Translate this page into:
Development of “Biosearch System” for biobank management and storage of disease associated genetic information
⁎Corresponding author at: Center of Excellence in Genomic Medicine Research, King Abdulaziz University, Jeddah, Saudi Arabia. skarim1@kau.edu.sa (Sajjad Karim)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
Objective
Databases and softwares are important to manage modern high-throughput laboratories and store clinical and genomic information for quality assurance. Commercial softwares are expensive with proprietary code issue while academic versions have adaptation issue. Our aim was to develop an adaptable in-house software that can stores specimen and disease-associated genetic information in biobank to facilitate translational research.
Methods
Prototype was designed as per the research requirements and computational tools were used to develop software under three tiers; Visual Basic and ASP.net for presentation tier, SQL server for data tier, and Ajax and JavaScript for business tier. We retrieved specimens from biobank using this software and performed microarray based transcriptomic analysis to detect differentialy expressed genes (DEGs) with FC ±2 and P-value <0.05 in triple negative breast cancer cases. Ingenuity pathway analysis tool was used to predict canonical molecular pathways associated with disease. Overall performance and utility of software was evaluated by JMeter software, CRUD function test and set of feedback questioners.
Results
We developed “Biosearch System”, a web-based software enabling management of biobank samples (tissue, blood, FTTP slides) and their extracts (DNA, RNA and proteins) with clinical and experimental details. The client satisfaction feedback was excellent with score 4.7/5. We identified a total of 1181 DEGs including both upregulated (IFI6, LEF1, FANCI, CASC5, PLXNA3 etc.) and down-regulated (ADH1B, LYVE1, ADH1C, ADH1B, ADIPOQ, PLIN1, LYVE1 etc.) genes in triple negative breast cancer. Pathway analysis of DEGs revealed significant activation of interferon signaling (z-score 2.646) and kinetochore metaphase signaling pathway (z-score 2.138) in cancer.
Conclusion
Biosearch System is a user friendly LIMS for collection, storage and retrieval of specimen and clinical information. It is secure, efficient, and very convenient in sample tracking and data analysis. We illustrated its utility in transcriptomic study of breast cancer. Additionally, it can facilitate and speed up any genomic study and translational research publications.
Keywords
Biosearch system
LIMS database
Biobank
Genomics
Microarray
Bioinformatics
- LIMS
-
Laboratory Information Management Systems
- CEGMR
-
Center of Excellence in Genomic Medicine Research
- MRN
-
Medical Record Number
- DEGs
-
Diferentialy expresed genes
- BS
-
Biosearch System
Abbreviations
1 Introduction
Recent high-throughput technological advancements in next generation sequencing and microarray have generated huge data at relatively lower cost (Diamandis, 2009; Fehniger and Marko-Varga, 2011; Glenn, 2011; Merdad et al., 2014). Biobanks are established for long-term storage and conservation facility for biological specimens along with their demographic, clinical and experimental information to support scientific investigation using bioinformatics tools (Artene et al., 2013). Once the samples size grow in thousands then manual methods fail in eficient handling and samples information can also be lost. Quality becomes another big concern with big data. Therefore, need of a robust software arises to manage biobank and laboratory information for assuring quality of results with approved ethical guideline and personal integrity ((ISBER) 2012; Bredenoord et al., 2011; Kang et al., 2013; Voegele et al., 2007). Softwares are helpful in managing the data flow cycle comprising of collection, storage, analysis and report generation to facilitate quick and easy retrieval of information and speed up biomarker and therapeutic discoveries (Melo et al., 2010).
Collection of significant cohort size with crucial factors like number and type of samples, clinical information, pathological finding, follow-up data etc. is a time taking process but strongly recommended to facilitate comprehensive translational research (Betsou et al., 2010; Hewitt, 2011; Huang et al., 2011; Riegman et al., 2008). Bioinformatics softwares are either available as commercial with proprietary code and high cost or academic/open source with complexity that are difficult to adopt with other laboratories (Greely, 2007; Huang, Arkin, and Chandonia, 2011; Kauffmann and Cambon-Thomsen, 2008; Minamikumo, 2012; Prilusky et al., 2005). We, therefore, developed and implemented a software to support the investigators to access all clinical and experimental disease associated information required for basic and translational research. Herein, we discuss the application of our in-house developed software for genomic analysis specifically for breast cancer transcriptomics and can be used further for any diseases.
2 Materials and methods
Software was developed using three-tier architecture model: (i) Presentation tier – an interactive web browser for end user’s computer, (ii) Data tier – a SQL Server Management Studio that manages the storage and DB-server, and (iii) Business tier – acting as bridge between the rest two and collects data from the presentation tier, checks for validations and finally sends them to the data tier and vice versa. The BS follows standard guidelines like legislation agreement, standardized architecture, workflow and dedicated staff members, and technical procedures to record and access clinical data as described below:
2.1 Legal requirements
Software has been designed in accordance with the Saudi Arabian approved regulation for genomic medicine research. Patients were informed prior and written consents were taken by their doctors for their samples to be sent to the biobank for research. We provided consent forms to patients approved by the ethics committee. Additionally, the software stores data in coded format to hide identity of patients during any research presentation and publications. In order to ensure bioethical safety, we followed Saudi national committee of bioethics guidelines (Royal decree No. M/59, dated 14/9/1431H – 24/8/2010).
2.2 Software and hardware architecture
Prototype is an early model building process to test a proposed concept to enhance precision by system analysts and users. Database software design started by designing prototype; (i) first low fidelity prototype (paper-based prototype) and then (ii) the high-fidelity prototype (computer-based prototype). Collected information is categorized into logical groups or entities like sample, storage condition, specimen request, approval system, project type, diagnosis, and disease type and an entity-relationship diagram was built to show numerical relationship among different entities. Software was built on the servers with following specifications: Windows Server 2012 R2 Enterprise Edition service packs 2, .NET 4.5, IIS 8.5, VS 2012, Microsoft SQL Server 2008 R2, ASP.NET, and AJAX Control Kit 4.5. A web browser was used for graphical user interface. Database software has been developed to support the high-performance hardware system and is compatible with common web-browsers. To develop a robust and reliable software following features have been included: (i) web-based application for wide access of database, (ii) sample labelling before enrolling into database, (iii) security system with restricted access permission to authorized person as per role, and (iv) added disaster management system to cope up with any natural disaster.
2.3 System structure
To manage the laboratory services the software, consist of following six basics functions: (i) Control sample data (add, edit, confirm transfer, retrieve, and search), (ii) Show log book, (iii) Sample status, (iv) Show box content, (v) Add new (diagnostics, extraction, hospital, sample type, project), and (vi) Request sample. Samples stored in biobank are provided to researchers as per project requirements with proper justification and ethical approval. The requested sample is first checked by Biobank staff then forwarded to Lab Manager and finally goes to Director for final approval. Researchers can track the processing steps while software updates the decision by email. After delivery of approved specimen, requisite volume is automatically subtracted from biobank stock.
2.4 Database structure
Normal relational database is used for the system. Specific information is stored in specific tables like SampleInfo table contains the samples information, ProductInfo table contains products (DNA, RNA) type of sample information, PatientInfo table contains patients related information, SampleStorage table contains the storage information (refrigerators, shelf, box container) and UsersInfo table contains user related information. The type of relation is one-to-many (one patient::multi-samples, one sample::multi-products, one refrigerator::multi-samples etc.).
2.5 Workflow and biobank organizational structure
The software can only be used with authorized username and password, and after successful login on web-based client PC, users are allowed to work on next layer features which lists- add new data, update/edit (add or change), follow up the patients; retrieve data for analysis and interpretation. BS also deals in storage, quality, quantity, distribution, and maintenance of specimen (tissue, blood, serum etc.) and its derivatives (DNA, RNA, protein, plasma etc.). As per the existing CEGMR biobank organizational structure, assigned users have different level of rights for using our software. Researchers are end users of biobank; they request samples based on their active research projects. Dedicated biobank staffs examine the request and update the status to supervisor, who approve or reject the request with valid reason. However, final decision is taken by the director, who has authority to reconsider the request or reverse the supervisors' decision. Once approved by director, biobank delivers requested items within a week. Reminders for pending tasks are mailed to concerned staffs till final decision comes. This system enables full access to the researchers with the biobank information by managing samples, the notification for request approval or rejection, and providing overall summary of the biobank’s inventory to researchers.
2.6 Technical procedure for linking clinical information to biobank specimen
Software encompasses all the relevant information for all deposited patients’ samples for systematic clinical research. We use medical record number (MRN) as primary key and biobank number as secondary key to connect two sections of database. Patients’ samples are stored at biobank with a unique allocated biobank number which consists of 10 digits and its format is like sample type (XX), serial number (XXXX), year (XX) and extraction type (XX). Sample types in our database includes AM (amniotic fluid), BL (blood), BO (bone marrow), CO (cord blood), CS (cervix swab), LN (lymph node), PC (product of conception), PL (paraffin embedded tissue with lymph node), PN (normal paraffin embedded tissue), PT (tumor paraffin embedded tissue), TM (tumor tissue) and TN (normal tissue). Depending on research needs the extraction is done from raw samples and the extraction type includes D (DNA), R (RNA), P (protein), etc. For a peripheral blood received with serial number 1252 in year 2014 and DNA is extracted from it then our assigned biobank number will be “BL-1252-14D”. This nomenclature system provides a clue about samples, however, patient confidentiality is protected as per current security regulations. To guarantee quality of samples, the biobank has ascertained a standard policies of quality management system. Samples are stored in liquid nitrogen, −80 °C, −20 °C, or 4 °C refrigerators depending on its type and requirement. Vials containing sample or extracted products are stored at assigned area so that all aliquots can be retrieved in the BS from the defined physical location.
2.7 Evaluation of software features and performance efficiency
We evaluated the efficiency of BS by performance test using (i) JMeter software, (ii) CRUD function and (ii) User feedback report. To verify the efficiency (speed, scalability and stability) of the BS, performance test was done by JMeter software by running different number of users (500, 100, 50, 10, 1), with 50 loop and 10 ramp up periods. Create, Read, Update, and Delete (CRUD) function using different SQL statement was also used for performance testing.
We also evaluated the BS by user’s feedback on 1 to 5 scale for its features and efficiency with specific questions like- is it easy to reach the website application? Does website application loads quickly? Are the fonts easy to read on various screen resolutions? Is the color used appropriate and comfortable to eye? Are the content logically separated and appear in appropriate way? Is the website application easy to use? Is navigation easy? Are all buttons (internal and external) valid and active? Is copy and paste feature allowed? Is autocomplete feature allowed? Is clickable icons work smoothly on single click? Is the website application free from server-side errors? Are you able to search, retrieve and edit data (samples description, patient information, project details etc.) easily? Do you get alert message for missing data? Is data printed in appropriate table format? Is help desk easy to contact for any issue in website application?
2.8 Transcriptomic analysis of breast cancer using specimen from Biosearch System
We retrieved breast cancer samples and healthy controls using software to conduct transcriptomic profiling using Affymetrix platform. Partek GS v6.7 (Partek, USA) was used for data analysis. Imported data was normalized with robust multiarray averaging process and ANOVA was applied to generated DEG gene list using p value <0.05 and FC >2. PCA was done for high dimensional visualization. Unsupervised hierarchical clustering was done for significant DEG as a similarity matrix.
2.9 Molecular pathway analysis
Pathways for DEGs were analyzed by IPA software (Ingenuity, USA) in triple negative breast cancer cases. IPA predicts molecular networks, canonical pathways associated with uploaded DEG with p-value and fold change cut-off (FC).
3 Results
In the past thirteen years (2007–2020) we collected significantly high number of specimens (n = 25,396) and their derivatives (n = 27,882) with number growing every year (Fig. 1, Table 1). Therefore, we established a disease-oriented biobank and developed Biosearch System (BS), an in-house database software to manage biobank and store disease associated genomic information.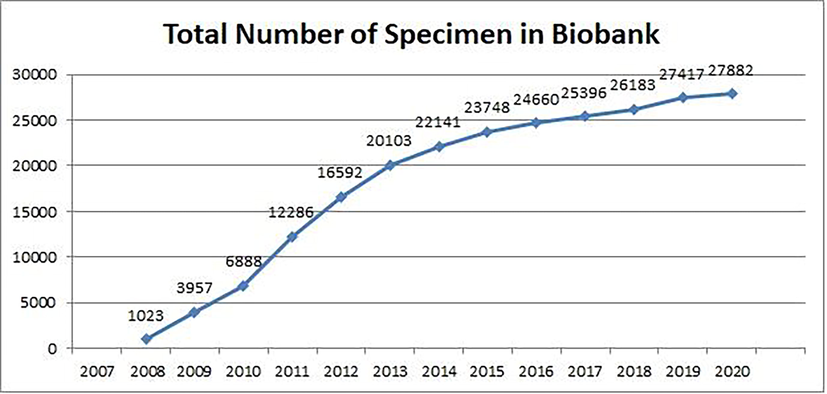
Collection and storage of specimen and disease associated information at CEGMR/KAU biobank. Pictorial graph showing progressive growth of specimen/derivatives from zero in 2007 to twenty-seven thousand till 2020.
Derivative Extraction Name
Abbreviation
Number
Percentage
DNA
D
22212
80.54611061
RNA
R
4276
14.83588925
MicroRNA
N
47
0.09367844
Protein
P
21
0.072861009
Plasma
M
348
1.207411005
Serum
S
288
0.999236694
Cell
C
243
0.843105961
miRNA of Plasma
PN
44
0.024287003
DNA of Plasma
PD
118
0.041634862
RNA of Plasma
PR
22
0.041634862
miRNA of Serum
SN
35
0.020817431
DNA of Serum
SD
125
0.03816529
RNA of Serum
SR
102
0.041634862
Total
27,882
100%
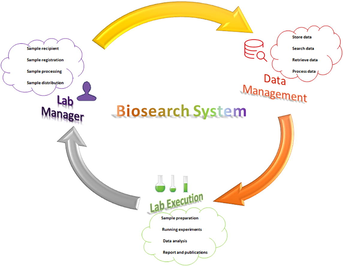
A prototype of an entity-relationship diagram was developed to depict numerical relationship among different entities like sample, storage condition, specimen request, approval system, project type, diagnosis, and disease type (Fig. 2). We established a communication between BS host server using SQL Server Management Studio and employed ASP.NET and JavaScript/Ajax toolkit for end users (Fig. 3). It was divided into 3 main parts: (i) data and specimen acquisition, (ii) data management and (iii) specimen distribution/request (Fig. 4). It classifies the user in four groups (biobank technicians, researchers, lab manager and director) and hierarchal permission is given accordingly. Administrator of BS has power to do all function, including addition of new users and granting permission according to hierarchy (Table 2). Final decision to accept or reject requested samples. Can search and retrieve biobank data. Checking and commenting on the requested samples Either add new abbreviation for extraction type, diagnostic type, hospital name, sample type etc or request the same to administrator. Edit user permission Add new project with its Id, period, type of diagnostic and title. Specify the required sample for project to provide accordingly Request sample Search and retrieve data based on sample id, sample type, extraction type, nationality, hospital name Allowed to add and edit sample information such as sample id, sample type, diagnostic type, add new vial, edit extraction Keep track on sample in and out Can determine the storage location (refrigerator number, refrigerator shelf, box container, coordinates) and actual status (extraction date, quantity, concentration, purity, availability) of samples requested by researchers. Can extract the excel file or csv file of all data using show logbook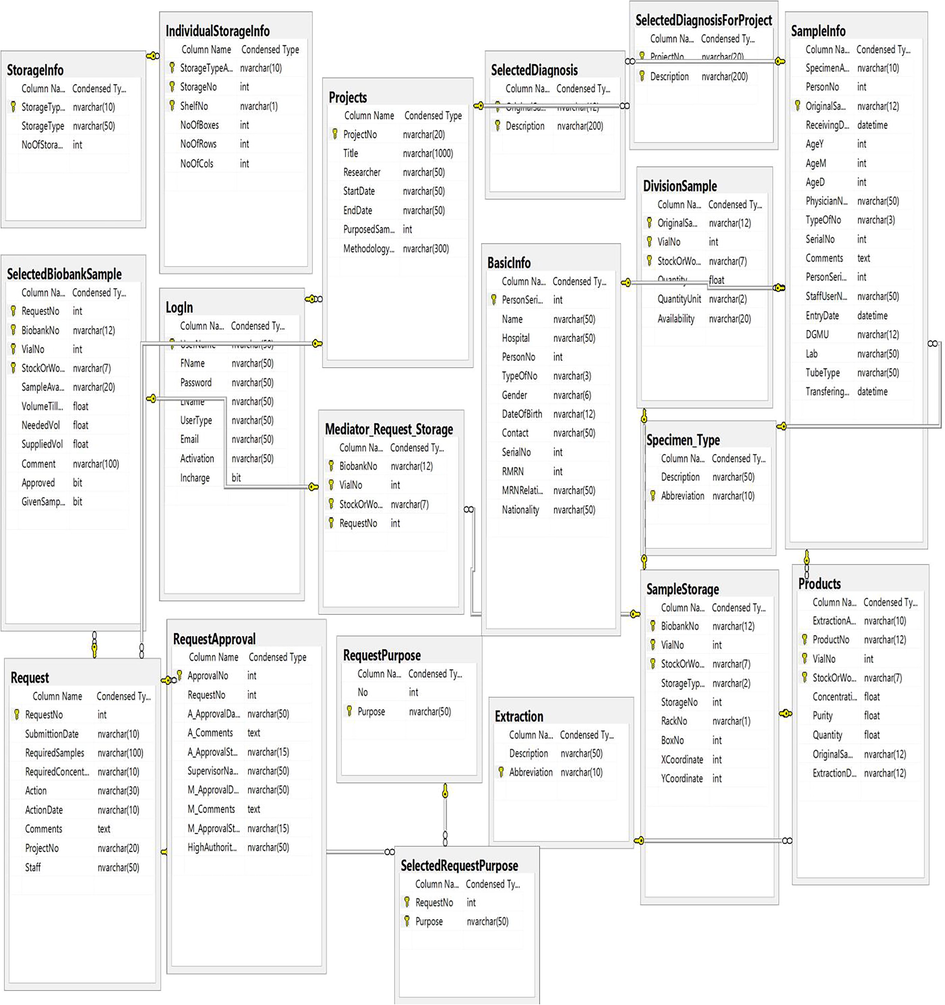
Entity-relationship diagram. An entity-relationship diagram for database structure used for Biosearch System depicting numerical relationship among different entities like sample, storage condition, specimen request, approval system, project type, diagnosis, and disease type.
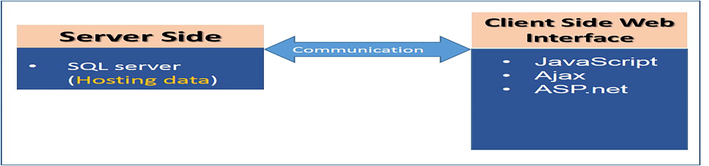
Black box of system structure. The relationship and the communication process between server side and client side; where Ajax and javascript sends client request to the SQL server side and ASP.net shows the processed response back at client end using the graphical user interface.
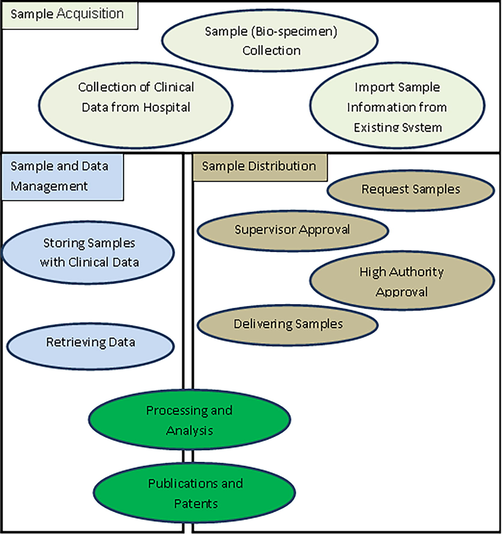
Biological sample life cycle. Sequential flow of each biobank specimen starting from collection, processing of sample, storage, maintenance, request, retrieval, distribution, and utilization.
User type
Job of user
Director
Lab manager
Researcher
Biobank team
BS is supported by a data warehouse and query tools interface that can be used efficiently to search and short list patients as per a particular criterion without disclosing their privacy. Based on evaluation form analysis we found that it is robust, secure, flexible, efficient, and user-friendly database. Robust, as it is platform independent, compatible with any version of window. Scripts are free from error, so no hanging of system. Secure, as in addition to appropriate permission strategy for different users and password protection, KAU network security strategy was also incorporated to transfer data safely, where each user has his own id and password to access specific computer and the password is stored in database in encrypted way. Each machine that has access to the BS must has its own firewall and anti-virus Trend Micro office scan agent provided by KAU IT deanship.
The system takes backup at regular interval (monthly backup) on KAU servers, so any loss of data from the system can be easily restored thereby saving the date from any disasters. Flexible, as new features/modules can easily be added by authorized users to customize according to laboratory and researcher requirements. Efficient, as it has been managing huge amount of clinical and experimental data of CEGMR competently since 2014. User-friendly, as simple icons, buttons, drop-down lists etc. are convenient for users and they can easily add or retrieve data using simple icons with available multiple options.
Our performance and utility of software evaluation with the help of real time feedback from users was excellent with satisfaction score was 4.7/5. Results of performance test of search, insert, update, and delete function are also satisfactory (Table 3). BS is efficient in term of number of transactions and number of users to maintain the quality of data and permits pliability in the workflow. Samples – The number of samples with the same label. Avg (ms) – The average elapsed time of a set of results in milli second. Min – The lowest elapsed time for the samples with the same label. Max – The longest elapsed time for the samples with the same label. Std. Dev. – the Standard Deviation of the sample elapsed time. Error % – Percent of requests with errors. Throughput – the Throughput is measured in requests per second/minute/hour. Received KB/sec – The throughput measured in Kilobytes per second. Sent KB/sec – The throughput measured in Kilobytes per second. Avg. Bytes – average size of the sample response in bytes.
Search function: performance test results
# User
Sample
Avg (ms)
Min
Max
Std. Dev.
Error %
Throughput
Received KB/s
Sent KB/s
Avg. Bytes
500 users
25,000
1489
99
10,950
972.25
0.0
1040.15
248
152
1185.01
100 users
5000
285
18
2190
189.85
0.0
210.03
48.29
29.99
235.01
50 users
2500
142
26
334
61.63
0.0
154.90
37.36
22.06
236.04
10 users
500
30
23
58
3.51
0.0
49.29
11.09
7.04
235.99
1 user
50
31
25
53
4.38
0.0
35.64
8.01
5.05
237.99
Insert Function: Performance Test Results
500 users
2500
1492
3
11,489
1075
0.0
1019
230.12
263.40
1234.01
100 users
5000
287
3
2299
213.92
0.0
205.04
48.04
57.93
236.99
50 users
2500
130
13
265
59.03
0.0
155.01
34.97
43.86
239.48
10 users
500
11
11
23
1.20
0.0
52.02
11.99
14.56
239.99
1 user
50
11
11
19
1.32
0.0
79.93
18.40
22.81
237.92
Update Function: Performance Test Results
500 users
25,000
1747
5
14,855
2025
0.0
856.20
278.03
232.8
1678
100 users
5000
351
4
2972
405.01
0.0
171.78
54.97
45.99
337.06
50 users
2500
422
21
901
161.01
0.0
85.56
27.37
22.05
333.82
10 users
500
21
20
37
2.32
0.0
50.40
16.06
14.06
334.09
1 user
50
21
21
60
5.98
0.0
45.44
14.60
12.24
334.12
Delete Function: Performance Test Results
500 users
25,000
1425
4
11,870
1095.02
0.0
1034.07
249.02
233.50
1200.01
100 users
5000
285
4
2380
221.01
0.0
207.01
49.07
47.05
241.08
50 users
2500
118
12
300
59.07
0.0
159.05
36.12
36.02
241.02
10 users
500
13
12
30
1.35
0.0
52.05
13.08
11.62
239.92
1 user
50
13
12
21
2.042
0.0
77.92
19.03
17.63
241.01
BS defers from other LIMS based on its customization approach for CEGMR laboratory and is flexible enough to be adjusted for any changes in future to suit the needs of projects and researcher such as new equipment, procedure, and/or software to complement or improve the workflow without changing the core code. For example: essential customization needed for research purpose is to link SMARTGENE software, a system for diagnostic lab, with BS system and exchange data with researchers consent. It can be used for any laboratory with minor modifications (QC, R&D, and analytical service) whereas most of the commercial software are a bit rigid. Its flexibility extends to data quality by achieving through the computerization (calculations, statistics) and automation of processes, and procedures to minimizing manual lab tasks such as data entry. For instance, the system makes sure entering data occurs in a standardized way using drop down pre-defined list for most processes. Presently BS is not available as open source because of institutional policy but we encourage the interested researchers to request for codes on individual basis.
BS is efficient to maintain the quality of data and permits pliability in the workflow and it had synchronized the biobank system of CEGMR/KAU. Current state of the CEGMR biobank is as follows: (1) Infrastructures: biobank is well equipped with liquid nitrogen cylinder (−196 °C), deep freezer (−80 °C and −20 °C), refrigerator and UPS system. (2) Personnel: presently fourteen dedicated staff are working hard for smooth running of biobank unit, and (3) Patient’s samples management system is fully functional and regularly handling the dispatch, collection, processing, and storage of thousands of specimens. The BS can be accessed by authorized person using the login username only (Fig. 5). Our clinical database contains the following clinicopathological parameters: CEGMR code for patient, receiving date, hospital MRN number, name, date of birth, age, sex, nationality, disease, date of diagnosis, status, filing date, histology, sites, grade, size, lymph node status, invasion, margin status, immunohistochemical data, family history, medication, follow up etc. Similarly, biobank management system contains specimen and their derivative related information: type of specimen, receiving date, extraction date, storage of specimen and its derivatives, quality and quantity record, handling request of researchers, distribution, and maintenance of specimen.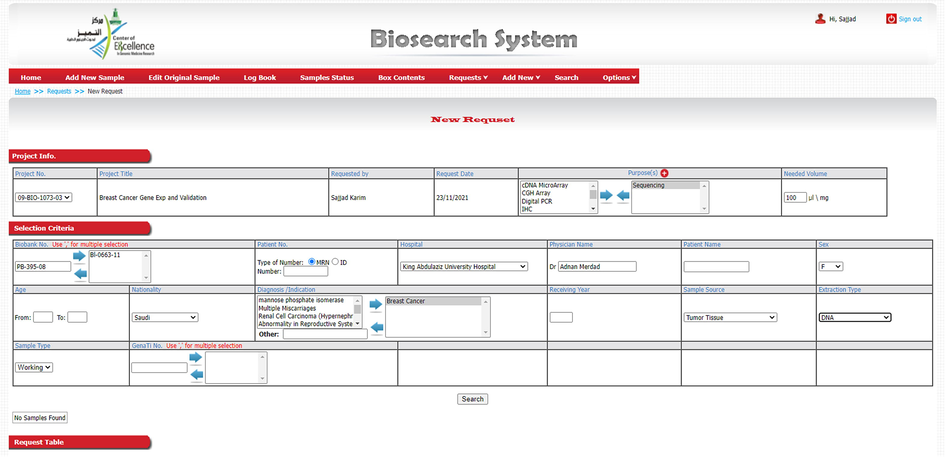
A glimpse of Biosearch System. Screenshot of the new sample addition page of BS allowing authorized researchers to access samples and their disease associated genomic and clinical information.
3.1 Identification of DEGs for breast cancer
We conducted genome wide expression study to understand the molecular phenomenon leading to triple negative breast cancer and found 1181 differentially expressed genes with P-value <0.05 and FC 2. The upregulated genes for TNBC were IFI6, LEF1, CCR8, FANCI, TRIM59, CASC5, and PLXNA3 while down regulated genes were ADH1B, LYVE1, ADH1C, ADH1B, FIGF, ADIPOQ, PLIN1, and LYVE1. Hierarchical clustering of top 135 DEGs showing distinct pattern for genes in triple negative BC and control samples (Fig. 6).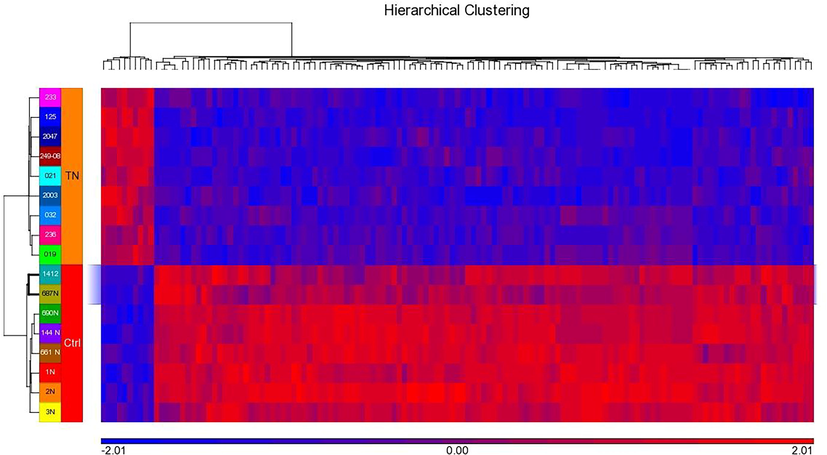
Hierarchical clustering of DEGs. Unsupervised clustering showing expression pattern of genes in triple negative BC. Blue and red colors indicating down and up-regulated genes. Row and Column represents samples and DEGs respectively.
3.2 Pathways associated with triple negative breast cancer (TNBC)
Molecular pathway analysis revealed more than hundred canonical pathways where most activated pathways were (i) interferon signaling pathways (z-score = 2.646) with participating genes IFI6, FNG, IRF1, IRF9, MED14, MX1, PSMB8, STAT1, STAT2, and (ii) kinetochore metaphase signaling pathway (z-score = 2.138) with following associated genes; BUB1, BUB1B, CDK1, CENPL, KIF2C, KNL1, KNTC1, NUF2, PLK1, PPP1CA, PPP1R14B, PTTG1, RAD21, REC8 and TTK (Fig. 7).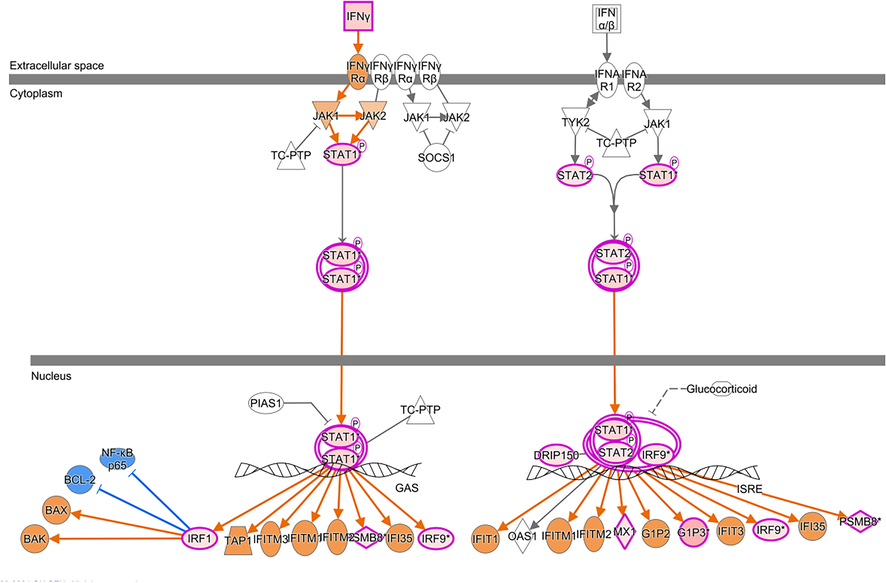
Interferon Signaling. Overall, the pathway is predicted to be upregulated and participating differentially expressed genes are IFI6, FNG, IRF1, IRF9, MED14, MX1, PSMB8, STAT1 and STAT2.
4 Discussion
Today’s biobanks are much more than just sample repository. They store a huge amount of clinical and experimental data related to specimen (Calleros et al., 2012). However, efficient data management is a bottleneck in the genomic medicine research process. It has been observed on many occasions that researchers face difficulties in data collection, maintenance, follow up studies and sometime publish the work with missing data (Greely, 2007; Kauffmann and Cambon-Thomsen, 2008; Minamikumo, 2012). Translational genomic research is based on a secure database and biobank system. A well-designed software provides the possibility of finding significant associations among stored information and facilitates diagnostics and therapeutics research (Lemmon et al., 2011; Zerhouni, 2005). BS is used to collect, store, distribute and maintain the specimen data, as well as its relevant clinical and experimental information to support ongoing research that can lead to discovery of novel cancer biomarkers and therapeutics targets (Karim et al., 2016; Karim et al., 2019; Merdad et al., 2014; Merdad et al., 2015; Mirza et al., 2015; Mirza et al., 2014; Rasool et al., 2021; Rasool et al., 2020; Schulten et al., 2016; Subhi et al., 2020; Sultan et al., 2021).
We used SQL server management studio, Visual Basic, .NET and Ajax control kit to develop interactive database, and ASP.NET for better code management, clean code structure and fast web applications. The first prototype of BS was ready in 2010 and after feasibility test and successful trial run, we released the present final version with minor modifications. BS is web-based tool accessible from off-and-on-campus. High level of protection allows only authorized researchers to access database. The security concerns like threat of hacking, virus attack etc were taken care of without any compromise (Bjugn and Hansen, 2013; Mintzer et al., 2013; Rogers et al., 2011; Simeon-Dubach et al., 2013). Bioethical safety was another big concern and needed proper care in establishing biobank and developing database ((ISBER) 2012; Bredenoord et al., 2011; Hansson 2009; McGuire et al., 2008; Rotimi and Marshall, 2010).
BS makes the job easier for everybody involved in research: (i) clinician and pathologist can provide clinicopathological information of each provided specimen from hospitals/clinics through online database system, (ii) biobank staff can store and maintain the specimen and extracted derivatives easily, and (iii) researchers can extract clinical information and request specific specimen/derivative as per requirement of on going project (Kang et al., 2013). Presently our data set is small, and we are collecting selected cases only, so the frequency of disease should not be used to represent society at large. However, the frequency of different type of cancer in our biobank is very much like national cancer registry. In future, this can pave the way for expanding this database in association with Saudi cancer registry or any other national level databases.
BS tool was tested by CEGMR staffs, and was found satisfactory. The users were comfortable to organize all sample related information and found it user friendly with simple icons, buttons, drop-down list etc. To ensure the safety of data, King Abdulaziz University network security was utilized and only user with approved permission and approved supporting with anti-virus were allowed to access. Periodic system backup strategy also ensured avoidance of any data loss due to unseen disaster. It is customized according to actual activities and workflow in CEGMR lab and is flexible enough to adopt new modules to add more features in future.
5 Conclusions
Biosearch system is a user’s friendly software to manage biobank specimen with clinical information to facilitate genomic medicine research leading to discovery of disease biomarkers and therapeutic targets. It is adoptable to new features and modules to add barcoding system, quality control system and reagent purchasing system in future.
CRediT authorship contribution statement
Sajjad Karim: Conceptualization, Data curation, Fund acquisition, Investigation, Project Administration, Software, Supervision, Writing - original draft. Adnan Merdad: Conceptualization, Fund acquisition, Validation, Writing - review & editing. Saddig Jastaniah: Conceptualization, Fund acquisition, Validation, Writing - review & editing. Sudhir Kumar: Conceptualization, Fund acquisition, Supervision. Adel Abuzenadah: Conceptualization, Project Administration, Resources, Validation, Writing - review & editing. Mohammed Al-Qahtani: Conceptualization, Project Administration, Resources, Validation, Writing - review & editing. Mona Alkharaz: Data curation, Formal analysis, Investigation, Methodology. Zeenat Mirza: Data curation, Investigation, Methodology, Software, Visualization, Writing - original draft. Mahmood Rasool: Data curation, Investigation, Validation, Writing - review & editing. Hend Noureldin: Formal analysis. Heba Abusamra: Formal analysis. Nofe Alganmi: Methodology, Resources, Software, Supervision, Visualization, Writing - review & editing.
Acknowledgement
We would like to thank Biobank and IT unit staffs of Center of Excellence in Genomic Medicine Research, AZIZ Supercomputing facilities at High Performance Computing Center and Deanship of Scientific Research, King Abdulaziz University for their help and technical support.
Disclosure of funding
This study was funded by Deanship of Scientific Research, King Abdulaziz University (2-117-1434-HiCi).
Ethics approval and consent to participate
Ethical committee approved this study (Reference Number: 08-CEGMR-02-ETH) of CEGMR, KAU.
Availability of data and materials
Datasets (.CEL file) submitted to NCBI’s (GEO) (accession number GSE36295.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 2012 best practices for repositories collection, storage, retrieval, and distribution of biological materials for research international society for biological and environmental repositories. Biopreserv. Biobank. 2012;10:79-161.
- [Google Scholar]
- Biobanking in a constantly developing medical world. Sci. World J.. 2013;2013:343275
- [Google Scholar]
- What are the biggest challenges and opportunities for biorepositories in the next three to five years? Biopreserv. Biobank. 2010;8:81-88.
- [Google Scholar]
- Disclosure of individual genetic data to research participants: the debate reconsidered. Trends Genet.. 2011;27(2):41-47.
- [Google Scholar]
- Start-up of a clinical sample processing, storage and management platform: organisation and development of the REDinREN Biobank. Nefrologia. 2012;32:28-34.
- [Google Scholar]
- Next-generation sequencing: a new revolution in molecular diagnostics? Clin. Chem.. 2009;55:2088-2092.
- [Google Scholar]
- Fehniger, Thomas E., György A. Marko-Varga, 2011. Clinical proteomics today, J. Proteome Res., 10: 3-3.
- Field guide to next-generation DNA sequencers. Mol. Ecol. Resour.. 2011;11:759-769.
- [Google Scholar]
- The uneasy ethical and legal underpinnings of large-scale genomic biobanks. Annu. Rev. Genomics Hum. Genet.. 2007;8:343-364.
- [Google Scholar]
- Biobanking: the foundation of personalized medicine. Curr. Opin. Oncol.. 2011;23:112-119.
- [Google Scholar]
- WIST: toolkit for rapid, customized LIMS development. Bioinformatics. 2011;27(3):437-438.
- [Google Scholar]
- Current status, challenges, policies, and bioethics of biobanks. Genomics Inform.. 2013;11:211-217.
- [Google Scholar]
- Cyclin D1 as a therapeutic target of renal cell carcinoma- a combined transcriptomics, tissue microarray and molecular docking study from the Kingdom of Saudi Arabia. BMC Cancer. 2016;16:741.
- [Google Scholar]
- Molecular analysis of V617F mutation in Janus kinase 2 gene of breast cancer patients. Saudi J. Biol. Sci.. 2019;26:1123-1128.
- [Google Scholar]
- Tracing biological collections: between books and clinical trials. JAMA. 2008;299:2316-2318.
- [Google Scholar]
- Challenges in small screening laboratories: implementing an on-demand laboratory information management system. Comb. Chem. High Throughput Screen.. 2011;14:742-748.
- [Google Scholar]
- Research ethics and the challenge of whole-genome sequencing. Nat. Rev. Genet.. 2008;9:152-156.
- [Google Scholar]
- SIGLa: an adaptable LIMS for multiple laboratories. BMC Genomics. 2010;11(Suppl. 5):S8.
- [Google Scholar]
- Expression of matrix metalloproteinases (MMPs) in primary human breast cancer: MMP-9 as a potential biomarker for cancer invasion and metastasis. Anticancer Res.. 2014;34:1355-1366.
- [Google Scholar]
- Transcriptomics profiling study of breast cancer from Kingdom of Saudi Arabia revealed altered expression of Adiponectin and Fatty Acid Binding Protein 4: Is lipid metabolism associated with breast cancer? BMC Genomics. 2015;16(Suppl. 1):S11.
- [Google Scholar]
- Preparedness for a natural disaster: how Coriell planned for hurricane Sandy. Biopreserv. Biobank. 2013;11:216-220.
- [Google Scholar]
- Molecular interaction of a kinase inhibitor midostaurin with anticancer drug targets, S100A8 and EGFR: transcriptional profiling and molecular docking study for kidney cancer therapeutics. PLoS ONE. 2015;10(3):e0119765.
- [Google Scholar]
- Impact of S100A8 expression on kidney cancer progression and molecular docking studies for kidney cancer therapeutics. Anticancer Res.. 2014;34:1873-1884.
- [Google Scholar]
- HalX: an open-source LIMS (Laboratory Information Management System) for small- to large-scale laboratories. Acta Crystallogr. D Biol. Crystallogr.. 2005;61:671-678.
- [Google Scholar]
- Array comparative genomic hybridization based identification of key genetic alterations at 2p21-p16.3 (MSH2, MSH6, EPCAM), 3p23-p14.2 (MLH1), 7p22.1 (PMS2) and 1p34.1-p33 (MUTYH) regions in hereditary non polyposis colorectal cancer (Lynch syndrome) in the Kingdom of Saudi Arabia. Saudi J. Biol. Sci.. 2020;27(1):157-162.
- [Google Scholar]
- Mutational spectrum of BRAF gene in colorectal cancer patients in Saudi Arabia. Saudi J. Biol. Sci.. 2021;28(10):5906-5912.
- [Google Scholar]
- Biobankonomics: a taxonomy for evaluating the economic benefits of standardized centralized human biobanking for translational research. J. Natl. Cancer Inst. Monogr.. 2011;2011:32-38.
- [Google Scholar]
- Tailoring the process of informed consent in genetic and genomic research. Genome Med.. 2010;2:20.
- [Google Scholar]
- Microarray expression profiling identifies genes, including cytokines, and biofunctions, as diapedesis, associated with a brain metastasis from a papillary thyroid carcinoma. Am. J. Cancer Res.. 2016;6:2140-2161.
- [Google Scholar]
- Disaster and recovery: the importance of risk assessment and contingency planning for biobanks. Biopreserv. Biobank. 2013;11:133-134.
- [Google Scholar]
- Genetic relationship between Hashimoto‘s thyroiditis and papillary thyroid carcinoma with coexisting Hashimoto‘s thyroiditis. PLoS ONE. 2020;15:e0234566
- [Google Scholar]
- Alterations of transcriptome expression, cell cycle, and mitochondrial superoxide reveal foetal endothelial dysfunction in Saudi women with gestational diabetes mellitus. Endocr. J.. 2021;68(9):1067-1079.
- [Google Scholar]
- A Laboratory Information Management System (LIMS) for a high throughput genetic platform aimed at candidate gene mutation screening. Bioinformatics. 2007;23:2504-2506.
- [Google Scholar]
- US biomedical research: basic, translational, and clinical sciences. JAMA. 2005;294:1352-1358.
- [Google Scholar]







