Translate this page into:
Effect of insilico predicted and designed potential siRNAs on inhibition of SARS-CoV-2 in HEK-293 cells
⁎Corresponding author at: King Fahd Medical Research Center, King Abdulaziz University, Jeddah 21589, Saudi Arabia. ssohrab@kau.edu.sa (Sayed Sartaj Sohrab)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objectives
The COVID-19 was identified for the first time from the sea food market, Wuhan city, China in 2019 and the pathogenic organism was identified as SARS-CoV-2. Currently, this virus has spread to 223 countries and territories and known as a serious issue for the global human community. Many vaccines have been developed and used for immunization.
Methods
We have reported the insilico prediction, designing, secondary structure prediction, molecular docking analysis, and in vitro assessment of siRNAs against SARS-CoV-2. The online bioinformatic approach was used for siRNAs selection and designing. The selected siRNAs were evaluated for antiviral efficacy by using Lipofectamine 2000 as delivery agent to HEK-293 cells. The MTT assay was used for cytotoxicity determination. The antiviral efficacy of potential siRNAs was determined based on the Ct value of q-RT-PCR and the data analysis was done by Prism-GraphPad software.
Results
The analyzed data resulted in the selection of only three siRNAs out of twenty-six siRNAs generated by online software. The secondary structure prediction and molecular docking analysis of siRNAs revealed the efficient binding to the target. There was no cellular toxicity observed in the HEK-293 cells at any tested concentrations of siRNAs. The purification of RNA was completed from inoculated cells and subjected to q-RT-PCR. The highest Ct value was observed in siRNA 3 than the others. The results offered valuable evidence and invigorated us to assess the potency of siRNAs by using alone or in combination in other human cells.
Conclusion
The data generated from this study indicates the significance of in silico prediction and narrow down the potential siRNA' against SARS-CoV-2, and molecular docking investigation offered the effective siRNAs binding with the target. Finally, it is concluded that the online bioinformatics approach provided the prediction and selection of siRNAs with better antiviral efficacy. The siRNA-3 was observed to be the best for reduction of viral RNA in cells.
Keywords
In silico prediction
Designing
siRNAs
SARS-CoV-2
HEK-293 cells
- COVID-19
-
The new Coronavirus Disease 2019
- MFE
-
Minimum free energy
- PCR
-
Polymerase Chain Reaction
- RNAi
-
RNA interference
- SARS-CoV-2
-
Severe Acute Respiratory Syndrome Coranavirus-2
- siRNA
-
short interfering RNA
- WHO
-
World Health Organization
Abbreviations
1 Introduction
A new case of unusual pneumonia was observed in infected patients in Wuhan, China in late 2019. The infected patients developed the most common symptoms like, breathing problems, fever, and failure of the multi organ in severe cases. The causative agent was identified and sequenced and, based on the high sequence similarity and phylogenetic relationships with bat coronavirus (SARS-bat virus), it was finally designated as Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), and this disease was designated as COVID-19 in February 2020 and pandemic was declared on March 11, 2020. Currently, the SARS-CoV-2 has been reported from 223 countries with 379,303,864 cases, 5,693,527 deaths, and 299,352,192 recovery (last accessed on 1.2.2022–10.44 GMT-WHO 2022) and represented a serious threat to human health globally with approximately 2–5% mortality rate of infected individuals (Azoulay et al., 2020; Zhou et al., 2020). The virus is attached to the host cell by Spike (S) protein gene and enters in cells by angiotensin–converting enzyme 2 (ACE2) and affects the critical organs resulting in malfunction of airways, lungs, kidneys, gastrointestinal tract as well as the associated cardiovascular system which finally leads to loss of life. This virus falls under Coronaviridae family, with +ve sense single-stranded RNA with an approximately 26–32 kb genome. Currently, many genetic variants have emerged and spread from circulating virions which are found to be more deleterious in fast spread among human population. The first mutation of SARS-CoV-2 was detected in the S protein gene within, a few months of the evolution of the virus. The small mutations tend to impact the virus phenotypes, biology, pathogenicity, more virulence, infectivity, host adaptability, as well as transmissibility and/or antigenicity. Due to the dynamic nature of SARS-CoV-2, frequent mutations, and new variants, it is difficult to predict and develop effective antiviral therapeutics in near future. An extreme measure to be taken to minimize the patient mortality and loss of life.
Recently, a new variant known as Omicron has been identified from South Africa, but many cases have been reported globally. Currently, five variants of concern have been reported from different locations and they are designated as Alpha (B.1.1.7), Beta (B.1.351), Delta (B.1.617.2), Gamma (P.1) and Omicron (B.1.1.529) (WHO, 2021; Khan et al., 2022; Aleem et al., 2022). Currently, many vaccines have been developed and approved by WHO by using different platforms against multiple targets of SARS-CoV-2 and being used for vaccination of the human population globally, but still an effective broad spectrum antiviral therapeutics are urgently needed to protect the global human population. Many antiviral compounds and therapeutic molecules have been identified, designed, developed, and are being investigated at various stages. Many compounds have reached an advanced stage of investigation. In continuation of the antiviral therapy development, a new and advanced technology known as RNA interference (RNAi) can also be applied. This technology has been adopted for developing antiviral therapy against many viral and deadly diseases by regulating and silencing the expression of desired genes using siRNAs/shRNAs and degradation of corresponding mRNAs (Hasan et al., 2014; Sohrab et al., 2020, Sohrab et al., 2021a; Sohrab et al., 2021b; Sohrab et al., 2021c; El-Kafrawy et al., 2021). The basic criteria for computational prediction, filtration and selection of siRNAs have been discussed in many reports (Naito et al., 2009; Naito and Ui-Tei, 2012; Sohrab et al., 2018, Uludağ et al., 2020; Donia and Bokhari 2021). The efficacy of miRNA and siRNA are under the stage of investigation in more than 20 clinical trials. Some of them are found to be the most promising and can be considered against respiratory viruses (Chakraborty et al., 2017; Alyan Pharmaceuticals, 2020).
For SARS-CoV-2, the computational identification of siRNAs, aptamers/siRNAs chimera, siRNA-Nanoparticle, siRNA-peptide dendrimer formulation utilizing multiple targets have been reported in many published papers (Panda et al., 2021; Chen, et al., 2020; Bappy et al., 2021; Khanali et al., 2020; Shawan et al., 2021; Chowdhury et al., 2021; Idris et al., 2021; Pandey and Verma, 2021; Wu and Luo, 2021; Rohani et al., 2021; Khaitov et al., 2021; Sohrab et al., 2021a). But none of them were experimentally evaluated except one tested in Vero cells and found some promising results (Niktab et al., 2021). Recently, we also have in silico predicted, designed, and experimentally evaluated the siRNAs against MERS-CoV, and promising results were observed to inhibit the MERS-CoV proliferation and replication in different cell lines (Sohrab et al., 2020, Sohrab et al., 2021b, Sohrab et al., 2021c; El-Kafrawy et al., 2021). Recently, for SARS-CoV-2, many papers have reported about the designing and selection of siRNAs using many target genes. In another study, the leader protein sequences were used and total of four siRNAs were finally designed, but none of them were experimentally validated (Pandey and Verma 2021; Tolksdorf et al., 2021). So, by using the similar leader protein sequences of SARS-CoV-2/human/SAU/85791C/2020, we tried to predict, design, and evaluate the potential siRNAs in HEK-293 cells. We have used different concentrations of siRNAs and delivered them to grown HEK-293 cells using Lipofectamine 2000 followed by virus inoculation. The viral RNA was purified and used for q-RT-PCR to determine the reduction of viral RNA based on the Ct value in the HEK-293 cells. The results obtained from this work concluded that only one siRNA-3 has the highest antiviral potency to inhibit and reduce the viral RNA in HEK-293 cells.
2 Materials and methods
2.1 In silico prediction and selection of potential siRNAs
The leader protein gene sequences were selected from SIAU isolate (GenBank:MT630432) for target-specific prediction, design, and filtration of siRNAs. This tool applies combined utilization Ui-Tei, Amarzguioui, and Reynolds rules. Generally, the strict criteria should be followed for siRNAs designing and filtration as described in the published paper (Naito et al., 2009; Naito and Ui-Tei, 2012 Sohrab et al., 2018). The predicted siRNAs were further screened by using NCBI Human genomic and transcript database to avoid any similarities with human genome sequences as well as to filter any off-target sequences.
2.2 Molecular docking and secondary structure prediction of siRNAs
For the analysis of interaction patterns, the computer-assisted molecular docking and molecular dynamics simulation was performed using a guide strand of predicted siRNAs with leader protein sequences of SARS-CoV-2, to determine the effective binding to the target, using online software HNADOCK (He et al., 2019). The 3D model structure was generated for guide strand and leader protein sequences and visualized in PyMOL 2.5.2. Based on the lowest Tm value for guide strand and the highest binding score of leader protein sequences as target were considered for best docking analysis. We have selected the best siRNAs model from the top ten docked models generated and used for further study.
To analyze the interaction between predicted siRNA and target leader protein gene sequences based on the thermodynamics as well as the secondary structure of siRNA, the online software like RNAfold 2.4.18 (https://rna.tbi.univie.ac.at) and Mfold server (https://www.mfold.-rna.albany.edu/) was used. The guide and passenger strand sequences were exported to the given locations of program and secondary structure was predicted and generated. The predicted secondary structure was visualized and downloaded in Forna format (Kerpedjiev et al., 2015). The secondary structure of siRNAs provides vital information about the effective binding to the target as well as the possibility of any undesired formation of hairpin structure that affect the binding of siRNAs to the selected target.
2.3 Filteration and synthesis of siRNAs
The predcited siRNAs were filtered for chemical synthesis as per the results generated by online software; siRNA Direct 2.0, HNADOCK, and RNAfold 2.4.18. The synthesis of potential siRNAs was done by Iintegrated DNA Technologies and used for further analysis for antiviral potency against SARS-CoV-2 in HEK-293 cells.
2.4 Transfection of siRNAs and cytotoxicity assay
Before testing the effect of siRNAs on the reduction of virus multiplication and proliferation, we tested them on normal and non-infected HEK-293 cells and the cell viability was compared with normal cells. To perform the siRNAs transfection, cells were initially grown in 96 well plates using standard DMEM media at 37 °C with 5% CO2. The transfection was made in 60–80% confluent cells by using the reverse transfection method with Lipofectamine 2000 (Invitrogen, USA) as per manufactures instructions. Each experiment was performed in triplicates adding the negative and positive controls. The siRNAs were used at different concentrations (0.1–50 nM) made from original stock and gently mixed with Lipofectamine 2000 and 100 μl Opti-MEM medium. The siRNA-lipid complex mixture was further incubated at room temperatures for 30 min and then the mixture was gently delivered to grown cells and incubated finally for 72 h. The cytotoxic effect of each siRNAs was evaluated after 72 h post-transfection.
The cytotoxicity of each siRNAs was determined by using MTT Assay Kit (ThermoFisher Scientific-USA) as per the manufacture’s instruction. In brief, the transfected cells were replenished with freshly prepared DMEM media (100 μl/well) as well as 10 μl MTT (12 mM) and further incubated for 4 h at 37 °C. The absorbance was measured at 570 nm by using SpectraMax i3x imaging cytometer and cytotoxicity was calculated based on the mean OD value recorded from SpectraMax i3x using the standard formula for each siRNAs separately. We used Lipofectamine 2000 alone Lipofectamine and Opti-MEM, only Opti-MEM, and only cells as negative control in this study to evaluate their effect on grown cells.
3 Evaluation of the antiviral potency of siRNAs against SARS-CoV-2 in HEK-293 cells
3.1 siRNAs transfection and virus inoculation
The antiviral activity of predicted, designed, and selected siRNAs were evaluated in grown HEK-293 cells at different concentrations (0.1–50 nM). The 60–80% confluent (1 × 10−4) HEK-293 cells were transfected with siRNAs and virus inoculation was performed. The cells were further incubated and observed till full CPE in positive control as described (Sohrab et al., 2020). The cells were harvested after full CPE and the commercial QIAamp Viral RNA Mini Kit was used to purify the viral RNA from lysed cells.
3.2 Purification of viral RNA and quantitative-real-time PCR
The QIAamp Viral RNA Mini Kit (Qiagen) was used to extract and purify the RNA from harvested and lysed cells. The q-RT-PCR was used to determine the relative expression of the viral genes in HEK-293 cells. The purified RNA was for q-RT-PCR and the level of viral RNA was determined by using PowerCheck SARS-CoV-2 real-time-PCR kit (Kogenebiotech-Korea) following the manufacturer instructions. The data were statistically analyzed, and figures and graphs were generated by using GraphPad Prism-9.3.0.
4 Results
4.1 In silico designing, and filteration of siRNAs
The online software siDirect 2.0 generated twenty-six siRNAs from leader protein sequence of SARS-CoV-2/human/SAU/85791C/2020 but, we selected only three siRNAs as per the strict selection criteria followed with different scoring tools, functional, Tm, off-targets reduced threshold, and algorithm scores (Naito et al., 2009; Naito and Ui-Tei, 2012, Sohrab et al., 2018). The sequences of siRNAs (guide/passenger) and their target location in the viral genome with seed-duplex stability (Tm/°C) have been presented in Table 1.
S.No
Location of siRNAs in the viral genome (Start-End)
Target sequence
Predicted RNA oligo sequences (5′→3′) Guide/Passenger strand
Seed-duplex stability (Tm/°C) Guide/Passenger strand
Minimum Free Energy (MFE) in Centroid Secondary Structure (kcal/mol)
Docking score/Ligand RMSD (Å)
1
18–40
TGGTTTCAACGAGAAAACACACG
UGUGUUUUCUCGUUGAAACCA
GUUUCAACGAGAAAACACACG
13.3/14.9
−25.05
−285.03/153.01
2
154–176
GGCTTAGTAGAAGTTGAAAAAGG
UUUUUCAACUUCUACUAAGCC
CUUAGUAGAAGUUGAAAAAGG7.7/11.3
–22.11
−261.10/350.30
3
468–483
ATGAAGATTTTCAAGAAAACTGG
AGUUUUCUUGAAAAUCUUCAU
GAAGAUUUUCAAGAAAACUGG13.3/5.3
−21.17
−328.83/360.59
4.2 Secondary structure prediction of siRNAs and leader protein
The secondary structure of each siRNAs and leader protein sequences was predicted by using online software RNAfold 2.4.18. The results of the secondary structure prediction have been computed and visualized in Forna format with variable MFE structure drawing encoding base-pair probabilities and presented in Figs. 1A and 1B. A variable range of MFE of each siRNAs was observed. The colouring of base pairs denotes the probability of their binding efficiency. There was no probability and structural complexity observed in any of the predicted siRNAs. The structure showed the improved binding efficacy to the target region which can regulate and silence the function of the desired gene.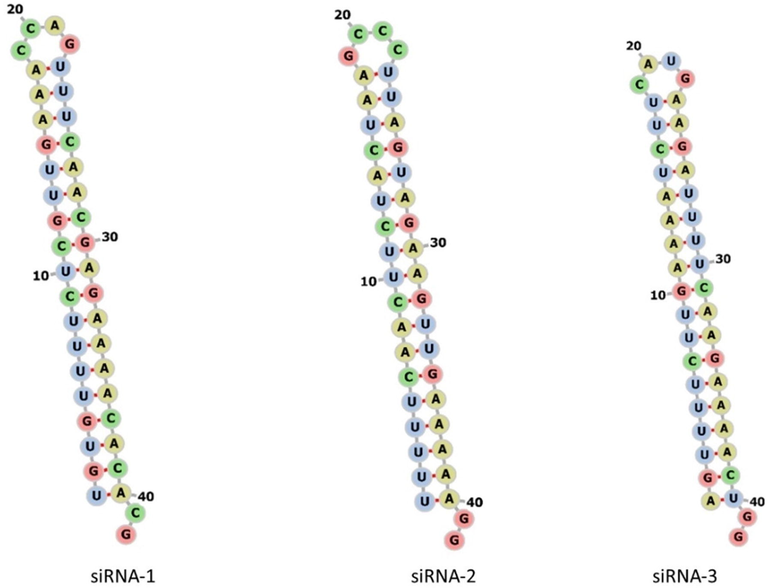
The possible folding and secondary structure prediction of in silico predicted siRNAs molecules computed using the online web server (siRNAs1-3).
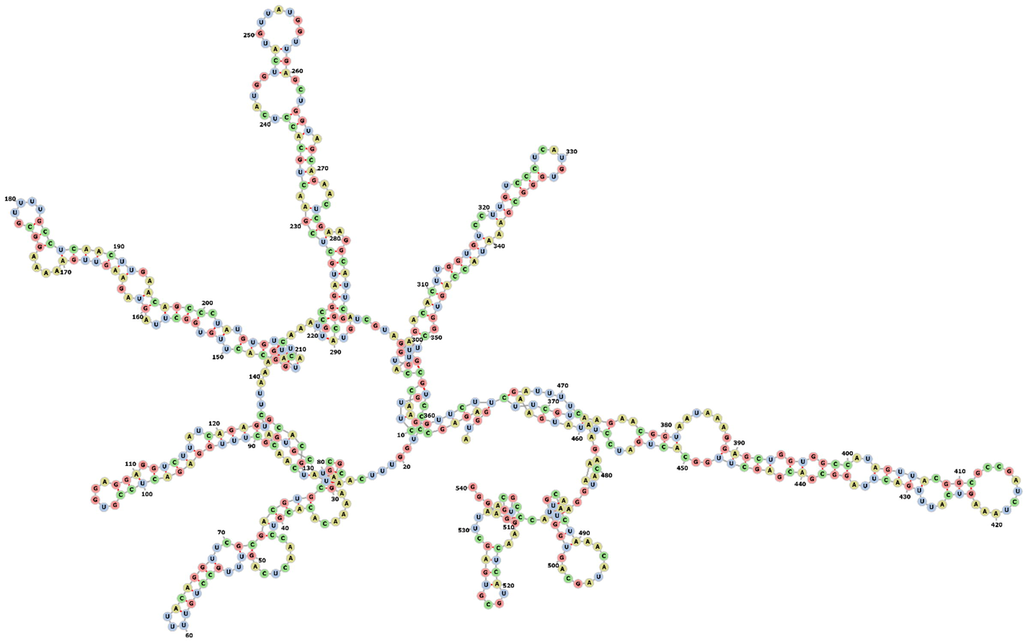
Secondary structure prediction of leader protein gene computed by using the online web server.
4.3 Molecular docking analysis
The molecular docking analysis was performed by HNADOCK online software to predict and reveal the efficient binding capacity to the target region. The software resulted in ten models and the best model was selected based on the docking score of siRNAs/target and visualized in PyMOL 2.5.2. software (Fig. 2). The docking root-mean-square deviation (RMSD) score ranged from −285.03/153.01 (Å)-siRNA-1, −261.10/350.30(Å)-siRNA-2, −328.83/360.59(Å)-siRNA-3 (Table 1). The root-mean-square deviation (RMSD) is used to define the average distance among the atoms. The highest binding score denotes the best binding efficiency with the target. The siRNA-3 showed the highest RMSD score −328.83/360.59(Å) with SARS-CoV-2-leader protein sequence, which confirms the best binding efficiency and is thereby expected to be the best siRNA for antiviral activity. The antiviral activity was confirmed by in vitro analysis in HEK-293 cells.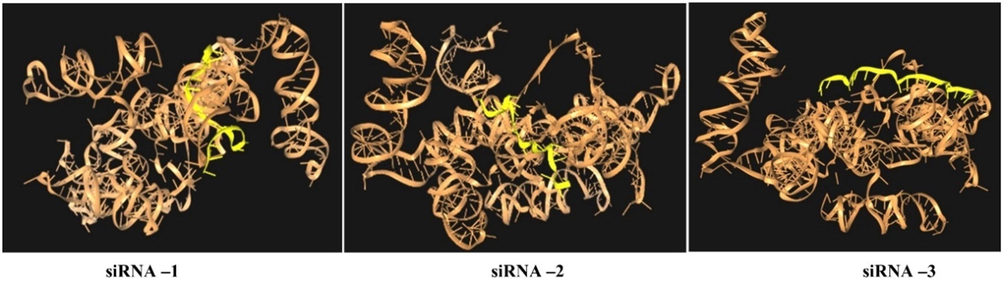
Molecular docking of predicted siRNAs (1–3) with SARS-CoV-2-leader protein gene and 3D interaction diagram of different docked complex with the target.
4.4 Transfection of siRNA and cytotoxicity assay
The selected siRNAs were used for evaluation of cytotoxicity in HEK-293 cells. The results showed the dose-dependent cytotoxicity of siRNAs in tested cells. The OD and CC50 value of each siRNA was variable at different concentrations of siRNAs (Table 2, Fig. 3). There was no significant cytotoxicity in HEK-293 cells observed in any siRNAs tested up to 50 nM concentration.
siRNAs Conc. (nM)
siRNA-1
siRNA-2
siRNA-3
50
1.51
1.50
1.58
25
1.40
1.43
1.41
10
1.21
1.27
1.25
5.0
1.22
1.24
1.21
1.0
1.33
1.31
1.36
0.5
1.24
1.23
1.25
0.25
1.31
1.30
1.35
0.1
1.27
1.25
1.24
CC 50
114.1
105.2
120.0
Lipofectamine 2000 in HEK-293 Cells
1.487
Lipofectamine + Opti-MEM in HEK-293 Cells
1.151
Only Opti-MEM in HEK-293 Cells
1.168
Only HEK-293 Cells (Negative control)
1.117
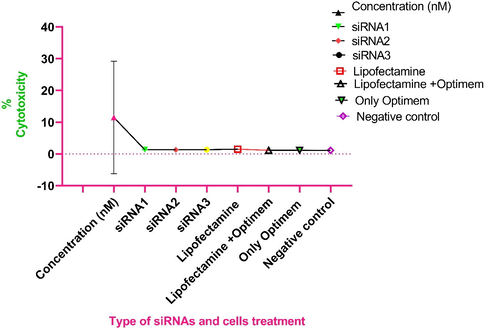
Cytotoxicity of different siRNAs-1–3 at various concentrations (0.1–50 nM) in HEK-293 cells. We used Lipofectamine alone, Lipofectamine and Opti-MEM, only Opti-MEM, and only cells as negative control in this study to evaluate their effect on grown cells.
4.5 Evaluation of viral load by quantitative RT-PCR
The determination of viral RNA level in HEK-293 cells was evaluated just after the transfection and virus inoculation. The inoculated cells were harvested, and viral RNA was purified after 72 h of post-inoculation with the full CPE observed in positive control. The Ct value of quantitative RT-PCR indicated the reduction of viral RNA at various concentrations of siRNAs (Table 3). The dose-dependent reduction of the level of viral RNA was observed in tested siRNAs. Interestingly, a significant reduction of viral RNA was reported in siRNA-3 than others at 5.0 nM and 10 nM respectively. The Ct value for each siRNAs have been shown as graphical representation in Fig. 4.
Conc. (nM)
siRNA-1
siRNA-2
siRNA-3
50
11.42
12.56
11.99
25
14.81
14.85
15.11
10
19.87
19.79
19.54
5.0
18.99
19.90
19.87
1.0
15.21
14.87
15.19
0.50
14.81
13.91
13.58
0.25
14.88
14.59
14.98
0.10
15.81
15.78
15.21
Negative control
44.20
45.69
48.36
Positive control
17.20
15.40
16.22
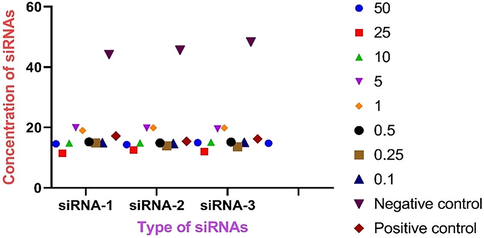
Graphical representation of Ct value of quantitative real-time PCR result of SARS-CoV-2 in HEK-293 cells. The siRNAs were delivered to the cells at various concentration such as 010, 0.25, 0.50, 1.0, 5.0, 10.0, 25 and 50 nM.
5 Discussion
A new disease known as COVID-19 was reported in December 2019 from China. The etiological agent was identified as SARS-CoV-2. As of 1.2.2022, this virus has spread to 223 countries and territories with 5,693,527 deaths. Currently, the pathogenic virus has been fully characterized and sequenced. A significant amount of information generated, and various vaccines have been developed by using different platforms and multiple targets including Spike (S) protein gene and being used for immunization. The S gene plays an important role in host cell attachment. Currently, many mutations have been identified especially in the S gene which favoured the introduction of many variants with different properties which attract the researchers to design and develop antivirals, therapeutic molecules, and targeted drugs against this virus for broad spectrum protection of the human population (Harvey et al., 2021; Greaneyet al., 2021; Khateeb et al., 2021; Starr et al., 2021).
The RNAi technology has been used against many viruses and significant results have been achieved. The nucleic acid-based treatments can be offered against SARS-CoV-2 as co-treatment due to the unique and high range of target sequence specificity and to achieve broad-spectrum resistance against multiple variants. This technology is being used to regulate by silencing of a particular gene/target using siRNA/shRNA molecules (Chakraborty et al., 2017). The pre-designed siRNAs against unique target sequences of the viral genome will regulate and terminate the virus replication and viral load in the infected cells. Currently, many siRNAs have been predicted, designed and their efficacy was evaluated against many viruses including MERS-CoV, SARS-CoV-1, SARS-CoV-2 and showed a significant inhibition of virus replication (Sohrab et al., 2018; Sohrab et al., 2020; Uludağ et al., 2020; Panda et al., 2021; Chen et al., 2020; Sohrab et al., 2021a; Sohrab et al., 2021b; El-Kafrawy et al., 2021; Khaitov et al., 2021; Idris et al., 2021; Pandey and Verma, 2021; Donia and Bokhari 2021; Bappy et al., 2021; Niktab et al., 2021; Shawan et al., 2021; Chowdhury et al., 2021; Wu and KQ, 2021; Rohani, et al., 2021; Khaitov et al., 2021). For designing of potential siRNAs molecule against a particular target gene, several challenges arise and need to be considered. These include siRNA delivery, stability, immunogenic response, off-target effect, possible cross-reactions with other sequences. To overcome these challenges, scientists have improved the model for selection and filtration of potential siRNAs in many published reports (Naito and Ui-Tei, 2012; Sohrab et al., 2018). Additionally, the secondary structure prediction as well as thermodynamics of RNA-RNA interaction of predicted siRNAs, play an important role in efficient binding to the target.
In this study, we have followed the strict criteria for prediction and filtration of siRNAs against the leader protein gene of SARS-CoV-2 by applying automated online software and evaluated in the HEK-293 cells. (Naito et al., 2009; Naito and Ui-Tei, 2012; Sohrab et al.,2018). The online software resulted in twenty-six siRNAs against the target, but only three siRNAs were selected for the evaluation of their efficacy in HEK-293 cells. The selected siRNAs were analyzed by secondary structure prediction and molecular docking simulation for their effective binding to the target. The secondary structure analysis results provide vital information about the probability efficient binding to the specific target. The secondary structure formation of a siRNA may inhibit the regulation and expression and cleavage of a particular target mediated by RNA-induced silencing complex (RSIC). Therefore, the secondary structure prediction and determination of minimum free energy are very critical for the efficacy of siRNAs. The most common algorithms were used by a web server for RNA secondary structure prediction by calculating the MFE (Bellaousov et al., 2013). The lower binding energy and MFE as well as with the free energy of the thermodynamic ensemble (TE) indicates the efficient binding of siRNAs, better potential of antiviral activity and significant inhibition of target gene. The minimum free energy of selected siRNAs ranged from −25.05-kcal/mol -siRNA-1, –22.11-kcal/mol-siRNA-2, −21.17-kcal/mol-siRNA-3. The docking RMSD score of each siRNAs (guide/passenger strand) ranged from −285.03/153.01 Å -siRNA-1, −261.10/350.30 Å- siRNA-2, −328.83/360.59 Å- siRNA-3. The higher docking score indicates the better binding to the target leader protein gene sequence of SARS-CoV-2. Based on the secondary structure and molecular docking analysis results, the siRNA-3 were expected to be the best as compared to other siRNAs.
The cytotoxicity and efficacy of siRNAs were evaluated in HEK-293 cells, and no significant cytotoxicity was observed in any tested siRNAs in HEK-293 cells at any concentration (0.1–50 nM). The reduction of viral RNA in treated cells was determined based on the Ct value of q-RT-PCR results. The high Ct value is correlated with the lower copy number of the target gene which further indicates the best reduction of viral RNA in the treated cells. The highest reduction of viral RNA was observed in siRNA3. Our evaluation results are supported by the findings of another research group (Carter et al., 2020). Recently, Chowdhury et al., (2021) designed eight siRNAs against NP genes and the S gene of SARS-CoV-2 by using an integrated bioinformatics approach while Chen et al., (2020) designed siRNAs against Orf1ab, Orf1b, S gene, Orf3a, M gene and N genes but the antiviral activity for designed siRNAs were not evaluated. Additionally, three different siRNAs were designed against E, M and N protein genes of SARS-CoV-1, and an 80% reduction of target gene expression was observed in treated cells (Shi et al., 2005). The results of our study should be evaluated further for the efficacy of selected siRNA in other cell lines either by using in combinations or alone as well as by using different delivery methods and transfection reagents soon study. Based on the overall results; it is obvious that the in silico designing, selection, filtration and experimental evaluation of selected siRNAs could be very useful for development of next-generation oligonucleotide-based therapeutics molecules against SARS-CoV-2 to protect the human population globally.
6 Conclusion
Based on the findings, it is concluded that the siRNAs could be considered as an alternative antiviral molecule against SARS-CoV-2. Additionally, more siRNAs can be designed and evaluated in multiple cell lines to get detail information against SARS-CoV.
Author contributions
SSS, SAE conceptualized, designed, and executed the experiments, SSS, SAE wrote the manuscript. EIA: Contributed to designing and execution of experiments and critically reviewed the manuscript. All authors provided critical feedback and analysis of manuscript. All authors reviewed the manuscript and approved.
Acknowledgements
The authors extend their appreciation to the Deputyship for Research & Innovation, Ministry of Education in Saudi Arabia for funding this research work through the project number IFPRC-204-141-2020 and King Abdulaziz University, DSR, Jeddah, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Alnylam Pharmaceuticals, 2020. Vir and Alnylam Expand Collaboration to Advance RNAi Therapeutics for the Treatment of Coronavirus Infection, Including Covid-19. Available online at: https://investors.alnylam.com/press-release?id= 24656.
- Emerging Variants of SARS-CoV-2 And Novel Therapeutics Against Coronavirus (COVID-19). In Stat Pearls: Stat Pearls Publishing; 2022.
- Increased mortality in patients with severe SARS-CoV-2 infection admitted within seven days of disease onset. Intensive Care Med.. 2020;46(9):1714-1722.
- [Google Scholar]
- Bappy et al., 2021. Designing potential siRNA molecule for the nucleocapsid(N) gene silencing of different SARS-CoV-2 strains of Bangladesh: Computational approach. Comput. Biol. Chem.. 92, 107486.
- Bellaousov et al., 2013. RNA structure: Web servers for RNA secondary structure prediction and analysis. Nucl. Acids Res., 41(Web Server issue), W471–W474. https://doi.org/10.1093/nar/gkt290.
- Therapeutic miRNA and siRNA: moving from bench to clinic as next generation meicine. Mol. Therapy. Nucleic Acids. 2017;8:132-143.
- [Google Scholar]
- Computational Identification of Small Interfering RNA Targets in SARS-CoV-2. Virol. Sinica.. 2020;35(3):359-361.
- [Google Scholar]
- A computational approach to design potential siRNA molecules as a prospective tool for silencing nucleocapsid phosphoprotein and surface glycoprotein gene of SARS-CoV-2. Genomics. 2021;113(1):331-343.
- [Google Scholar]
- RNA interference as a promising treatment against SARS-CoV-2. Int. Microbiol.. 2021;24(1):123-124.
- [Google Scholar]
- El-Kafray et al., 2021. In Vitro Inhibitory Analysis of Rationally Designed siRNAs against MERS-CoV Replication in Huh7 Cells. Molecules. (Basel, Switzerland), 26(9), 2610.
- Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun.. 2021;12(1)
- [Google Scholar]
- A Computational Approach for Predicting Role of Human MicroRNAs in MERS-CoV Genome. Advances in bioinformatics. 2014;967946
- [CrossRef] [Google Scholar]
- SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol.. 2021;19(7):409-424.
- [Google Scholar]
- HNADOCK: a nucleic acid docking server for modelling RNA/DNA-RNA/DNA 3D complex structures. Nucleic Acids Res.. 2019;47(W1):W35-W42.
- [CrossRef] [Google Scholar]
- A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Therapy. 2021;29(7):2219-2226.
- [Google Scholar]
- Khanali, J., Mohammadreza, A-K., Yasaman, A., Monire, J., Jafar, K., 2020. An aptamer/ siRNA Chimera against The SARS-CoV-2: A Dual Therapeutic Strategy for The Virus Neutralizing and RNA Interfering. Research Square. https://doi.org/10.21203/rs.3.rs-76450/v1.
- Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics (Oxford, England).. 2015;31(20):3377-3379.
- [Google Scholar]
- Silencing of SARS-CoV-2 with modified siRNA-peptide dendrimer formulation. Allergy. 2021;76(9):2840-2854.
- [Google Scholar]
- The Omicron (B.1.1.529) variant of SARS-CoV-2 binds to the hACE2 receptor more strongly and escapes the antibody response: insights from structural and simulation data. Int. J. Biol. Macromol.. 2022;200:438-448.
- [Google Scholar]
- Emerging SARS-CoV-2 variants of concern and potential intervention approaches. Critical Care (London, England).. 2021;25(1)
- [Google Scholar]
- siRNA design software for a target gene-specific RNA interference. Front. Genet.. 2012;3:102.
- [CrossRef] [Google Scholar]
- siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinf.. 2009;10(1)
- [CrossRef] [Google Scholar]
- Niktab et al., 2021. Design of advanced siRNA therapeutics for the treatment of COVID-19. Meta Gene. 29, 100910.
- Prediction of potential small interfering RNA molecules for silencing of the spike gene of SARS-CoV-2. Indian J. Med. Res.. 2021;153(1)
- [CrossRef] [Google Scholar]
- An in silico analysis of effective siRNAs against COVID‐19 by targeting the leader sequence of SARS‐CoV‐2. Adv. Cell Gene Ther.. 2021;4(2)
- [Google Scholar]
- Rohani et al., 2021. DisCoVering potential candidates of RNAi-based therapy for COVID-19 using computational methods. PeerJ. 9, e10505.
- Shawan et al., 2021. Designing an effective therapeutic siRNA to silence RdRp gene of SARS-CoV-2. Infect., Genetics Evol.: J. Mol. Epidemiol. Evolut. Genetics Inf. Dis., 93, 104951.
- Inhibition of genes expression of SARS coronavirus by synthetic small interfering RNAs. Cell Res.. 2005;15(3):193-200.
- [Google Scholar]
- Designing and evaluation of MERS-CoV siRNAs in HEK-293 Cell line. J. Infect. Public Health. 2020
- [CrossRef] [Google Scholar]
- In silico prediction and experimental validation of siRNAs targeting ORF1ab of MERS-CoV in Vero cell line. Saudi J. Biol. Sci.. 2021;28(2):1348-1355.
- [Google Scholar]
- in silico prediction and designing of potential siRNAs to be used as antivirals against SARS-CoV-2. Curr. Pharm. Des.. 2021;27(32):3490-3500.
- [Google Scholar]
- Designing and evaluation of MERS-CoV siRNAs in HEK-293 cell line. J. Infection Public Health. 2021;14(2):238-243.
- [Google Scholar]
- Design and Delivery of Therapeutic siRNAs: application to MERS-Coronavirus. Curr. Pharm. Des.. 2018;24(1):62-77.
- [Google Scholar]
- SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597(7874):97-102.
- [Google Scholar]
- Inhibition of SARS-CoV-2 replication by a small interfering RNA targeting the leader sequence. Viruses. 2021;13(10):2030.
- [Google Scholar]
- Prospects for RNAi therapy of COVID-19. Front. Bioeng. Biotechnol.. 2020;8
- [CrossRef] [Google Scholar]
- World Health Organization (2022): WHO, https://www.who.int/health-topics/coronavirus#tab=tab. Last accessed on 26.1.2022.
- World Health Organization (2021): https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern.
- Developing effective siRNAs to reduce the expression of key viral genes of COVID-19. International journal of biological sciences.. 2021;17(6):1521-1529.
- [Google Scholar]
- Coronavirus disease 2019 (COVID-19): a clinical update. Front. Med.. 2020;14(2):126-135.
- [Google Scholar]







