Translate this page into:
DNA barcodes of Saudi Arabian birds: Implications for species identification and diversity analysis
⁎Corresponding author at: Department of Biochemistry, College of Science, Bldg. 5, King Saud University, P.O. Box 2455, Riyadh 11451, Saudi Arabia. haseeb@ksu.edu.sa (Haseeb A. Khan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objectives
DNA barcoding using cytochrome c oxidase I (COI) gene is an effective tool for species identification with additional power of measuring molecular diversity and phylogenetic inference. In this study, we evaluated the performance of COI gene sequences for species identification and molecular diversity analysis of some Saudi Arabian birds.
Methods
We sequenced the 694 base pair segment of COI gene from 5 samples of white cheeked bulbul (Pycnonotus leucogenys), 4 samples of black scrub robin (Cercotrichas podobe), and 2 samples of crested lark (Galerida cristata), all belonged to the same Order. We also included all the COI sequences available in the GenBank for these birds with the aim of studying molecular diversity across geographies. For species identification, we enriched our dataset by including COI barcodes of other Saudi Arabian bird species, including Arabian partridge (Alectoris melanocephala), houbara bustard (Chlamydotis undulata macqueenii), green bee-eater (Merops orientalis), laughing dove (Streptopelia senegalensis), namaqua dove (Oena capensis) and collared dove (Streptopelia decaocto), representing different genera.
Results
White-cheeked bulbul from Saudi Arabia showed more diversity than the same birds from Iraq. The mean nucleotide difference and nucleotide diversity were 5.20 and 0.009 respectively with as Tajima’s D score of 1.6549 indicating the scarcity of rare alleles. The specimens of black scrub robin from Saudi Arabia and Djibouti showed mixed phylogeny with only two segregating sties and a near zero Tajima’s D score indicating no major selection. Out of 18 samples of crested lark, 13 birds from 4 different geographical regions showed identical sequences, while 3 birds from Russia and 1 from Cyprus grouped in a separate cluster and 1 crusted lark from Djibouti differed from all. The mean nucleotide difference and nucleotide diversity were 0.6993 and 0.0014, while a negative Tajima’s D of −1.1956 indicated the presence of rare alleles. Phylogenetic analysis of 9 different species of Saudi Arabian birds showed the discriminatory power of COI barcodes as all the different species grouped separately in specific clusters.
Conclusions
COI barcodes are not only indispensable tools for species identification but can also be used for analyzing molecular diversity and phylogenetic inference.
Keywords
COI barcoding
Saudi Arabian birds
Species identification
Molecular diversity
1 Introduction
Avian populations are at constant threat from factors such as declining natural habitat, climate change and poaching. For instance, the loss of natural habitats due to logging and other activities posed threat to the Asir magpie population in Saudi Arabia (BirdLife International 2017). The increase in tourism activities in these areas is also a threat to bird species. It is estimated that there are approximately 135 or fewer pairs of this species left in the wild (BirdLife International 2017). The avian community is also affected by overhunting practices, which negatively impact their conservation status. Many cases of illegal bird hunting in Saudi Arabia and surrounding regions have been reported which may lead to crucial conservation issues if not timely controlled (Brochet et al 2019). Houbara bustard, a game species in Saudi Arabia, has been extirpated from most of its natural range due to excessive hunting (Alwelaie 1994, Seddon et al 1995). The Lanner falcon is an Afro-tropical and Mediterranean polytypic species considered critically endangered in Arabian Peninsula (Alabdulhafith et al 2022).
The diversity of local flora and fauna of Saudi Arabia is largely influenced by its geographical variation and its location between Eurasia and Africa (Al Midfa et al 2011). This location is also an important route for migratory birds in the region (Al Midfaet al 2011). The conservation of biological diversity is therefore gaining an increasing global attention. Particularly, in Saudi Arabia, conservation actions have become a topic of focus, and if not effectively implemented, certain challenges could threaten the conservation status of local wildlife (Alatawi 2022). In fact, several wild species in Saudi Arabia require special attention to ensure their conservation and long-term survival (Alatawi 2022). Maintaining a healthy and biologically diverse ecosystem is crucial whereas the destruction of natural resources can have catastrophic effects. The Saudi environmental program embodies the ambitious goals of the Kingdom’s Vision-2030 to safeguard wildlife and natural habitats in the Kingdom. Moreover, utilization of modern technology such as molecular methods is crucially important for species identification, diversity analysis and captive breeding of wild animals (Arif and Khan 2009).
DNA barcoding is a molecular technique for identification of taxa to species level by utilizing a standard region of the cytochrome c oxidase I (COI) gene. Since this technique is based on molecular-level variation, it offers a greater accuracy and authenticity than the plumage-based subjective phylogeny of birds that tends to be problematic due to high levels of homoplasy in color pattern leading to a weaker phylogenetic signal (Arif et al 2011a).
A recent review on the multiple applications of DNA barcodes in avian evolutionary studies has showed that DNA barcodes offer high-quality data well beyond their main purpose of serving as a molecular tool for species identification (Barreira et al 2016). Bilgin et al (2016) made global phylogeographic comparisons to define four categories of bird species at a migratory hotspot, based on barcoding suitability, intraspecific divergence and taxonomy. Their findings provided a good example of how DNA barcoding can build upon its primary mission of species identification and use available data to integrate genetic variation observed at the local scale into a global framework.
Since its inception, COI barcoding has been used for identification and diversity analysis of birds from different regions including New Zealand (Tizard et al 2019), Brazil (Chaves et al 2015), Japan (Saitoh et al 2015), Korea (Kwon et al 2012), Netherlands (Aliabadian et al 2013) and North America (Hebert et al 2004). Moreover, COI barcoding could be of immense help in preventing illegal trading of eggs pertaining to endangered birds as it solved the identity the embryos' species in a criminal investigation of trading avian eggs at Brazilian airport (Gonçalves et al 2015). Another application of COI barcoding has been reported for identification of bird involved in the bird strike incident that could help the airport staff to ensure flying safety (Yang et al 2010).
Saudi Arabia's dry deserts, coastlines, and oases are home to over 500 different birds, of these, about 277 species are migratory birds and 223 nesting birds. The first COI barcode of a Saudi Arabian bird (Arabian partridge) was published in 2010 (Khan et al 2010). Subsequently the barcodes of green bee-eater (Arif et al 2011a), houbara bustard (Arif et al 2012) and doves (Khan and Arif 2013) from Saudi Arabia were also reported. In this study, we sequenced the COI barcodes of white-cheeked bulbul (Pycnonotus leucogenys), black scrub robin (Cercotrichas podobe), and crested lark (Galerida cristata) and evaluated these sequences for species identification and molecular diversity analysis. In order to evaluate the discriminatory power of a COI barcode, it is imperative that sufficient members of a genus be examined, rather than a random sampling of imprecisely defined close relatives, and taxa should be gathered from more than one geographic region (Hebert et al 2004). For this reason, we also included all the COI sequences available in the GenBank, for these three bird species as well as other birds from Saudi Arabia representing different genera.
2 Materials and methods
2.1 Bird samples
We collected 5 specimens of white cheeked bulbul (Pycnonotus leucogenys), 4 specimens of black scrub robin (Cercotrichas podobe), and 2 specimens of crested lark (Galerida cristata) from the Al Hayathem and Al Hair regions, of the Riyadh district of Saudi Arabia. All the three bird species belonged to the same order (Passeriformes) but differed at the family level.
The gene sequences resulting from this study were deposited into GenBank with the following accession numbers: white-cheeked bulbul (HQ168045- HQ168049), black scrub robin (HQ168050- HQ168053), and crested lark (HQ168060, HQ168061).
Besides the above sequences, we also included all the COI sequences available in the GenBank for these 3 species. Our final data set of COI barcodes was as follows: white-cheeked bulbul (total 11: 5 from Saudi Arabia, 6 from Iraq), black scrub robin (total 6: 4 from Saudi Arabia, 2 from Djibouti), and crested lark (total 18: 2 from Saudi Arabia, 5 from Afghanistan, 4 from Iraq, 3 from Russia, 1 each from Cyprus, Djibouti, Kazakhstan and Turkey).
For comparative evaluation and validation of COI barcodes for species identification, we also included 19 more sequences of 6 different Saudi Arabian bird species, including Arabian partdridge (Alectoris melanocephala), houbara bustard (Chlamydotis undulata macqueenii), green bee-eater (Merops orientalis), laughing dove (Streptopelia senegalensis), namaqua dove (Oena capensis) and collared dove (Streptopelia decaocto), representing different genera (Khan et al 2010, Arif et al 2011, 2012, 2013). The taxonomic classification of all these species is summarized in Table 1.
Birds*
Order
Family
Genus
Species
White cheeked bulbul
Passeriformes
Pycnonotidae
Pycnonotus
Leucogenys
Black scrub robin
Passeriformes
Muscicapidae
Cercotrichas
Podobe
Crested lark
Passeriformes
Alaudidae
Galerida
Cristata
Namaqua dove
Columbiformes
Columbidae
Oena
Capensis
Collared dove
Columbiformes
Columbidae
Streptopelia
Decaocto
Laughing dove
Columbiformes
Columbidae
Spilopelia
Senegalensis
Green bee-eater
Coraciiformes
Meropidae
Merops
Orientalis
Arabian partridge
Galliformes
Phasianidae
Alectoris
Melanocephala
Houbara bustard
Otidiformes
Otididae
Chlamydotis
Undulata
2.2 DNA extraction
The commercially available DNeasy Blood and Tissue Kit (Qiagen GmbH, Hilden, Germany) was used for the extraction of DNA from thigh muscle tissues of the birds. The protocol provided as kit insert was strictly followed. The extracted DNA was finally dissolved in 200 µL of elution buffer and stored at −20 °C until analyzed.
2.3 PCR amplification
The PCR primers, BirdF1 (TTCTCCAACCACAAAGACATT GGCAC) and BirdR1 (ACGTGGGAGATAATTCCAAATCCTG) were used for amplification of COI sequences, as reported earlier (Kerr et al 2007, Khan et al 2010). A total volume of 30 µL of PCR reaction mixture contained 15 µL of FideliTaq PCR Master Mix (USB Corporation, Cleveland, OH), with a final concentration of 200 µM of each deoxynucleotide and 1.5 mM MgCl2, 1 µM each primer, 2 µL of genomic DNA and the rest was adjusted with nuclease free distilled water. PCR amplification was carried out using a Veriti 96 well thermal cycler (Applied Biosystems). The conditions of PCR amplification were as follows: denaturation of DNA at 94 °C for 1 min followed by 6 cycles of 1 min at 94 °C, 1.5 min at 45 °C, and 1.5 min at 72 °C. The annealing temperature was then raised to 55 °C for the next 35 cycles while keeping the other conditions same as before. After completion of all the PCR cycles, a final extension step for 5 min at 72 °C was used.
2.4 DNA sequencing
Before sequencing of COI gene, all the PCR products were purified using MicroSpin S300 columns (GE Healthcare), according to manufacturer’s instructions. A BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) was used for sequencing of PCR products. The sequencing reactants contained BigDye Terminator Ready Reaction Mix (4 µL), forward or reverse primer (1 µL), PCR product (1 µL) and sterile deionized water (4 µL). The microplate was placed in a thermal cycler and PCR was performed with one cycle of incubation at 96 °C for 1 min followed by 25 cycles of 96 °C for 10 s, 50 °C for 5 s and 60 °C for 4 min and then holding the plate at 4 °C. The PCR products were purified by BigDye XTerminator before injecting into the capillaries of a 3130XL Genetic Analyzer (Applied Biosystems, USA). Each sample was sequenced in both the directions using forward and reverse primers for a greater accuracy.
2.5 Data analysis
The sequences were aligned by ClustalW (Larkin et al 2007) and the sequence alignment file was saved as Molecular Evolutionary Genetics Analysis (MEGA) format. Mean nucleotide differences, number of segregating sites, nucleotide diversity, percent divergence, and Tajima’ D scores were computed. The pre-aligned sequences were subjected to un-weighted pair group method with arithmetic mean (UPGMA) hierarchical clustering method (Sneath and Sokal 1973) for phylogenetic analyses using the evolutionary distances computed by maximum composite likelihood method. The analysis was performed using the MEGA4 software (Tamura et al 2007) and the bootstrap consensus trees inferred from 500 replicates were used to represent the evolutionary history of the taxa analyzed (Felsenstein 1985).
3 Results
Phylogenetic analysis of 11 samples of white-cheeked bulul differentiated the taxa into two major clusters, one of them grouped the birds from Saudi Arabia and the other comprised the birds from Iraq (Fig. 1A). Among 6 samples of white-cheeked bulbul from Iraq, 5 samples showed identical COI sequences whereas only two within-group variable sites were observed in 1 sample (Fig. 1B). The 5 samples of white-cheeked bulbul from Saudi Arabia were separated into three groups with sequence variations at one and three locations. The mean nucleotide difference across the 576-bp long partial sequence of COI was 5.20 and the nucleotide diversity was 0.009. There were 11 segregating sites and Tajima’s D was 1.6549 (Table 2). A positive Tajima’s D score indicates the scarcity of rare alleles, balancing selection and sudden population contraction.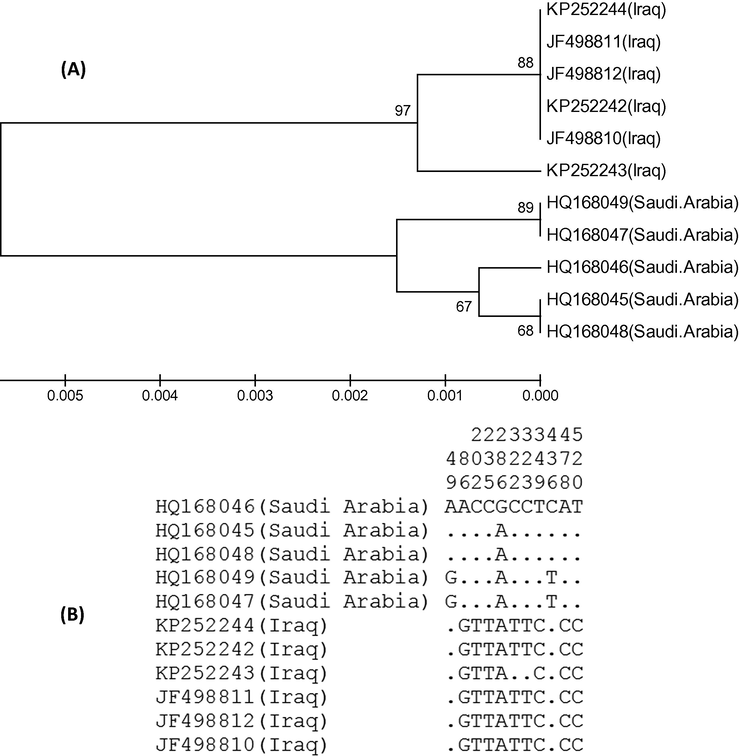
(A) Phylogenetic tree showing the relationship among the 11 specimens of white-cheeked bubuls from Saudi Arabia and Iraq. The COI gene sequences were aligned by ClustalW and subjected to UPGMA hierarchical clustering method for phylogenetic analyses using the MEGA4 software. The bootstrap consensus trees inferred from 500 replicates were used to represent the evolutionary history of the taxa analyzed. The optimal tree with the sum of branch length = 0.01480575 is shown. There were a total of 576 positions in the final dataset. (B) Haplogram of COI gene showing the variable sites among the 11 samples. The identical sites are represented by dots.
White cheeked bulbul
Black scrub robin
Crested lark
No. of samples
11
6
18
Mean base difference
5.2000
0.8666
0.6993
No. of segregating sites
11
2
4
Nucleotide diversity0.0090
0.0013
0.0014
% Divergence
0.9027
0.1331
0.1424
Tajima’s D
1.6549
−0.0500
−1.1956
The total 6 specimens of black scrub robin (4 from Saudi Arabia and 2 from Djibouti) showed mixed phylogeny (Fig. 2A). Two samples of black scrub robin showed identical COI sequences and clustered separately whereas 1 sample from Saudi Arabia was grouped with 2 samples from Djibouti. One sample of black scrub robin clustered individually (Fig. 2A). Altogether there were two haplotypes with sequence variation at position 203 and 613 (Fig. 2B). The mean nucleotide difference across 651-bp long segment of COI sequence was 0.8666 and nucleotide diversity was 0.0013 (Table 2). There were only two segregating sties and Tajima’s D score was close to zero (D = -0.05) indicating no major selection.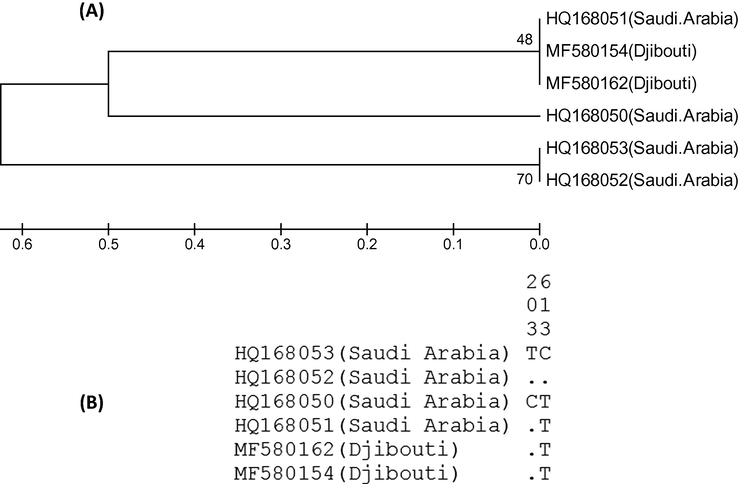
(A) Phylogenetic tree showing the relationship among the 6 specimens of black scrub robins from Saudi Arabia and Djibouti. The COI gene sequences were aligned by ClustalW and subjected to UPGMA hierarchical clustering method for phylogenetic analyses using the MEGA4 software. The bootstrap consensus trees inferred from 500 replicates were used to represent the evolutionary history of the taxa analyzed. The optimal tree with the sum of branch length = 1.75000000 is shown. There were a total of 651 positions in the final dataset. (B) Haplogram of COI gene showing the variable sites among the 6 samples. The identical sites are represented by dots.
Both the specimens of crested lark from Saudi Arabia had identical COI sequences with 100% match from 5 samples for Afghanistan, 4 samples from Iraq, 1 sample each from Turkey and Kazakhstan (Fig. 3A). Three specimens of crested lark from Russia and 1 specimen from Cyprus formed a separate cluster with identical sequences (Fig. 3A) and differed from the major cluster with only a single nucleotide difference at position 31 (Fig. 3B). There was only 1 sample of crested lark from Djibouti that showed the maximum difference and grouped separately in the phylogenetic tree (Fig. 3A). The mean nucleotide difference across 491 bases long segment of COI sequence was 0.6993 and nucleotide diversity was 0.0014 (Table 2). A negative Tajima’s D value of −1.1956 is indicative of rare alleles and population expansion after a recent bottleneck.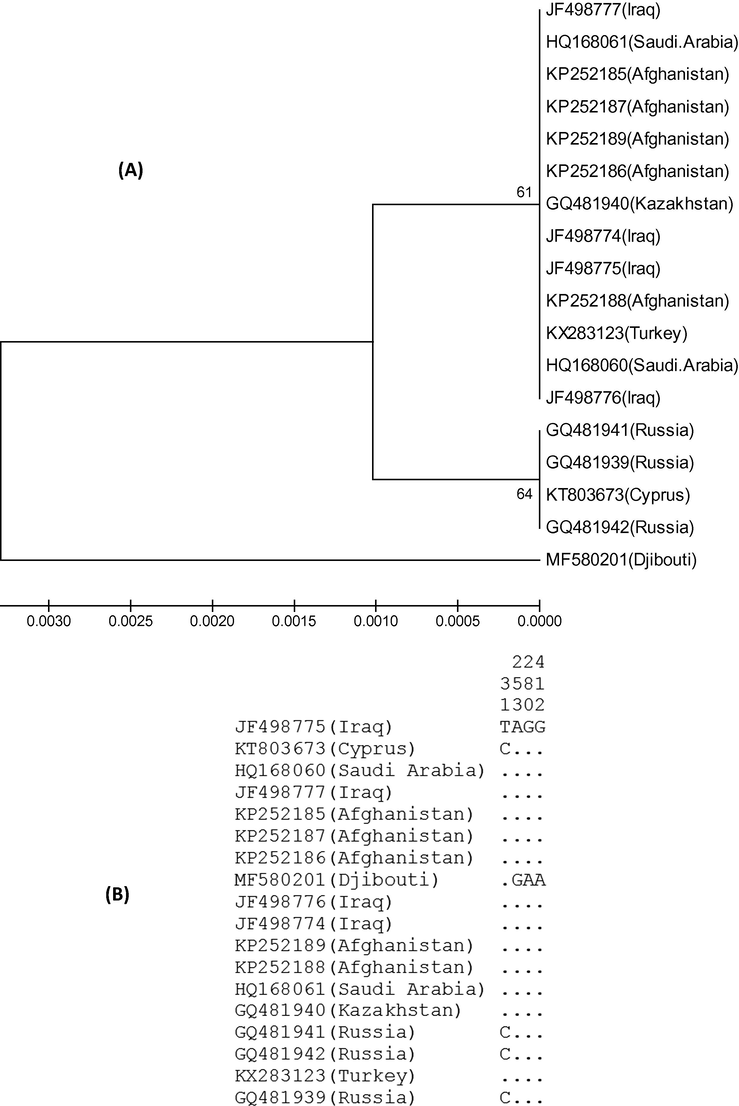
(A) Phylogenetic tree showing the relationship among the 18 specimens of crested larks from Saudi Arabia and 7 other countries. The COI gene sequences were aligned by ClustalW and subjected to UPGMA hierarchical clustering method for phylogenetic analyses using the MEGA4 software. The bootstrap consensus trees inferred from 500 replicates were used to represent the evolutionary history of the taxa analyzed.The optimal tree with the sum of branch length = 0.00760752 is shown. There were a total of 491 positions in the final dataset. (B) Haplogram of COI gene showing the variable sites among the 18 samples. The identical sites are represented by dots.
Phylogenetic analysis using COI sequences of 9 different species of Saudi Arabian birds showed the discriminatory power of COI barcodes as all the different species grouped separately in specific clusters (Fig. 4). White-cheeked bulbul, black scrub robin and crested lark, all from the order Passeriformes, formed a separate clad comprised of three distinct clusters for each species. All the three species of doves were grouped in a single clad of three clusters showing collared dove to be more closely related to laughter dove than namaqua dove (Fig. 4). Arabian partridge (Order: Galliformes) and houbara bustard (Order: Otidiformes) were grouped in a single clad with low bootstrap support; however, they were placed in individual clusters without any ambiguity (Fig. 4).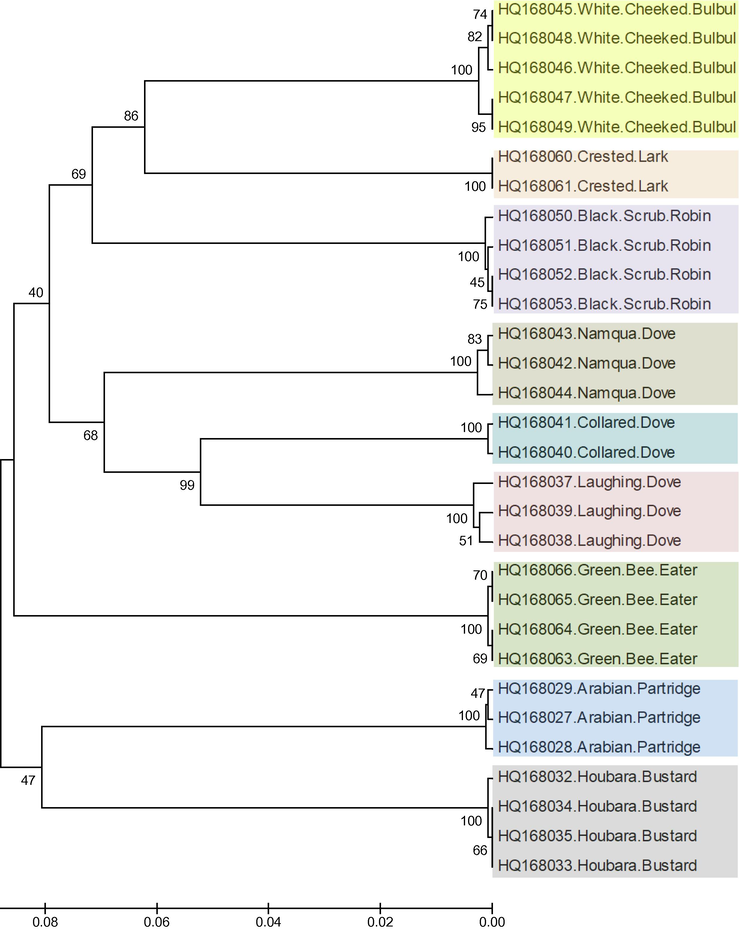
Phylogenetic tree showing the relationship among the 30 specimens of 9 different species of Saudi Arabian birds. The COI gene sequences were aligned by ClustalW and subjected to UPGMA hierarchical clustering method for phylogenetic analyses using the MEGA4 software. The bootstrap consensus trees inferred from 500 replicates were used to represent the evolutionary history of the taxa analyzed. The optimal tree with the sum of branch length = 0.69472345 is shown. There were a total of 682 positions in the final dataset. All the species were clearly differentiated by COI barcodes.
4 Discussion
Mitochondrial protein-coding genes are considered as useful markers for analyzing genetic diversity at lower taxonomic levels like family, genus, and species (Arif and Khan 2009, Arif et al 2011b). Under the same marker category, COI barcodes have the ability to identify taxa below the species level that may constitute separate conservation units (Rach et al 2008). Kerr et al (2007) have shown that COI barcoding can be effectively applied across the geographical and taxonomic expanse of bird species. Even the closely related sister species of birds can be identified reliably by COI barcodes (Tavares and Baker 2008). Kerr et al (2009) used COI barcodes for evaluation of intraspecific sequence divergences in eastern Palearctic birds. Pulgarín-R et al (2021) have reported the barcodes for low elevation bird species from tropical forests of northern Colombia with the goal to provide tools for species identification.
They obtained 26 COI barcode sequences from 18 species, 10 families and three orders and found that barcodes largely matched phenotypic identification (>90%) and helped in identification of several challenging passerine species. Colihueque et al (2021) used COI barcodes to differentiate among Chilean bird species by analyzing the gene sequences of 76 species comprised of 197 individuals. The COI barcodes correctly identified 94.7% of the species analyzed with mean intraspecific K2P distance of 0.3%. Their results suggested that bird species from Chile showed low levels of genetic structure and divergence; the small overlap between intra- and inter-specific distances implies that COI barcodes could be used as an effective tool to identify nearly all the Chilean bird species (Colihueque et al 2021).
McLaughlin et al (2023) identified potential cryptic species based on a comprehensive dataset of COI barcodes from 2,333 Panamanian birds across 429 species, representing 391 of the 659 resident land bird species of the country, as well as opportunistically sampled water birds. Using barcode identification numbers (BINs), they found putative cryptic species in 19% of land bird species, highlighting hidden diversity in the relatively well-described avifauna of Panama. Grealy et al (2021) compared three sampling methodologies to genetically identify 45 data-poor eggshell specimens, including a putatively extinct bird's egg. Their findings showed sufficient DNA for molecular identification can be obtained from even the tiniest eggshells without significant alteration to the specimen's appearance or integrity. This method was applied to confirm that a purported clutch of Paradise Parrot eggs collected 40 years after the species' accepted extinction date were falsely identified, laying to rest a five decades-old ornithological controversy.
This is probably the first study reporting the molecular diversity of white-cheeked bubul and black scrub robin across different geographical regions. Although white-cheeked bulbuls from Saud Arabia and Iraq grouped in two different clusters (Fig. 1), black scrub robin showed mixed phylogeny for the birds from Saudi Arabia and Djibouti (Fig. 2). Phylogenetic analysis of crested larks from different geographical regions separated the birds into two major clusters while a single specimen of crested lark from Djibouti placed separately (Fig. 3). These results are in agreement with a previous report showing two major groups of crested lark with divergence of approximately 1.1 Ma (Guillaumet et al 2005). The genus Galerida contains 7 species including G. cristata (crested lark) which is found in northern Africa and in parts of western Asia and China. Guillaumet et al (2008) suggested that phenotypic variation in lark species did not correlate with the history of populations, but was strongly influenced by current ecological conditions; the desert-adapted plumage evolved at least three times and variation in body size was mainly driven by interspecific competition, while the adaptive response to competition was more prominent in arid areas.
Our results also demonstrated the discriminatory power of COI barcodes for species identification as all the 9 different species of Saudi Arabia birds appeared as separate clusters of individual species (Fig. 4). Based on the general patterns of sequence variation, the identification system on the Barcode of Life Data (BOLD) delivers species identification if the query sequence shows a tight match, 1% divergence, to a reference sequence (Ratnasingham and Hebert 2007). Our group was the first to report the COI barcodes of Arabian partridge and Philby’s rock partridge (Khan et al 2010) and houbara bustard (Arif et al 2012). Phylogenetic analysis using 619 bp nucleotide segment of COI placed all the four specimens of houbara bustard in a single clade that was clearly separated from other two individuals (Otis tarda and Tetrax tetrax) of the same family (Arif et al 2012). The sequences from the two samples of Philby’s rock partridge were found to be identical whereas only 3 within-species variable sites were observed in the three samples of Arabian partridge (Khan et al 2010). Genetic data showd that patterns of speciation and population diversification of Przewalski’s rock partridge was affected by the stability of climate, natural selection, and human intervention (Chen et al 2006). In a previous study, we compared the COI gene segments of green bee-eater (Merops orientalis) with European bee-eater (Merops apiaster), showing the phylogenetic separation of Merops apiaster (resident) and Merops orientalis (migratory) into two distinct clusters (Arif et al 2011a). Marks et al (2007) have also reported the grouping of the bee-eaters into two well-supported clusters defined on ecological behavior, sedentary or migratory birds. Regarding the COI gene-based phylogeny of doves, the genus-level classification was well resolved for 9 of the 11 genera, it was only partially resolved for 2 genera, Streptopelia and Turtur; this could be the result of polyphyletic nature of several avian genera (Khan and Arif 2013).
COI barcode data from a large number of different species of North American birds have shown that each species has a unique COI barcode (Hebert et al 2004). Yoo et al (2006) used COI barcodes used for authentic discrimination of a large number of Korean birds. Using the COI barcodes of 39 species from 12 genera of Corvidae, the average genetic distance between the species was found to be 22-times higher as compared to the average genetic distance within species (Huang and Ruan 2018). COI gene data not only discriminated each species but also provided good evidence for the monophyly of the Corvidae while the members of Cyanopica and Pyrrhocorax were the first to split from the Corvidae lineage. Huang et al (2016) analyzed the COI barcodes of 32 species from 17 genera belonging to the family Ardeidae and observed that average genetic distance between species was 34-fold higher than the average genetic distance within species. Each bird species possessed a barcode distinct from that of other bird species except for Egretta thula and E. garzetta, which shared one barcoding sequence. Among the 163 species of neotropical bats, 98.8% possessed distinct sets of COI haplotypes making them easily recognizable, while only a single case was observed with shared haplotypes (Clare et al 2011). Kerr et al (2009) highlighted the importance of COI barcodes in avian research for refining avian taxonomy and to provide an invaluable tool for species assignment when morphological differences are difficult to measure. However, some studies have suggested that the COI sequences need to be cautiously selected as a DNA barcode for identifying bird species (Kwon et al 2012). Although the universal primers for the standard COI barcode (648-bp) have been validated, the PCR success rate for amplification of a mini-barcode region (131-bp) was only 80% for mammals, 45% for reptiles and 57% for birds, indicating the limited utility of universal primers for mini-barcoding (Arif et al 2011c).
In conclusion, our results showed that mitochondrial cytochrome c oxidase subunit I (COI) barcodes are not only indispensable tools for species identification but are fairly applicable for analyzing molecular diversity and phylogenetic inference. The sequencing of COI gene segment is a straightforward protocol using the universal primers. However, only few COI barcodes are currently available for Saudi Arabian birds. Considering the recent trend in climate change, environmental pollution and habitat crisis, it is crucially important to keep on record and utilize the COI barcodes of Saudi Arabian birds for their preservation by monitoring their diversity and population structure.
Acknowledgments
We sincerely thank Prof. Mohammad Shobrak for providing birds samples. This study was supported by Researchers Supporting Project Number (RSPD2023R 770), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Ten years of conservation workshops for the fauna of Arabia 2000–2009. Zool. Middle East.. 2011;54:7-12.
- [Google Scholar]
- Predicting the potential distribution of a near-extinct avian predator on the Arabian Peninsula: implications for its conservation management. Environ. Monit. Assess.. 2022;194:535.
- [Google Scholar]
- Conservation action in Saudi Arabia: Challenges and opportunities. Saudi J. Biol. Sci.. 2022;29:3466-3472.
- [Google Scholar]
- DNA barcoding of Dutch birds. Zookeys.. 2013;365:25-48.
- Protected areas in Saudi Arabia: Sustainable use of natural resources. GeoJournal. 1994;34:383-392.
- [Google Scholar]
- Molecular markers for biodiversity analysis of wildlife animals: A brief review. Anim. Biodivers. Conserv.. 2009;32:9-17.
- [Google Scholar]
- Cytochrome c oxidase subunit I barcoding of the green bee-eater (Merops orientalis) Genet. Mol. Res.. 2011;10:3992-3998.
- [Google Scholar]
- DNA marker technology for wildlife conservation. Saudi J. Biol. Sci. 2011;18:219-225.
- [Google Scholar]
- Limited efficiency of universal mini-barcode primers for DNA amplification from desert reptiles, birds and mammals. Genet. Mol. Res.. 2011;10:3559-3564.
- [Google Scholar]
- DNA barcodes of Asian Houbara Bustard (Chlamydotis undulata macqueenii) Int. J. Mol. Sci.. 2012;13:2425-2438.
- [Google Scholar]
- The multiple applications of DNA barcodes in avian evolutionary studies. Genome. 2016;59:899-911.
- [Google Scholar]
- DNA barcoding of birds at a migratory hotspot in Eastern Turkey highlights continental phylogeographic relationships. PLoS One. 2016;11:e0154454.
- [Google Scholar]
- BirdLife International, Pica asirensis. The IUCN Red List of Threatened Species 2017: e.T103727136A119432544.
- A preliminary assessment of the scope and scale of illegal killing and taking of wild birds in the Arabian Peninsula. Iran and Iraq. Sandgrouse.. 2019;41:154-175.
- [Google Scholar]
- Barcoding Neotropical birds: assessing the impact of nonmonophyly in a highly diverse group. Mol. Ecol. Resour.. 2015;15:921-931.
- [Google Scholar]
- Genetic structure of Przewalski’s rock partridge (Alectoris magna) populations in the Longzhong Loess Plateau. China. Biochem Genet.. 2006;44:209-221.
- [Google Scholar]
- Neotropical bats: estimating species diversity with DNA barcodes. PLoS One. 2011;6:e22648.
- [Google Scholar]
- Revealing the biodiversity of Chilean birds through the COI barcode approach. Zookeys.. 2021;1016:143-161.
- [Google Scholar]
- Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783-791.
- [Google Scholar]
- DNA Barcoding Identifies Illegal Parrot Trade. J. Hered.. 2015;106(Suppl 1):560-564.
- [Google Scholar]
- Genetic barcoding of museum egg shell improves data integrity of avian biological collections. Sci. Rep.. 2021;11:1605.
- [Google Scholar]
- Phenotypic variation in Galerida larks in Morocco: the role of history and natural selection. Mol. Ecol.. 2005;14:3809-3821.
- [Google Scholar]
- Climate-driven diversification in two widespread Galerida larks. BMC Evol. Biol.. 2008;8:32.
- [Google Scholar]
- DNA barcoding and phylogenetic relationships of Ardeidae (Aves: Ciconiiformes) Genet. Mol. Res.. 2016;15(3)
- [CrossRef] [Google Scholar]
- DNA barcodes and insights into the phylogenetic relationships of Corvidae (Aves: Passeriformes) Mitochondrial DNA A DNA Mapp Seq Anal.. 2018;29:529-534.
- [Google Scholar]
- Comprehensive DNA barcode coverage of North American birds. Mol. Ecol. Notes. 2007;7:535-543.
- [Google Scholar]
- Filling the gap - COI barcode resolution in eastern Palearctic birds. Front. Zool.. 2009;6:29.
- [Google Scholar]
- COI barcodes and phylogeny of doves (Columbidae family) Mitochondrial DNA. 2013;24:689-696.
- [Google Scholar]
- DNA barcodes of Arabian partridge and Philby's rock partridge: Implications for phylogeny and species identification. Evol. Bioinform Online.. 2010;6:151-158.
- [Google Scholar]
- Low resolution of mitochondrial COI barcodes for identifying species of the genus Larus (Charadriiformes: Laridae) Mitochondrial DNA. 2012;23:157-166.
- [Google Scholar]
- Molecular phylogenetics of the bee-eaters (Aves: Meropidae) based on nuclear and mitochondrial DNA sequence data. Mol. Phylogenet. Evol.. 2007;45:23-32.
- [Google Scholar]
- McLaughlin, J.F., Aguilar, C., Bernstein, J.M., et al. Comparative phylogeography reveals widespread cryptic diversity driven by ecology in Panamanian birds. bioRxiv. 2023. 2023.03.15.530646.
- Character–based DNA barcoding allows discrimination of genera, species and populations in Odonata. Proc. Biol. Sci.. 2008;275:237-247.
- [Google Scholar]
- BOLD: The Barcode of Life Data System (www.barcodinglife.org) Mol. Ecol. Notes. 2007;7:355-364.
- [Google Scholar]
- DNA barcoding reveals 24 distinct lineages as cryptic bird species candidates in and around the Japanese Archipelago. Mol. Ecol. Resour.. 2015;15:177-186.
- [Google Scholar]
- Restoration of houbara bustard populations in Saudi Arabia: developments and future directions. Oryx. 1995;29:136-142.
- [Google Scholar]
- Sneath PHA, Sokal RR. Numerical Taxonomy. 1973; Freeman, San Francisco.
- MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol.. 2007;24:1596-1599.
- [Google Scholar]
- Single mitochondrial gene barcodes reliably identify sister-species in diverse clades of birds. BMC Evol. Biol.. 2008;8:81.
- [Google Scholar]
- DNA barcoding a unique avifauna: an important tool for evolution, systematics and conservation. BMC Evol. Biol.. 2019;19:52.
- [Google Scholar]
- Using DNA barcodes to identify a bird involved in a birdstrike at a Chinese airport. Mol. Biol. Rep.. 2010;37:3517-3523.
- [Google Scholar]







