Translate this page into:
Effect of guggulsterone, a sterol identified in Commiphora gileadensis (Becham), on the dengue virus enzymes: Pharmacokinetics, molecular docking and molecular dynamics simulations studies
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objectives
Dengue infection is a serious public health problem in several regions of the world due to the lack of effective and appropriate therapy mainly for severe infection. Therefore, the development of agents with inhibitory properties targeting the dengue virus (DNV) replication is of utmost significance.
Methods
Guggulsterone, a sterol reported in a local medicinal tree, Commiphora gileadensis, was investigated in silico for its inhibitory potency of dengue virus. Interaction between this guggulsterone and NS5 RNA dependent RNA polymerase, dengue methyltransferase, NS3 protease-helicase, and dengue virus type 2 envelope glycoprotein were determined using molecular docking and molecular dynamics simulations study. Guggulsterone's ADME and toxicity were predicted in silico as well.
Results
Our data revealed that guggulsterone has the lowest docking energy (−5.5 kcal/mol) with dengue NS5 RNA-dependent RNA polymerase. While the interaction of guggulsterone with dengue virus type 2 envelope glycoprotein exhibited the highest docking energy (−3.4 kcal/mol), and was the most stable complex during molecular dynamic simulation. Guggulsterone was predicted to be a probable inhibitor of dengue virus type 2 envelope glycoprotein. ADME prediction showed no violation of Lipinski and Veber rules of guggulsterone. The tested compound may inhibit CYP2C19 and CYP2C9 and cannot inhibit CYP1A2, CYP2D6, and CYP3A4. Guggulsterone was shown to have no hepatotoxicity, cytotoxicity, or mutagenicity effect.
Conclusions
It can be concluded from this study that guggulsterone may be applied as a natural compound for the prevention or treatment of dengue infections. More in vitro and in vivo testing is needed to validate the effectiveness of this natural compound.
Keywords
Guggulsterone
Dengue virus
Molecular docking
Molecular dynamics
ADME and toxicity profile
1 Introduction
Dengue virus (DNV) is the primary cause of dengue fever, which affects both children and adults in tropical areas worldwide (Adawara et al., 2020). Dengue viruses are the causative agents of dengue fever (DF), dengue haemorrhagic fever (DHF), and dengue shock syndrome (DSS) respectively (Gubler 1998). DF is a tropical and subtropical disease that affects over a hundred countries. Female mosquitoes, Aedes albopictus or Aedes aegypti, spread the viruses to people after a blood meal (Fatima et al., 2011). Three to fourteen days after an infective mosquito bite, symptoms may arise. Hight fever, acute headache, extreme bone pain, muscle pain, and heavy rashes are some of the signs of dengue infection (Nouroz et al., 2021). Currently, available dengue vaccines are ineffective in protecting people of all ages and against all serotypes. Medicinal plants are reported to be a good alternative to antiviral drugs against dengue fever (Ansori et al., 2021; Saleh and Kamisah 2021). Molecular docking was applied to detect the interaction between natural compounds, NS3 and NS5 protease of the dengue virus (Suganya and Mahendran 2016). Recently, Adawara et al., (2020), using density functional theory (DFT) and molecular docking investigation, demonstrated the anti-dengue potential of three compounds from the leaves of Isatis tinctoria. According to Nag and Chowdhury (2020), binding free energy calculation and molecular dynamic simulation showed that an alkaloid identified in black pepper (piperine), inhibits the Methyl-transferase of dengue viruses. Similarly, Ul Qamar et al. (2014) tested more than 2000 flavonoids compounds from various medicinal plants and found that six flavonoids have a strong antiviral potential and blocked the Asn-130 glycosylation site of NS1 glycoprotein involved in RNA replication of dengue virus.
Commiphora gileadensis (L.), known as bechan, or balessan, is a tree growing in Saudi Arabia. The plant is used in traditional medicine to treat headaches, urinary retention, constipation, liver diseases, and inflammatory disorders (Al-Howiriny et al., 2004). In addition, C. gileadensis essential oil exhibited a significant activity toward drug-resistant Staphylococcus aureus (Orchard et al., 2017). Additionally, Bouslama and colleagues confirmed that guggulsterone, identified in C. gileadensis leaves, has virucidal properties against coxsackievirus, herpes simplex virus, respiratory syncytial virus, and adenovirus type 5 (Bouslama et al., 2019). More recently, Mishra et al., (2021) demonstrated in silico that guggulsterone could interact with SARS-CoV-2 main protease with lower binding energy (−9.67).
The present paper aimed to evaluate the potential antiviral activity of guggulsterone against dengue virus, targeting four proteins (NS5 RNA dependent RNA polymerase, dengue methyltransferase, NS3 protease-helicase, and dengue virus type 2 envelope glycoprotein), using molecular docking and molecular dynamics simulations analysis. In addition, the physicochemical, pharmacokinetic properties and toxicity of the tested compound were predicted using SwissADME and ProTox-II webservers.
2 Methods
2.1 Natural compound selection and preparation
The antiviral activity of guggulsterone has been documented against different viruses, including herpes simplex virus type 2 and SARS-CoV2 (Bouslama et al., 2019; Mishra et al., 2021). In this study, guggulsterone was selected for in silico evaluation of the antiviral activity on four dengue proteins, the structures of guggulsterone and a reference antiviral drug (ribavirin) were obtained from the PubChem database (Kim et al., 2016). The compounds (guggulsterone and ribavirin) were prepared for docking using LigPrep of Maestro (Schrodinger maestro 2020–3). Ligands energy was minimized with the OPLS3e force field, then protonation and tautomeric states were generated at pH 7 ± 2 using Epik (Greenwood et al., 2010).
2.2 Protein models preparation
Four proteins from the dengue virus were selected as a receptor for docking experiments, and their 3D structures were obtained from the RCSB PDB database (Berman et al., 2002). These proteins include: NS5 RNA dependent RNA polymerase domain (PDB id: 2J7W), dengue methyltransferase (PDB id: 1L9K), NS3 protease-helicase (PDB id: 2VBC) and dengue virus type 2 envelope glycoprotein (PDB id: 1OK8) (Nag and Chowdhury, 2020). Prior docking proteins structures were subjected to protonation step, removal of water atoms beyond 3 Å of heteroatoms, and energy minimization step achieved by OPLS3e at pH 7 ± 2. Maestro interface (Protein preparation, SiteMap and Receptor Grid generation) were utilised for active site prediction and protein preparation.
2.3 Molecular docking
Prior docking procedure the docking protocol was validated by redocking the co-crystalized ligand of the dengue methyl transferase (1L9K), then RMSD was calculated for superposition of ligands using Maestro. After preparing the selected proteins and ligands, molecular docking was used to study the interaction of guggulsterone and dengue target proteins using Precision (SP) docking of Maestro interface, docking score, the energy of van der Waals (evdW), and glide hydrogen bond was calculated.
The protein was set as rigid while ligand was set as flexible, the van der Waals radii scaling factor was set at 0.8 and partial charge cutoff at 0.15. Post docking energy minimization step was done, and only 55 poses per ligand were included. After docking, the PLIP webserver and Maestro were used to examine and visualize residues of the protein associated with ligand interaction from the docking process.
2.4 Molecular Dynamics simulation
Molecular Dynamics (MD) simulation was proven to be a valuable method for analysing the physical foundations of biomolecule structure and functional properties (Khan et al., 2022). In this study, the stability of the complex of guggulsterone and the four selected proteins have been studied by MD simulation for 50 ns. The simulation process was achieved using the Desmond package in the Schrödinger Maestro interface (Release, 2020). The system was prepared by using TIP4P water model, and orthorhombic box shape was assigned with distances 10 Å and force filed OPLS4 (Lu et al., 2021). Then ions were neutralized by additions of required charges. During MD simulation at each 100 ps interval, snapshots were recorded, the nose-hover thermostat method was specified with a relaxation time of 1 ps and 2 fs time step. The system was minimized at 2000 iterations, NPT temperature was at 300 °K, then Maestro was used for visualization of trajectories. Then Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF) were generated.
2.5 Calculation of binding free energy
The relative binding free energy (ΔG_bind) was calculated for the complex of protein and ligands during MD simulation. The ΔG_bind of the trajectories generated from simulation were calculated by Schrodinger Prime MM/GBSA using thermal_mmgbsa.py script with step size 10 (around 100 conformations were used as input for prime) (Alnajjar et al., 2020).
2.6 ADME and toxicity profiles prediction
In order to select a Drug-like molecule, absorption, tissue distribution, metabolic effects (ADME), and toxicity profiles should present favourable parameters and fill with Lipinski's rule of 5 (Lipinski et al., 1997). Physicochemical and pharmacokinetic properties of guggulsterone were estimated using Swiss ADME (Daina et al., 2014). The ProTox-II web server (Banerjee et al., 2018) was also used to explore hepatotoxicity, immunotoxicity, carcinogenicity, cytotoxicity and mutagenicity. Values were estimated after comparing the properties of a selected molecule with those of 95% of known recognized medicines (Saxena et al., 2019).
3 Results and discussion
3.1 Validation of the docking protocol and active site prediction
For validation of docking protocol, the co-crystalized ligand (S-adenosyl-l-homocysteine) of dengue methyl transferase (1L9K) was removed and redocked in the protein binding site, the redocked ligand gave an RMSD of 0.6407 Å, which indicates the validity of the used docking protocol (Jamal et al., 2017). The superposition of co-crystallized ligands and redocked is shown in Fig. 1. Schrodinger SiteMap was used to assess the druggability of binding pockets. Two proteins were of highly druggability (druggability score [D score] > 1.0) (2VBC: 1.105 and 2J7W: 1.026), while only one (1OK8: 0.663) was poorly druggable (Yazdani et al., 2021). The active site of dengue methyl transferase (1L9K) was estimated according to the site of the co-crystalized ligand.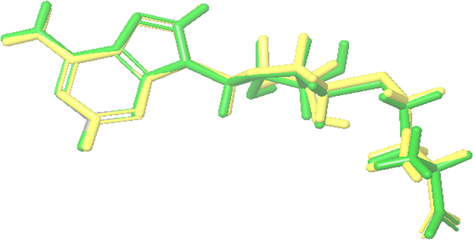
Superposition of co-crystalized (green) and redocked (yellow) ligands.
3.2 Molecular docking analysis
Molecular docking studies were done to examine the best conformation of guggulsterone (ligand) on selected dengue virus target proteins, and the binding affinity scores are shown in Table 1. The docking results revealed that guggulsterone has a binding affinity ranging from −5.485 to −3.422 kcal/mol. The in silico activity of guggulsterone on dengue target proteins is aligned with a previous in vitro study in which they showed the activity of guggulsterone on the dengue virus (Chen et al., 2021).
Target proteins
(PDB id)Ligands
Docking score
kcal/molGlide evdw
kcal/molGlide energy
kcal/mol
NS5 RNA dependent RNA polymerase (PDB id: 2J7W)
Guggulsterone CID6439929
−5.485
−26.64
−27.57
Ribavirin CID37542.1
−6.288
−26.643
−38.397
Methyl-transferase (PDB id: 1L9K)
Guggulsterone CID6439929
−4.724
−32.014
−32.246
Ribavirin CID37542.1
−5.742
−19.198
−39.233
NS3 protease-helicase (PDB id: 2VBC)
Guggulsterone CID6439929
−4.162
−28.74
−30.987
Ribavirin CID37542.1
−6.206
−17.393
−38.98
Dengue virus type 2 envelope glycoprotein (PDB id: 1OK8)
Guggulsterone CID6439929
−3.422
−24.178
−25.107
Ribavirin CID37542.1
−6.4
−25.015
−42.407
The guggulsterone showed the best binding affinity to NS5 RNA dependent RNA polymerase (RdRp) domain complex (PDB id: 2J7W), the docking score was −5.5 kcal/mol, Van der Waals energy (evdw) −27.5 kcal/mol, and glide energy −26.64 kcal/mol. The docking score of guggulsterone was near to the score of reference antiviral drug ribavirin (−6.2 kcal/mol), which is known as an inhibitor to dengue inosine 5′-monophosphate dehydrogenase inhibiting polymerases enzymes (Benarroch et al., 2004). We also found that TRP447 is associated with stabilization of docking reaction via formation of one strong hydrogen bond with small distance (2 Å) of hydrogen to acceptor atom (H-A) and between donor and acceptor atom (D-A) 2.8 Å (Table 2 and Fig. 2A and B). The previously published paper revealed that the TRP447 is located at the catalytic domain of dengue RdRp domain (Yap et al., 2007); the inhibition of such residue could block the enzyme activity. Also, as shown in Table 3, four hydrophobic interactions were formed as a result of guggulsterone binding to RdRp active site, TRP302, PHE354, VAL358, and GLN602, residues from 341 to 361 were located at the conserved region of βNLS region of NS5, which is associated with NS3 helicase interaction (Yap et al., 2007).
Index
Residue
AA
Distance H-A
Distance D-A
(Å)Donor Angle
Donor Atom
Acceptor Atom
NS5 RNA
dependent RNA polymerase (PDB id: 2J7W)1
477A
TRP
2.06
2.83
130.77
2659 [Nar]
9174 [O2]
Methyl-transferase (PDB id: 1L9K)
1
181A
LYS
2.33
2.96
119.32
2716 [N3 + ]
4094 [O2]
NS3 protease-helicase (PDB id: 2VBC)
–
–
–
–
–
–
–
–
Dengue virus type 2 envelope glycoprotein (PDB id: 1OK8)
1
7A
SER
3.2
3.74
114.28
99 [Nam]
5948 [O2]
2
355A
ASN
2.06
2.79
127.49
5319 [Nam]
5948 [O2]
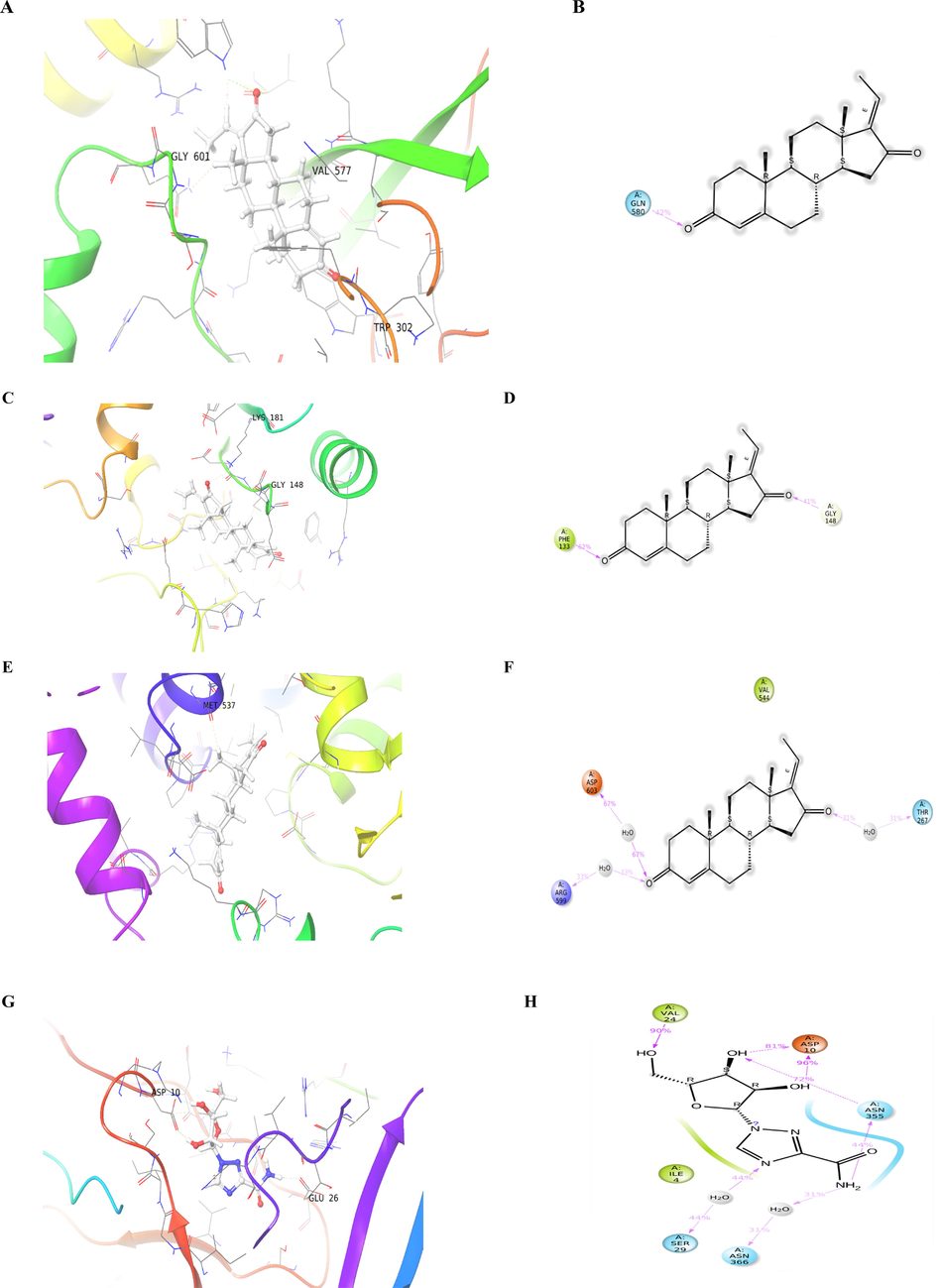
Interaction of the guggulsterone compound with: dengue NS5 RNA dependent RNA polymerase: A. 3D interaction during molecular docking and B. 2D interaction of ligand during MD simulation; dengue methyl-transferase: C. 3D interaction during molecular docking and D. 2D interaction of ligand during MD simulation; dengue NS3 protease-helicase: E. 3D interaction during molecular docking and F. 2D interaction of ligand during MD simulation; and Dengue virus type 2 envelope glycoprotein: G. 3D interaction during molecular docking and H. 2D interaction of ligand during MD simulation.
Index
Residue
AA
Distance
(Å)Ligand Atom
Protein Atom
NS5 RNA
dependent RNA
polymerase
(PDB id: 2J7W)1
302A
TRP
3.58
9192
487
2
354A
PHE
3.5
9190
1163
3
358A
VAL
3.53
9192
1223
4
602A
GLN
3.77
9196
4628
Methyl-transferase
(PDB id: 1L9K)1
105A
LYS
3.62
4112
1551
2
146A
ASP
3.96
4114
2179
3
147A
ILE
3.86
4113
2193
NS3 protease-helicase
(PDB id: 2VBC)1
544A
VAL
3.66
7107
5738
Dengue virus type
2 envelope glycoprotein
(PDB id: 1OK8)1
24A
VAL
3.71
5955
347
2
31A
VAL
3.47
5970
441
3
355A
ASN
3.46
5958
5316
The interaction of dengue methyltransferase (PDB id: 1L9K) and guggulsterone produced −4.724 kcal/mol docking energy, one hydrogen bond was formed with LYS181 with a small distance of 2.33 Å, and three hydrophobic interactions were formed with LYS105, ASP146, and ILE147. This finding agree with Geoffrey et al. (2020) who found various compounds targeting the dengue virus protein and displayed hydrogen bonds with LYS181, ASP146, and LYS105 (Fig. 2C and D).
The NS3 protease-helicase (PDB id: 2VBC) and guggulsterone have been shown −4.1 docking score (Table 1), which may be due to one hydrophobic interaction (Fig. 2E and F) with the residue VAL544 with a distance of 3.66 Å (Tables 3).
The interaction of dengue virus type 2 envelope glycoprotein (PDB id: 1OK8) and guggulsterone generate a docking score of −3.4 kcal/mol, glide energy of −25.1 kcal/mol, and Van der Waals energy (evdw) −24.1 kcal/mol suggesting its potency to interact with dengue virus by blocking major the envelope glycoprotein. Two hydrogen bonds were formed with SER7 and ASN355; their distances were 3.2 Å and 2.06 Å, respectively (Tables 1 and 2). Additionally, three hydrophobic interactions were generated with dengue major envelope protein E residues VAL24, VAL31, and ASN355; their distances were 3.71 Å, 3.47 Å, and 3.46 Å respectively (Table 3 and Fig. 2G and H). Geoffrey et al. (2020) reported a good binding affinity of piperine to different proteins of dengue virus, including major envelope protein E; and they found that Ile4A, Arg9A, Val24A, Val31A, Lys284A were associated with this interaction. In agreement with these findings, it was found that guggulsterone strongly interacted with VAL24.
3.3 Molecular dynamic (MD) simulation
Since we set the protein rigid during docking, MD simulation will compute atom movement during a period of time by applying Newton’s equation of motion (Adcock and McCammon, 2006). The stability of the protein–ligand complex was assessed with MD simulation, therefore the four docking complexes were run at 50 ns simulation time. As presented in Fig. 3A, the root mean square deviation (RMSD) plot showed that the guggulsterone is not stable with dengue NS5 RNA dependent RNA polymerase (2J7W) and protein backbones fluctuated from the beginning of simulation until the end. Most of the observed interactions lasted <20% of simulation time except GLN580, in which one hydrogen bond last for 50% (Fig. 4A); which might be the reason for the instability of the complex.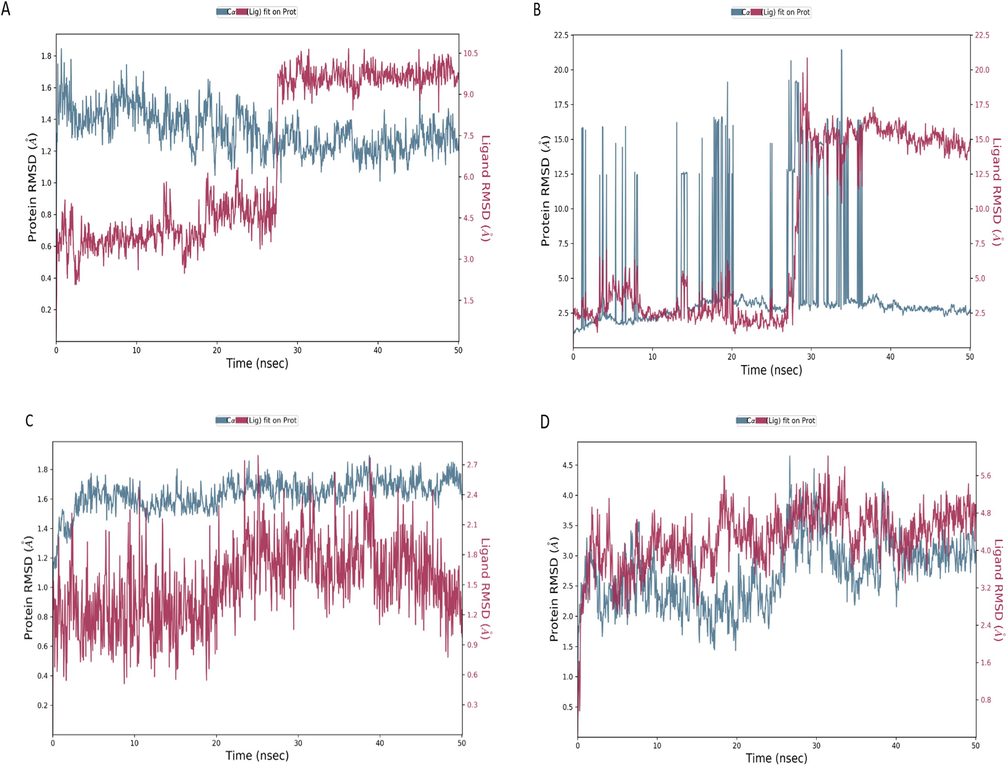
Root mean square deviations (RMSDs) trajectories of the dengue protein residues (A = 2J7W, B = 1L9K, C = 2VBC, D = 1OK8) and guggulsterone complexes run at 50 ns, the protein backbones carbon alpha atoms showed in in blue color and the ligand in red color.
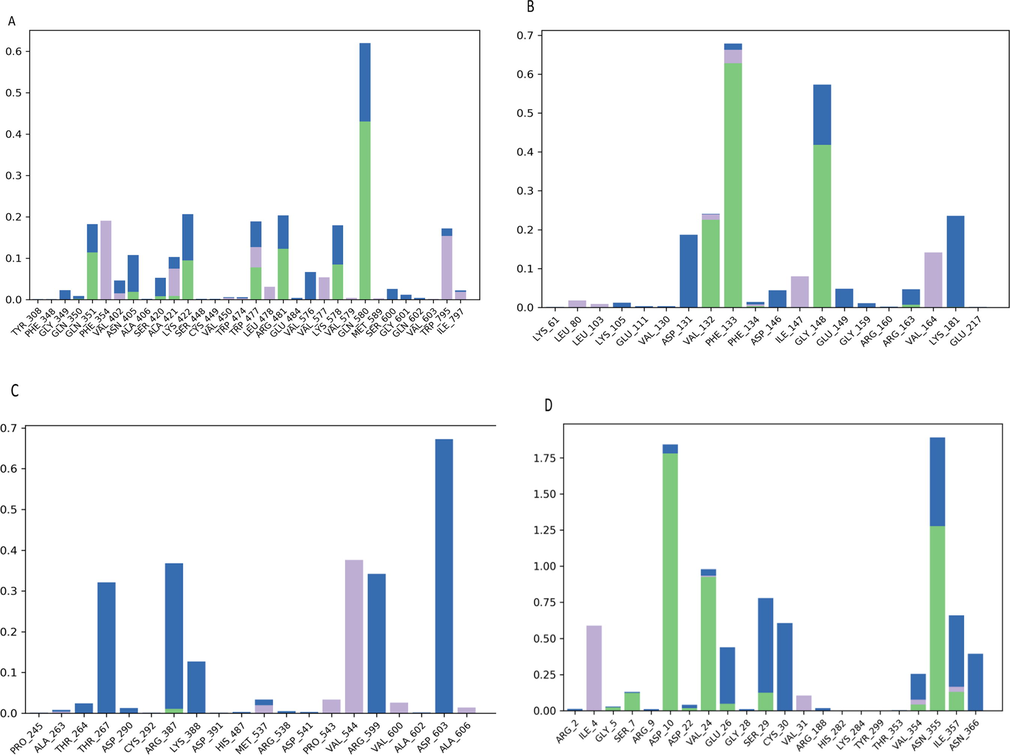
The histogram shows the percentage of bonds lasting during MD simulation of the complex of Dengue protein residues (A = 2J7W, B = 1L9K, C = 2VBC and D = 1OK8) and guggulsterone. The green color indicated the H-bonds, the blue color showed the water bridges, and the violet color demonstrated the hydrophobic interaction.
Fig. 3B show that the protein backbones of dengue methyltransferase (PDB id: 1L9K) is more flexible, most of the flexibility was observed at a region between 2 and 8, 13–20, and 27–45 ns of trajectories. These trajectories are part of the flexible region β2 and β3 which are part of the AdoMet-binding pocket (Zhao et al., 2015). Instead of high protein flexibility, the guggulsterone is still stable in protein backbones from the beginning until 50 ns of simulation time. A stable hydrogen bonds with protein residues (PHE133) in more than 60 % of simulation time, and the presence of water bridges in more than 50 % of simulation time, with GLY 148 (Fig. 4B) were also noticed.
Fig. 3C showed the complex of the NS3 protease-helicase (PDB id: 2VBC) and guggulsterone, the ligand RMSD fluctuation observed from 1 ns until the end of the simulation, the ligand separated from protein backbones in the range of 0.4 to 1.8 Å, which fallen in the permissible range of 1–3 Å (Choudhary et al., 2020), and indicate the stability of the complex.
According to Fig. 4C, a water bridges with ASP 603 (more than 60 % of simulation time) and a hydrophobic interaction with VAL 544 were observed.
The RMSD plot in Fig. 3D show the complex of dengue virus type 2 envelope glycoprotein (PDB id: 1OK8) and guggulsterone. The stability of the complex was the best during MD simulation; but the docking score was the lowest one (−3.4 kcal/mol); this could be due to the considering protein as rigid during docking, which may cause failure in assessing binding affinity in some proteins (Radwan and Mahrous, 2020). The complex remains stable (Fig. 3D) and aligned during the whole simulation time (50 ns); a slight deviation within the acceptable range is observed from 10 to 20 ns. This stability could be due to the presence of stable hydrogen bonds with protein residues (ASP10, VAL24, ASN355) in more than 80 % of simulation time, and the presence of water bridges in more than 60 %, with SER29, CYS30, and ILE357 (Fig. 4D).
As shown in Fig. 5 (A–D), the RMSF showed the fluctuation interacting residues during MD simulation. The interaction of guggulsterone and dengue virus NS5 RNA dependent RNA polymerase domain ((PDB id: 2J7W) showed 80 % (25) of interacting residues were at stable region (Fig. 5A), the interaction of the ligand occurred with 22 (100 %) stable residues of dengue methyltransferase (PDB id: 1L9K), 20 (100 %) stable residues of NS3 protease-helicase ((PDB id: 2VBC) and 24 (100 %) residues of dengue virus type 2 envelope glycoprotein ((PDB id: 1OK8) (Fig. 5B–D).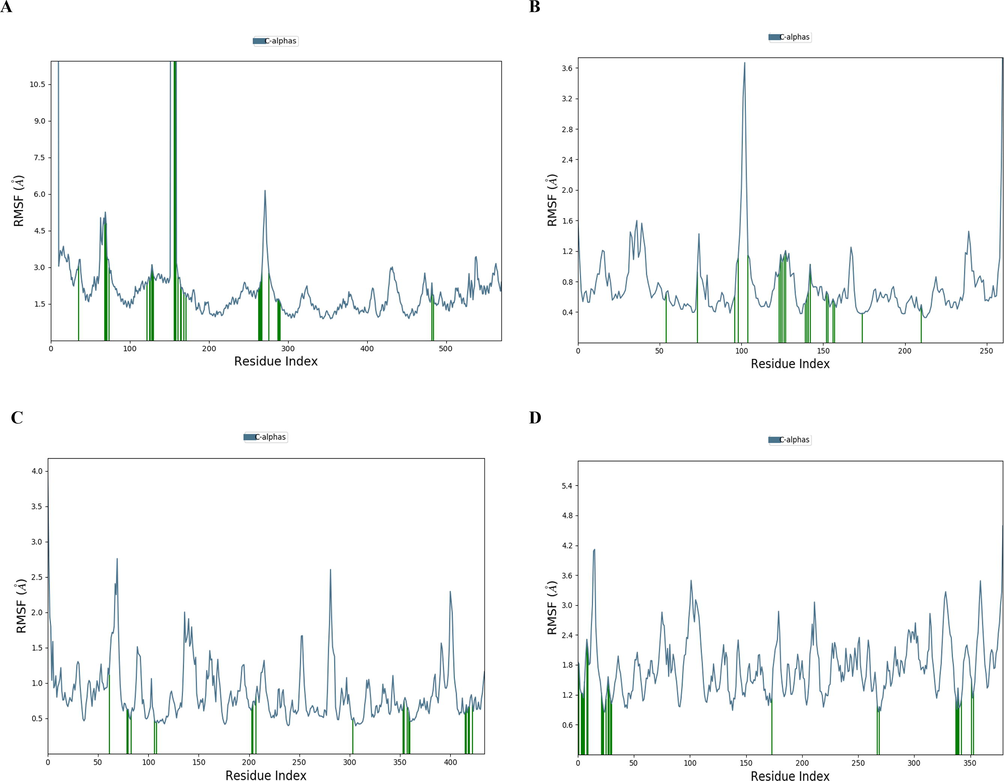
The RMSF lasting during MD simulation of the complex of dengue protein residues (A = 2J7W, B = 1L9K, C = 2VBC and D = 1OK8) and guggulsterone. The green coloured lines indicate the interacting residues.
3.4 Calculation of binding free energy
MM/GBSA analysis was used for the estimation of relative binding energies of ligands binding to protein's active sites, usually more negative values indicate better stability (Dash et al., 2019). As shown in Table 4, the dengue virus type 2 envelope glycoprotein ((PDB id: 1OK8) showed the lowest ΔG_bind (−53.24) in a good range −59.0159 to −47.1885 kcal/mol, this finding is aligned with MD simulation in which the ligand was the most stable in protein backbones. While the complex of guggulsterone and dengue virus NS5 RNA dependent RNA polymerase domain ((PDB id: 2J7W) showed higher fluctuation during MD simulation and thermodynamic energy calculation (high standard deviation 7.26).
Target proteins
(PDB id)dG(NS) Average
Standard Deviation
Range
NS5 RNA
dependent RNA
polymerase (PDB id: 2J7W)−41.9647
7.26
−58.9277 to −23.3482
Methyl-transferase
(PDB id: 1L9K)−44.8435
3.85
−55.4281 to −35.9026
NS3 protease-helicase
(PDB id: 2VBC)−34.8695
2.04
−39.4669 to −29.72429
Dengue virus type 2 envelope glycoprotein (PDB id: 1OK8)
−53.2440
2.84
−59.0159 to −47.1885
3.5 ADME and toxicity profiles prediction
As presented in Table 5, the pharmacokinetics, drug-likeness and toxicity profiles of guggulsterone were estimated in silico. Results showed that guggulsterone follows Lipinski’s rule of five (molecular weight (MW) < 500 Da, number of hydrogen bond donors (HBDs) < 5, LogP < 5, and number of hydrogen bond acceptors (HBAs) < 10). Also, no violation of Veber and collaborators rules (number of rotatable bonds (NBR) < 10 and polar surface area (PSA) < 140 Å2) was noticed (TPSA value of 34.14 Å2), which may facilitate the cross of guggulsterone toward cell membranes (Veber et al., 2002).
Physicochemical properties
Formula
C21H28O2
Molecular weight
312.45 g/mol
Num. heavy atoms
23
Num. arom. heavy atoms
0
Fraction Csp3
0.71
Num. rotatable bonds
0
Num. H-bond acceptors
2
Num. H-bond donors
0
Molar Refractivity
93.54
TPSA
34.14 Å2
Pharmacokinetics
GI absorption
High
BBB permeant
Yes
P-gp substrate
No
CYP1A2 inhibitor
No
CYP2C19 inhibitor
Yes
CYP2C9 inhibitor
Yes
CYP2D6 inhibitor
No
CYP3A4 inhibitor
No
Log Kp (skin permeation)
−5.41 cm/s
Druglikeness
Lipinski
Yes; 0 violation
Ghose
Yes
Veber
Yes
Egan
Yes
Muegge
Yes
Bioavailability Score
0.55
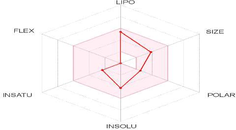
Bioavailability (Radar plot)
LIPO: Lipophilicity
FLEX: Flexibility
POLAR: Polarity
INSAT: Insaturation
INSOLU: Insolubility
SIZE: Size
Oral toxicity prediction (Probability)
Hepatotoxicity
Inactive (0.67)
Immunotoxicity
Active (0.98)
Carcinogenicity
Active (0.56)
Cytotoxicity
Inactive (0.79)
Mutagenicity
Inactive (0.99)
The blood–brain barrier (BBB) permeability and glycoprotein P (P-gp) substrate were used to predict distribution. The BBB is a structure separating the central nervous system (CNS) from peripheral tissue, controlling the passage of nutrients and materials to maintain homeostasis (Domínguez-Villa et al., 2021). It appeared from this investigation that guggulsterone was highly absorbed in the gastrointestinal (GI), cannot be a substrate of P-gp, and passed the blood–brain barrier (BBB).
Based on the bioavailability index, radar plot provides a graphical insight about the drug likeness of a tested molecule (Ali and Shabeer, 2021). The pink area is the acceptable range for oral bioavailability of each parameter (Mendie and Hemalatha, 2022). As shown in Table 5, the radar plot of guggulsterone falls in the pink region, indicating that it may be regarded as drug-like. Furthermore, FLEX and POLAR are two characteristics that define a compound's bioavailability. FLEX is defined by rotatable bonds (rotatable bonds greater than ten indicate limited oral bioavailability), whereas polarity is determined by topological polar surface (TPSA). The result showed that the number of rotatable bonds was 0 and TPSA 34.14 Å2.
Metabolism was also estimated by determining the effect of guggulsterone on inhibition of the main cytochromes (CYP1A2, CYP2C19, CYP2C9, CYP2D6 and CYP3A4) among the P450 superfamily. CYP2C19 is involved in the detoxification of potential carcinogens and CYP2C9 is the major enzyme implicated in drugs metabolism. According to Manikandan and Nagini (2018), the inactivation of CYPs may increase toxicity or decrease the efficiency of a potential drug. Guggulsterone was predicted to likely inhibit CYP2C19 and CYP2C9 and cannot inhibit CYP1A2, CYP2D6, and CYP3A4.
The liver is involved in the biotransformation of a variety of xenobiotics and drugs. Our results revealed that guggulsterone doesn’t induce hepatotoxicity, cytotoxicity, and mutagenicity but may cause immunotoxicity and carcinogenicity.
4 Conclusion
Computational techniques such as molecular docking, molecular dynamics, ADME and toxicity profiling, were widely employed in drug development.
In this study, we efficiently demonstrated that guggulsterone may be considered as a potent inhibitor of the dengue virus proteins including NS5 RNA dependent RNA polymerase, dengue methyltransferase, NS3 protease-helicase and dengue virus type 2 envelope glycoprotein. Guggulsterone was found to have the lowest docking energy (−5.5 kcal/mol) with dengue NS5 RNA-dependent RNA polymerase domain. While the interaction of guggulsterone with the dengue virus type 2 envelope glycoprotein exhibited the highest docking energy (−3.4 kcal/mol), but it was the most stable complex during molecular dynamic simulation. Guggulsterone is predicted to be a significant inhibitor of major envelope protein E of dengue virus. Concerning the ADME and toxicity prediction, no violation of Lipinski and Veber rules was recorded. Moreover, low values of toxicity were predicted for the tested compound. Guggulsterone may inhibit CYP2C19 and CYP2C9, and do not induce hepatotoxicity, cytotoxicity, and mutagenicity. More in-vitro testing is required to validate the effectiveness of this compound on dengue major envelope protein E.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Anti-Dengue potential, Molecular Docking Study of Some Chemical constituents in the leaves of Isatis tinctoria. Chem. Rev. Lett.. 2020;3(3):104-109.
- [Google Scholar]
- Molecular dynamics: survey of methods for simulating the activity of proteins. Chem. Rev.. 2006;106(5):1589-1615.
- [Google Scholar]
- Hepatoprotective properties of Commiphora opobalsamum (“Balessan”), a traditional medicinal plant of Saudi Arabia. Drugs Exp. Clin. Res.. 2004;30(5–6):213-220.
- [Google Scholar]
- Identifying clinically significant novel drug candidate for highly prevalent Alzheimer's disease. Indian J. Chem. Technol.. 2021;28:618-623.
- [Google Scholar]
- Molecular docking, molecular dynamics, and in vitro studies reveal the potential of angiotensin II receptor blockers to inhibit the COVID-19 main protease. Heliyon. 2020;6(12):e05641
- [Google Scholar]
- In vitro antiviral activity of Pinus merkusii (Pinaceae) stem bark and cone against dengue virus type-2 (DENV-2) Res. J. Pharm. Technol.. 2021;14(7):3705-3708.
- [Google Scholar]
- ProTox-II: a webserver for the prediction of toxicity of chemicals. Nucl. Acids Res.. 2018;46(W1):W257-W263.
- [Google Scholar]
- A structural basis for the inhibition of the NS5 dengue virus mRNA 2′-O-methyltransferase domain by ribavirin 5′-triphosphate. J. Biol. Chem.. 2004;279(34):35638-35643.
- [Google Scholar]
- The protein data bank. Acta Crystall. Sect. D: Biol. Crystall... 2002;58(6):899-907.
- [Google Scholar]
- Virucidal effect of guggulsterone isolated from Commiphora gileadensis. Planta Med.. 2019;85(16):1225-1232.
- [Google Scholar]
- (E)-guggulsterone inhibits dengue virus replication by upregulating antiviral interferon responses through the induction of heme oxygenase-1 expression. Viruses.. 2021;13(4):712.
- [Google Scholar]
- In silico identification of potential inhibitors of key SARS-CoV-2 3CL hydrolase (Mpro) via molecular docking, MMGBSA predictive binding energy calculations, and molecular dynamics simulation. PLoS One. 2020;15(7):e0235030
- [Google Scholar]
- iLOGP: a simple, robust, and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J. Chem. Inf. Model.. 2014;54(12):3284-3301.
- [Google Scholar]
- Structure-based identification of potent VEGFR-2 inhibitors from in vivo metabolites of a herbal ingredient. J. Mol. Model.. 2019;25(4):1-15.
- [Google Scholar]
- Synthesis, molecular docking, and in silico ADME/Tox profiling studies of new 1-aryl-5-(3-azidopropyl) indol-4-ones: Potential inhibitors of SARS CoV-2 main protease. Bioorg. Chem.. 2021;106
- [Google Scholar]
- Serotype and genotype analysis of dengue virus by sequencing followed by phylogenetic analysis using samples from three mini outbreaks-2007-2009 in Pakistan. BMC Microbiol.. 2011;11(1)
- [Google Scholar]
- Geoffrey, B., A. Sanker, R. Madaj, et al., 2020. A program to automate the discovery of drugs for West Nile and Dengue virus—programmatic screening of over a billion compounds on PubChem, generation of drug leads and automated in silico modelling. J. Biomol. Struct. Dyn. 1-9.
- Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J. Comput.-Aided Mol. Des.. 2010;24(6-7):591-604.
- [Google Scholar]
- An integrative in-silico approach for therapeutic target identification in the human pathogen Corynebacterium diphtheriae. PLoS One. 2017;12(10):e0186401
- [Google Scholar]
- Structure-based identification of potential SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn.. 2022;40(8):3595-3608.
- [Google Scholar]
- PubChem substance and compound databases. Nucleic Acids Res.. 2016;44(D1):D1202-D1213.
- [Google Scholar]
- Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev.. 1997;23(1-3):3-25.
- [Google Scholar]
- OPLS4: Improving force field accuracy on challenging regimes of chemical space. J. Chem. Theory Comput.. 2021;17(7):4291-4300.
- [Google Scholar]
- Cytochrome P450 structure, function and clinical significance: a review. Curr. Drug Targets. 2018;19(1):38-54.
- [Google Scholar]
- Molecular docking of phytochemicals targeting GFRs as therapeutic sites for cancer: an in silico study. Appl. Biochem. Biotechnol.. 2022;194(1):215-231.
- [Google Scholar]
- Natural compounds as potential inhibitors of SARS-CoV-2 main protease: An in-silico study. Asian Pac. J. Trop. Biomed.. 2021;11(4):155.
- [Google Scholar]
- Piperine, an alkaloid of black pepper seeds can effectively inhibit the antiviral enzymes of Dengue and Ebola viruses, an in silico molecular docking study. Virus Dis.. 2020;31(3):308-315.
- [Google Scholar]
- In silico exploitation of antiviral phytochemicals against dengue. Pak. J. Bot.. 2021;53(1):309-319.
- [Google Scholar]
- The in vitro antimicrobial activity and chemometric modelling of 59 commercial essential oils against pathogens of dermatological relevance. Chem. Biodivers.. 2017;14(1):e1600218
- [Google Scholar]
- Docking studies and molecular dynamics simulations of the binding characteristics of waldiomycin and its methyl ester analog to Staphylococcus aureus histidine kinase. PLoS One. 2020;15(6):e0234215
- [Google Scholar]
- 3: Desmond molecular dynamics system, DE Shaw research, New York, NY, 2020. Schrödinger, New York, NY: Maestro-Desmond Interoperability Tools; 2020.
- Potential medicinal plants for the treatment of dengue fever and severe acute respiratory syndrome-coronavirus. Biomolecules. 2021;11(1):42.
- [Google Scholar]
- Multiple e-pharmacophore modelling pooled with high-throughput virtual screening, docking and molecular dynamics simulations to discover potential inhibitors of Plasmodium falciparum lactate dehydrogenase (PfLDH) J. Biomol. Struct. Dyn.. 2019;37(7):1783-1799.
- [Google Scholar]
- molecular docking studies of selected medicinal plant compounds against NS5 & NS3 protein of dengue virus: a comparative approach. Int. J. Pharm. Bio. Sci.. 2016;7(3):1135-1144.
- [Google Scholar]
- Molecular docking based screening of plant flavonoids as dengue NS1 inhibitors. Bioinformation. 2014;10(7):460.
- [Google Scholar]
- Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem.. 2002;45(12):2615-2623.
- [Google Scholar]
- Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol.. 2007;81(9):4753-4765.
- [Google Scholar]
- Genetic variability of the SARS-CoV-2 pocketome. J. Proteome Res.. 2021;20(8):4212-4215.
- [Google Scholar]
- Flexibility of NS5 methyltransferase-polymerase linker region is essential for dengue virus replication. J. Virol.. 2015;89(20):10717-10721.
- [Google Scholar]







