Translate this page into:
Detection of some haemorrhagic fever viruses in wild shrews collected from different habitats in Saudi Arabia: First record in the Middle East
⁎Corresponding author. faleanizy@ksu.edu.sa (Fadilah Sfouq Aleanizy)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Shrews (family Soricidae) are tiny mole-shaped mammals belong to the order Eulipotyphla. The main objective of this study is to screen wild specimens of shrews for specific RNA viruses cause hemorrhagic fevers. Wild specimens of shrews were collected from rural areas in Kingdom of Saudi Arabia.
Collection of shrews were carried out from the traps then identified using the classical morphological keys. Specimens were dissected then extraction of single stranded RNA of shrews was performed from internal organs including lungs, livers, kidneys, and stomach using QiagenRNeasy Mini Kit. RT-PCR was utilized for screening of Crimean- Congo hemorrhagic fever virus (CCHFV), Rift Valley Fever Virus (RVFV), and Chikengunya virus (CHIKV), and Sindbis virus (SINV). The results revealed SINV, CHIKV, and CCHFV were all found in the internal viscera of shrews in four different groups. This demonstrated that the viruses were propagating and spreading throughout the tissues of the shrews.
Conclusions
Depending to our knowledge results of this study constitute first record in Kingdom Saudi Arabia and significantly highlight some of the neglected wild reservoirs of arboviruses; therefore future studies should focus on evaluating other hosts, including bats.
Keywords
Shrews
Soricidae
Haemorrhagic Fever
Viruses in Wild Shrews
1 Introduction
Shrews are tiny, mole-shaped, closely morphologically similar to mice, insectivorous mammals in the family Soricidae, order Soricomorpha. There are at least 385 species in this family, with a nearly global range (Sasaki, et al., 2014; Sasaki et al., 2015). Shrews (Tupaiabelangerichinensis) have demonstrated success as an experimental animal in many cases where a tiny omnivorous non-rodent species is required and can be investigated in many fields of preliminary clinical research such as virology (Shang, 2015; Gu et al., 2020; Zhou et al., 2015; Yang, 2015; Li et al., 2016). There is no available data to prove the distribution of shrews in Middle East.
Shrews may transmit many viruses like Borna Disease Virus (Schulze et al., 2020; Malbon et al., 2021; Hilbe et al., 2006), Bufavirus (Sasaki et al., 2015), and Hantavirus (Schmidt et al., 2016; McElhinney et al., 2016; Vial et al., 2016). Also, some published researches demonstrated that shrew could harborsome hepatitis viruses (Wang and Meng, 2021; Sun et al., 2013).
Many studies conducted in Switzerland (Tsoleridis et al., 2016), Zambia (Sasaki et al., 2014; Sasaki et al., 2015) and China (Sasaki et al., 2014; Gong et al., 2015) revealed that laboratory diagnosis of viruses relies on histological techniques, immunohistologic (IHC) testing, reverse transcription– polymerase chain reaction (RT-PCR), and recently TaqMan real-time RT-PCR (Kabuga, 2021). The polymerase chain reaction (PCR) assay amplifies DNA; PCR has totally revolutionized the detection of RNA and DNA viruses. PCR is a swift, specific, and sensitive technique so it is valuable for results confirmation. PCR has also the credit to detect molecular parts of various infectious agents (Kabuga, 2021; Kendall and Riley, 2000; Kumar et al., 2021).
Many papers revealed the distribution of four hemorrhagic fever viruses that included in this study in KSA but no available data regarding hanta virus distributed in KSA.
It is recommended to use PCR as the best method for the detection of viral DNA or RNA present at very low levels in biological samples, and it allows the molecular diagnosis and study of most acute and chronic viral infections (Kumar et al., 2021).
This information gave the justification to do this research which targeted detecting Shrews-transmitted arboviruses using real- time PCR technique. This is the first study that we are aware of.in Kingdom of Saudi Arabia that examined wild shrews’viscera for the presence of arboviruses RNAs using Real- Time PCR technique. The study also identified the natural habitats of shrews in Saudi Arabia.
2 Materials and methods
2.1 Study setting
Riyadh is the metropolis of Saudi Arabia and Riyadh region. It is localized in the center of the Arabian Peninsula. The climate of the city is hot and temperatures during the summer months are extremely high may approaches over 43 °C. The overall climate is arid and it gets very little rain, especially in the summer, although it gets a lot of rain in March and April. Arabian camels or dromedaries (the most common domestic animals), sheep, goats, and donkeys are examples of domesticated animals.
2.2 Study population
Internal visceral organs (Lungs, liver, kidney, and stomach) obtained from 63 wild shrews collected from rural area 55 km west to Riyadh.
2.3 Study design
This is a baseline study conducted for the first time in the Middle-East region for screening of certain viruses in shrews. Baseline surveys conducted in Riyadh (Capital Saudi Arabia) between January - March 2016. It will follow more studies in the near future to get a better understanding of the correlations of shrews and arboviruses transmission as well the kinetics of the virus replications.
2.4 Sample size
Number of shrews included in this study was determined according to the availability of shrews in the study site. Total of 63 shrews were trapped during the survey period. Each shrews specimen was divided into 24 pools (4 viscera each of which was examined for 6 viruses). Therefore the overall sample size was calculated as follows: 63 shrews × 24 pools = 1521 sample.
2.5 Ethical endorsement
This article does not include human participants. Ethical clearance gained from the IRB board in Princess Nourah Bint Abdurrahman University.
2.6 Samples processing
Shrews were captured from three traditional wet paces in the rural area around 55 km west and south districts of to Riyadh city using food-baited traps. The shrews were kept in wire cages then transferred to a lab, fed on small fish and kept for additional examination. Collected specimens were transported to a laboratory for research, Faculty of Science, Princess NourahBint Abdurrahman University in their traps. Specimens were then anthesized to death using formalin and shrews were morphologically sorted out according to Wilson and Reeder; Sonenshine and Roe (Rizzoli, 2014; Reeder, 2011).
Shrews were then dissected using dissection tools. Four groups were prepared from internal viscera of shrews as follows: lungs, livers, kidneys, and stomach. Homologous groups represented each organ were pooled separately. Pools were preserved in normal saline in labeled cryovials in – 80 °C freezer for 10 days. Frozen organs were then dissolved then triturated separately using tissue Dissociator instrument (Miltenyi biotech, Germany) in certain volumes of Virus Transport Medium (VTM). Specific programs in the instrument were selected for each sample (Lungs, liver, kidney, and stomach), respectively. No ethical approval from an animal ethics council was necessary because the study did not utilize laboratory animals and was conducted as part of a natural disease inquiry.
Certain volumes of VTM were added to each group based on the pool size as below: 10–50 specimens (2.0 ml), 100 specimens (3.0 ml), 150 specimens (4.0 ml), 200 specimens (5.0 ml), and 300 specimens (8.0 ml). The Leibovitz medium supplemented with 10% fetal calf serum, antibiotics, and fungicide. The supernatant of each sample pool has been transferred into labeled cyovial and preserved in – 80 °C freezer for 4 days till RNA extraction.
2.7 Primers (Synthetic genes) preparation, RNA extraction and cDNA synthesis
Manufacturers' instructions were followed for preparation of synthetic genes (Table 1). Total RNA was extracted from the four groups of shrews organs; lung, liver, kidney and stomach serum samples. Total RNA extraction was performed, as directed by the manufacturer with QiagenRNeasy Mini Kit and QiagenRNAeasy Columns (Qiagen, Hilden, Germany) and eluted in 40 µl of nuclease-free water. It was utilized immediately or stored at −70° C. The preferred QPCR method was carried out in 20 µl reaction mixtures containing 1 µl RNA template, 25 µM of each primer, 10 µl of KAPA master mixture (KAPA SYBR® FAST qPCR Kits) (Mohamed et al., 2013). The samples were firstly incubated at 42 °C for 30 min for RT step: 95 °C for 3 min, as a denaturation step. Around 40 cycles were conducted; each included a Denaturation step at 95 °C for 3 s, then temperature minimized to 58 °C for annealing of RVFV & CHIKV, CCHFV, and to 56 °C for SINBV. For each reaction, negative and positive controls were included to verify the specificity of the chosen primer pairs.
Organs
viruses
No
Stomach
Kidney
Liver
Lung
Pool 4
Pool 3
Pool 2
Pool 1
RVFV
1
Pool 8
Pool 7
Pool 6
Pool 5
CHIK E1
2
Pool 12
Pool 11
Pool 10
Pool 9
CCHFV
3
Pool 16
Pool 15
Pool 14
Pool 13
INKV
4
Pool 20
Pool 19
Pool 18
Pool 17
SINV
5
Pool 24
Pool 23
Pool 22
Pool 21
Pan Hanta
6
2.8 RT- PCR procedures
Primers were synthesized aiming for i. primers having an nearly equivalent Tm, ii.minimising primer-primer and primer-primer interactions, while iii.still operating within a conserved stretch. All oligonucleotides were checked for primer-primer annealing as well as self-annealing loops, utilizing a cutoff of –5 kCal/Mol for the Gibbs free energy [ΔG] of oligonucleotide interactions. Their predicted Tm and oligonucleotide interactions were checked at. The primer-primer interactions were studied using OligoAnalyzer 3.0 (http://207.32.43.70/biotools/oligocalc/oligocalc.asp,&http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/). Primers were obtained from DNA Technology A/S (Risskov, Denmark) (Kabuga, 2021; Mohamed et al., 2013; Rasmuson et al., 2016; Nahla Mohamed, 2015). QRT-PCR procedures were evaluated at two different locations using different real-time PCR instruments; ABI Prism 7900HT Sequence Detection System 2.0; Applied Biosystems and Mastercycler® eprealplex, real-time thermal cycler (Eppendorf AG, Germany). Two frequently used in-house PCR kits (Power SYBR Green One step RT-PCR Master Mix (KAPA Biosystems, Boston, MA), were tested along with primers (Table 2) purchased from DNA Technology(Denmark). For evaluation, isolation of the synthetic genes plasmid using standard procedures, and used at selected dilutions to be a templates for the above primer pairs. The PCR kits were used under the same basic circumstances as the manufacturer suggested. Temperature gradient PCRs were used to investigate the annealing temperatures individually, starting at 52 °C and ending at 62 °C, with 2 °C increments.
GC- Content
Vol for 100 pmol/µl
Sequence (5″->3″)
Oligo name
No
50%
187
AAGGCAAAGCAACTGTGGAG (20)
RVFV 233F
1
55%
207
TGAGTGGCTTCCTGTCACTG (20)
RVFV 388R
2
36.8%
200
CATGCAAAACAGAATTTGC (19)
CHIK E1 FW
3
44.4%
187
TAGGCAGTTACAGTGATG (18)
CHIK E1 RV
4
42.9%
212
AGGTTTCCGTGTCAATGCAAA (21)
CCHFV (S-gene) fw
5
50%
231
TTGACAAACTCCCTGCACGAGT(22)
CCHFV(S-gene) rv
6
42.3%
365
ACAAGATCTTCTTTGAGTAGTCCAGC(26)
SINV FW
7
52.4%
201
SINF RV
SINF RV
8
55%
164
GAGAGTGGCAGGTGGAGATT(20)
INKV NS FW
9
50%
205
AAAGCCGGTGGATGGTAAGA(20)
INKV NS RV
10
45.2%
280
TGCWGATGZACRAAATGGTC(21)
PANHANTA-F2
11
43.2%
199
GCATCATCWGARTGATGZGCAA(22)
PANHANTA-R2
12
3 Results
There were 63 commensal wild shrews collected, with 44 Rattus norvegicus (27 males and 17 females), 11 Rattus rattus (5 males and 6 females), and 8 Suncus murinus (5 males and 3 females). Among the 63 shrews collected, 58 were discovered to host endo-parasites, revealing an overall prevalence of 94.1%. There were no significant differences between the various shrew species or sexes (p > 0.05).
3.1 Screening of CCHV in the visceral organs of shrews
CCHFV has been detected inside the four pools represented lungs, livers, kidneys, and stomach of shrews (Fig. 1). CT value for the positive samples was approximately 30. CT value for the negative control (RNA free water) was zero.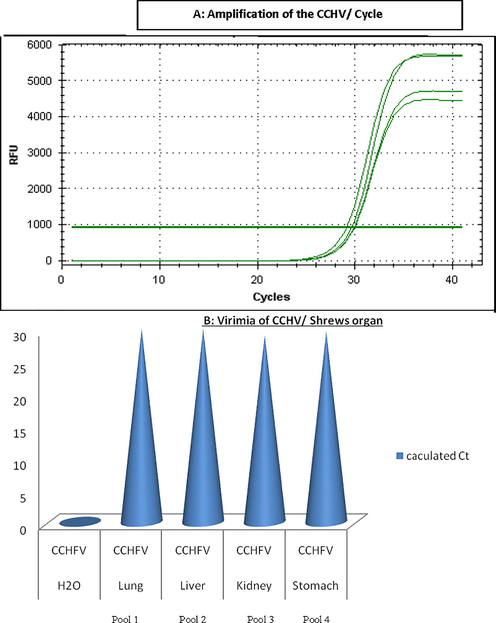
Calculated Ct value for CCHV- positive shrews organs.
3.2 Screening of RVFV in the visceral organs of shrews
Fig. 2 showed that RVFV has been found inside the four pools represented lungs, livers, kidneys, and stomach of shrews. CT value for the stomach pool was the highest 25. CT value for the negative control (RNA free water) was zero.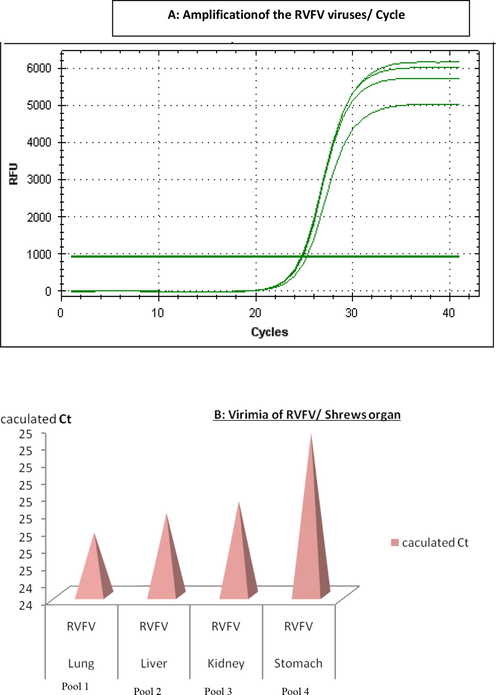
Calculated Ct value for RVFV- positive shrews organs.
3.3 Screening of SINDV in the visceral organs of shrews
As shown in Fig. 3, SINDV has been detected inside the four pools represented lungs, livers, kidneys, and stomach of shrews. CT value for the positive pools was approximately 25. CT value for the negative control (RNA free water) was zero.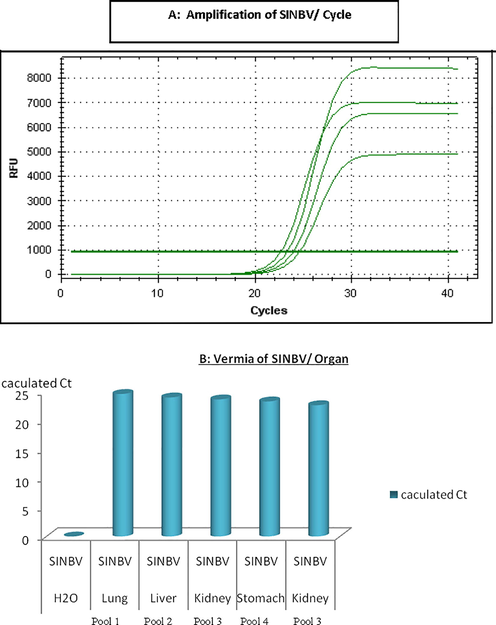
Calculated Ct value for SINBV- positive shrews’ organs.
3.4 Screening of CHIKV in the visceral organs of shrews
CHIKV has been detected inside the four pools represented lungs, livers, kidneys, and stomach of shrews. CT value for the positive pools 48 (Fig. 4). CT value of the negative control (RNA free water) was zero (see Figs. 5 and 6).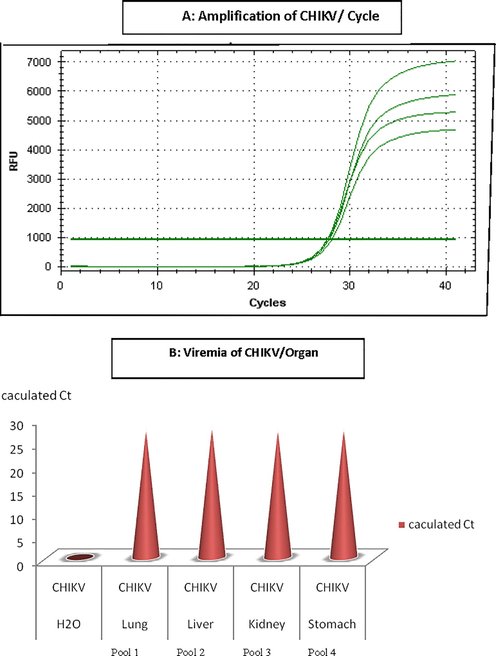
Calculated Ct value for CHIKV- positive shrews’ organs.

Anthesized shrews ready for dissection.
Dissection of shrews to extract the internal organs.
3.5 Screening of ALKV in the visceral organs of shrews
RVFV and ALKV were not detected inside the four pools represented lungs, livers, kidneys, and stomach of shrews. CT value for these negative pools was zero. CT value of the negative control (RNA free water) was zero.
4 Discussion
The shrew intestinal virome had a significant percentage of insect viruses. In this research, presence of nucleic acids of certain viruses has been examined inside organs of wild shrews. Shrews were collected from rural areas 55 Km west to Riyadh, Capital of Saudi Arabia. Results of this study may give a clue to the diets of shrews as a branch of the insectivorous bats (Schmidt et al., 2016; McElhinney et al., 2016; Ge, et al., 2012).
Hemorrhagic fever viruses are important pathogens affecting both humans and animals. In humans, arboviral infections are correlated to respiratory flu- like disease, symptoms range from moderate upper respiratory tract symptoms to more severe life-threatening symptoms, associated with hemorrhages. In humans and some animals they may associate with encephalitis and other neurological disorders (Vial et al., 2016; Tsoleridis et al., 2016; Rasmuson et al., 2016; Limongi, et al., 2016; Adouchief, et al., 2016).
Real- Time PCR has been used for screening the viruses inside shrew’s organs because it is efficient as a confirmation test. and it is a fast, specific, and sensitive diagnostic technique (Kabuga, 2021; Mohamed et al., 2013; Joshi and Deshpande, 2011).
Although it remains unclear whether shrews can be incriminated as actual transmitters of the novel viruses described in this study, the detection of CCHFV, RVFV, CHIKV, and SINDV using RT- PCR from some tissues upholds the assumption of replication in the organs of shrews and coincides with previous studies conducted in Europe (Hilbe et al., 2006; Vial et al., 2016; Tsoleridis et al., 2016; McElroy et al., 2009; Lo Presti et al., 2012; Wang, 2012).
In the present study shrews organs tissues were used as a confirmation and proof markers to documents the shrews could be a carrier reservoir and the majority of the tissues examined showed no signs of pathological lesions. These findings corroborate the theory that shrews can be infected with these viruses for a long time and thus serve as reservoirs and vectors, as suggested in a previously reported study (Hilbe et al., 2006; Lin et al., 2014). Furthermore, these animals are useful model for studying hosts, flow the kinetics of those viruses and pathogen virulence factors that allow persistent viral co-existence in apparently healthy carriers. The influence of other biases including the small size of the population, limited geography, and the host ranges of the identified viruses require to be considered.
According to our knowledge findings of this study constitute the first record in the Middle East. Despite this, the probability that the identified viruses came from common prey can’t be excluded. Xeno-diagnosis of these viruses in shrews as well as screening of the viruses in other hosts; particularly bats are highly suggested in the forthcoming researches. Phylogenic studies to study the link between the identified viruses and human viruses were not carried out, despite the fact that they are necessary. More epidemiological research is required to obtain a better understanding of the distribution and host specificity of these viruses.
5 Conclusions
The findings of this study are the first evidence of SINV, CHIKV, and CCHFV present in Saudi shrews. Detection of the five viruses in the internal organs viscera of shrews is achieved via Real Time PCR which indicated successful viral maintenance in the reservoir population and even fatal transmission to susceptible accidental hosts such as other animals and humans. Further investigations to determine whether shrews are widely recommended as an alternate mammalian model for studying viral pathogenesis.
Acknowledgement:
This research was funded by the Deanship of Scientific Research at Princess Nourah bint Abdulrahman University through the Fast-track Research Funding Program.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Sasaki, M., et al., 2014. Molecular epidemiology of paramyxoviruses in Zambian wild rodents and shrews. J. Gen. Virol. 95(Pt 2), 325-330.
- Distinct lineages of bufavirus in wild shrews and nonhuman primates. Emerg. Infect. Dis.. 2015;21(7):1230-1233.
- [Google Scholar]
- The formation and extinction of fear memory in tree shrews. Front. Behav. Neurosci.. 2015;9:204.
- [Google Scholar]
- Response of the gut microbiota during the Clostridioides difficile infection in tree shrews mimics those in humans. BMC Microbiol.. 2020;20(1)
- [CrossRef] [Google Scholar]
- The position of tree shrews in the mammalian tree: Comparing multi-gene analyses with phylogenomic results leaves monophyly of Euarchonta doubtful. Integr. Zool.. 2015;10(2):186-198.
- [Google Scholar]
- Chronic hepatitis B virus infection and occurrence of hepatocellular carcinoma in tree shrews (Tupaia belangeri chinensis) Virol J. 2015;12:26.
- [Google Scholar]
- Herpes simplex virus 1 infection of tree shrews differs from that of mice in the severity of acute infection and viral transcription in the peripheral nervous system. J. Virol.. 2016;90(2):790-804.
- [Google Scholar]
- Schulze, Vanessa, et al., 2020. Borna disease outbreak with high mortality in an alpaca herd in a previously unreported endemic area in Germany. Transboundary Emerg. Dis. 67(5), 2093-2107.
- MALBON, Alexandra J., et al., 2021. New World camelids are sentinels for the presence of Borna disease virus. Transboundary Emerg. Dis.
- Shrews as reservoir hosts of borna disease virus. Emerg. Infect. Dis.. 2006;12(4):675-677.
- [Google Scholar]
- Hantavirus (Seoul virus) in pet rats: a zoonotic viral threat. Vet. Rec.. 2016;178(7):171-172.
- [Google Scholar]
- Molecular method for the detection of Andes hantavirus infection: validation for clinical diagnostics. Diagn. Microbiol. Infect. Dis.. 2016;84(1):36-39.
- [Google Scholar]
- Hepatitis E virus: Host tropism and zoonotic infection. Curr. Opin. Microbiol.. 2021;59:8-15.
- [Google Scholar]
- Metabolomic analysis of key regulatory metabolites in hepatitis C virus-infected tree shrews. Mol .Cell Proteomics. 2013;12(3):710-719.
- [Google Scholar]
- Discovery of novel alphacoronaviruses in European rodents and shrews. Viruses. 2016;8(3):84.
- [CrossRef] [Google Scholar]
- Nairobi sheep disease virus RNA in ixodid ticks, China, 2013. Emerg Infect Dis. 2015;21(4):718-720.
- [Google Scholar]
- Cell culture demonstrates superior sensitivity over one step real time RT PCR and nested VP1 amplification for Enteroviruses. J. Virol. Methods. 2021;287:113994
- [Google Scholar]
- Development and evaluation of reverse transcription loop-mediated isothermal amplification for rapid and real-time detection of Kyasanur forest disease virus. Int. J. Infect. Dis. 2021
- [CrossRef] [Google Scholar]
- Ixodes ricinus and Its Transmitted Pathogens in Urban and Peri-Urban Areas in Europe: New Hazards and Relevance for Public Health. Front Public Health. 2014;2:251.
- [Google Scholar]
- Reeder, W.a., 2011. Classification of Recent Mammalia based on Wilson & Reeder.
- Development and evaluation of a broad reacting SYBR-green based quantitative real-time PCR for the detection of different hantaviruses. J. Clin. Virol.. 2013;56(4):280-285.
- [Google Scholar]
- Cytotoxic immune responses in the lungs correlate to disease severity in patients with hantavirus infection. Eur. J. Clin. Microbiol. Infect. Dis.. 2016;35(4):713-721.
- [Google Scholar]
- A novel approach for detection of sindbis viral RNA –with QPCR. Int. J. Sci.: Basic Appl. Res. (IJSBAR). 2015;24(4):415-424.
- [Google Scholar]
- Ge, X., et al., 2012. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 86(8), 462046-30.
- Limongi, J.E., et al., 2016. Hantavirus pulmonary syndrome and rodent reservoirs in the savanna-like biome of Brazil's southeastern region. Epidemiol. Infect., 144(5), 1107-1116.
- Adouchief, S., et al., 2016. Sindbis virus as a human pathogen-epidemiology, clinical picture and pathogenesis. Rev. Med. Virol.
- Joshi, M. and Deshpande, J.D, 2011. Polymerase chain reaction: methods, principles and application. Int. J. Biomed. Res. 5, 81‐97
- Development of a RVFV ELISA that can distinguish infected from vaccinated animals. Virol. J.. 2009;6:125.
- [Google Scholar]
- Lo Presti, A., et al., 2012. Origin, evolution, and phylogeography of recent epidemic CHIKV strains. Infect. Genet. Evol. 12(2), 392-398.
- A depression model of social defeat etiology using tree shrews. Dongwuxue Yanjiu. 2012;33(1):92-98.
- [Google Scholar]
- Lin, J., et al., 2014. Phylogenetic affinity of tree shrews to Glires is attributed to fast evolution rate. Mol. Phylogenet. Evol. 71, 193-200.







