biotic stresses (MacDonald and D'Cunha, 2007). PPO catalyzes the oxidation of phenols to highly reactive quinone compounds that are toxic to pathogens, metabolism of toxic oxygen and ROS molecules and regulates the phenylpropanoid pathway (Huang et al., 2018). Similarly, Singh et al. (2016) reported that seed biopriming with T. asperellum BHU8 resulted into increase in PAL level by 57.19%, whereas, PPO activity by 21.75% and PO activity by 105.1% compared to uninoculated control. Our result was also consistent with the observations and results reported for T. hamatum UoM13 induced resistance against downy mildew pathogen where PAL expression was 3.65-folds higher in T. hamatum UoM 13 treated seedlings compared to the uninoculated control seedlings (Siddaiah et al., 2017). Moreover, in our results, the activities of defense-related enzymes like SOD, CAT, PO in Fol + Th treated plants were significantly higher compared to Fol challenged plants as well as uninoculated control. The increase in activitiy of the antioxidative defense enzymes in Th + Fol treated leaf tissues could be associated with enhanced expression of SOD as well as CAT genes or increased transcript abundance to scavenge the ROS molecules. SOD activity might be attributed to nullify the effects of toxic oxygen free radicals and their immediate conversion into H2O2. In contrast, increased CAT activity under stressed environment could be correlated with rapid breakdown, and therefore, preventing the level of accumulated H2O2. Increased SOD activity in plants primed with Th could be correlated with much more active ROS system and increased antioxidative defense mechanism against the Fol infection in tomato.
5 Conclusion
The present study showed that T. harzianum BHU LMMT induced systemic resistance in tomato against the Fusarium wilt pathogen and unraveled the critical role defense enzymes in disease resistance. Trichoderma root colonization stimulated the defense machinery of the plant for rapid establishment of SAR following the Fol challenged condition. The increased expression of plant defense-related enzymes and proteins demonstrated the successful establishment and manifestation of SAR mediated plant defense against the Fol pathogen. This clearly represent that microbial biopriming with T. harzianum could be useful against vascular wilt pathogen.
Acknowledgments
The authors would extend their appreciation to the Researchers Supporting Project number (RSP-2021/306), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Oxidative damage, antioxidant mechanism and gene expression in tomato responding to salinity stress under in vitro conditions and application of iron and zinc oxide nanoparticles on callus induction and plant regeneration. BMC Plant Biol.. 2021;21(1):597.
- [CrossRef] [Google Scholar]
- Dissection of Trichoderma longibrachiatum-induced defense in onion (Allium cepa L.) against Fusarium oxysporum f. sp. cepa by target metabolite profiling. Plant Sci.. 2016;246:128-138.
- [CrossRef] [Google Scholar]
- Trichoderma: Beneficial role in sustainable agriculture by plant disease management. In: Egamberdieva D., Ahmad P., eds. Plant Microbiome: Stress Response. Microorganisms for Sustainability. Vol Volume 5. Singapore: Springer Nature; 2018. p. :105-126.
- [CrossRef] [Google Scholar]
- Polyphenolic extract of InsP 5-ptase expressing tomato plants reduce the proliferation of MCF-7 breast cancer cells. PLoS ONE. 2017;12(4):e0175778.
- [Google Scholar]
- Expression of pathogenesis-related proteins in transplastomic tobacco plants confers resistance to filamentous pathogens under field trials. Sci. Rep.. 2019;9:2791.
- [CrossRef] [Google Scholar]
- Genome-wide identification and evaluation of reference genes for quantitative RT-PCR analysis during tomato fruit development. Front. Plant Sci.. 2017;8
- [CrossRef] [Google Scholar]
- A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull.. 1987;19:11-15.
- [Google Scholar]
- Properties of phenylalanine ammonia-lyase from marrow cotyledons. Plant Sci.. 2002;162(1):1-7.
- [CrossRef] [Google Scholar]
- Superoxide dismutases. In: Meister A., ed. Advances in Enzymology and Related Areas of Molecular Biology. Vol Vol. 41. John Wiley Sons Inc; 1974. p. :35-97.
- [CrossRef] [Google Scholar]
- Selected isolates of Trichoderma gamsii induce different pathways of systemic resistance in maize upon Fusarium verticillioides challenge. Microbiol Res.. 2020;233:126406
- [CrossRef] [Google Scholar]
- New spectrophotometric assay for polyphenol oxidase activity. Anal. Biochem.. 1993;215(1):59-65.
- [CrossRef] [Google Scholar]
- Constitutive activation of NF-κB causes resistance to apoptosis in human cutaneous T cell lymphoma HuT-78 cells: autocrine role of tumor necrosis factor and reactive oxygen intermediates. J. Biol. Chem.. 1998;273(22):14008-140014.
- [CrossRef] [Google Scholar]
- Association of enhanced peroxidase activity with induced systemic resistance of cucumber to Colletotrichum lagenarium. Physiol. Plant Pathol.. 1982;20(1):73-76.
- [CrossRef] [Google Scholar]
- The ascorbate peroxidase APX1 is a direct target of a zinc finger transcription factor ZFP36 and a late embryogenesis abundant protein OsLEA5 interacts with ZFP36 to co-regulate OsAPX1 in seed germination in rice. Biochem. Biophys. Res. Commun.. 2018;495:339-345.
- [CrossRef] [Google Scholar]
- Host–pathogen interaction in Fusarium oxysporum infections: where do we stand? Mol. Plant Microbe Interact.. 2018;31(9):889-898.
- [CrossRef] [Google Scholar]
- Enhancement of the expression of defense genes in tomato against Ralstonia solanacearum by N-octanoyl-L-homoserine lactone. Afr. J. Microbiol. Res.. 2017;11(5):194-203.
- [CrossRef] [Google Scholar]
- A modern view of phenylalanine ammonia lyase. Biochem. Cell Biol.. 2007;85(3):273-282.
- [CrossRef] [Google Scholar]
- Superoxide dismutase enhances tolerance of freezing stress in transgenic alfalfa (Medicago sativa L.) Plant Physiol.. 1993;103(4):1155-1163.
- [CrossRef] [Google Scholar]
- Comparative evaluation of biochemical changes in tomato (Lycopersicon esculentum Mill.) infected by Alternaria alternata and its toxic metabolites (TeA, AOH, and AME) Front. Plant Sci.. 2016;7:1408.
- [CrossRef] [Google Scholar]
- Antagonistic assessment of Trichoderma spp. by producing volatile and non-volatile compounds against different fungal pathogens. Arch. Phytopathol. Plant. Protect.. 2017;50(13–14):629-648.
- [CrossRef] [Google Scholar]
- Isolation, characterization and toxicological potential of tenuazonic acid, alternariol and alternariol monomethyl ether produced by Alternaria species phytopathogenic on plants. Sci. Rep.. 2017;7:8777.
- [CrossRef] [Google Scholar]
- Monographic contribution on Trichoderma. Associated Publishing Company; 2002.
- Association of β-1,3-glucanase activity and isoform pattern with systemic resistance to blue mould in tobacco induced by stem injection with Peronospora tabacina or leaf inoculation with tobacco mosaic virus. Physiol. Mol. Plant Pathol.. 1991;39:25-39.
- [CrossRef] [Google Scholar]
- Trichoderma isolates inhibit Fusarium virguliforme growth, reduce root rot, and induce defense-related genes on soybean seedlings. Plant Dis.. 2020;104(7):1949-1959.
- [CrossRef] [Google Scholar]
- Effect of Trichoderma harzianum on maize rhizosphere microbiome and biocontrol of Fusarium stalk rot. Sci. Rep.. 2017;7:1771.
- [CrossRef] [Google Scholar]
- Elicitation of resistance and associated defense responses in Trichoderma hamatum induced protection against pearl millet downy mildew pathogen. Sci. Rep.. 2017;7:43991.
- [CrossRef] [Google Scholar]
- Trichoderma asperellum spore dose depended modulation of plant growth in vegetable crops. Microbiol. Res.. 2016;193:74-86.
- [CrossRef] [Google Scholar]
- Biological control agents: diversity, ecological significances, and biotechnological applications. In: Singh J., Yadav A., eds. Natural Bioactive Products in Sustainable Agriculture. Singapore: Springer; 2020. p. :31-44.
- [CrossRef] [Google Scholar]
- Enhanced tomato disease resistance primed by arbuscular mycorrhizal fungus. Front. Plant Sci.. 2015;6:786.
- [CrossRef] [Google Scholar]
- Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot.. 2013;64(5):1263-1280.
- [CrossRef] [Google Scholar]
- Activation of defense response in tomato against Fusarium wilt disease triggered by Trichoderma harzianum supplemented with exogenous chemical inducers (SA and MeJA) Braz. J. Bot.. 2017;21:1-14.
- [CrossRef] [Google Scholar]
- Synergistic effects of plant defense elicitors and Trichoderma harzianum on enhanced induction of antioxidant defense system in tomato against Fusarium wilt disease. Bot. Stud.. 2017;58:44.
- [CrossRef] [Google Scholar]
- Efficiency of microbial bio-agents as elicitors in plant defense mechanism under biotic stress: a review. Curr. Res. Microb. Sci.. 2021;2:100054
- [CrossRef] [Google Scholar]
- Breeding wheat for resistance to Fusarium head blight in the global North: China, USA, and Canada. Crop J.. 2019;7(6):730-738.
- [CrossRef] [Google Scholar]







