Translate this page into:
Description of a new species of Tetmemena (Ciliophora, Oxytrichidae) using classical and molecular markers
⁎Corresponding author. helserehy@ksu.edu.sa (Hamed A. El-Serehy)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Tetmemena saprai n. sp. was isolated from fresh-water samples collected from Okhla Bird Sanctuary, Delhi, India and described based on its morphology, morphogenesis and molecular markers, namely the small-subunit (SSU) rRNA gene and internal transcribed spacers (ITS1-5.8S-ITS2). The morphological features of T. saprai n. sp. are as follows: colourless; rigid body with no cortical granules; water expelling vesicle about 15 µm in size, located below the adoral zone of membranelles near the left body margin; body size about 125–140 × 50–60 μm in live and about 100 × 45 μm in protargol preparations, with body length : width ratio should be 2:1; undulating membranes in a Stylonychia-pattern; adoral zone about 44 μm in length with (on average) 45 adoral membranelles; 18 frontal-ventral-transverse cirri; one right and one left marginal row; six dorsal rows including two dorsomarginals, of which the fourth row is shortened anteriorly; three caudal cirri which are not equidistant; two macronuclear nodules; 2–4 micronuclei. A detailed morphogenetic study revealed the oral primordium to originate near the leftmost transverse cirrus, and the involvement of five parental cirri (three frontals and two ventrals) in the formation of streaks I–VI for the proter and opisthe. The genetic distance between T. saprai n. sp. and its congeners T. pustulata and T. vorax (S. vorax) varied from between 1% and 2% in the SSU rRNA gene and 1% and 6% in the ITS1-5.8S-ITS2 sequence. The secondary structures of the ITS1 and ITS2 RNA transcripts of four different species of genus Tetmemena (T. saprai n. sp., T. pustulata, T. vorax and T. bifaria) were also compared, revealing that the ITS1 region was more variable than ITS2. The morphological and morphogenetic characterisation, and phylogenetic analyses based on the molecular markers, confirm that the present species is a distinct species of the genus Tetmemena belonging to the subfamily Stylonychinae. This study also reveals that the macronuclear ITS region can be a suitable candidate for species identification.
Keywords
Oxytrichid ciliates
New species
Ciliature
Taxonomy
SSU rRNA gene
Molecular markers
1 Introduction
In the late 1990s Berger divided the family Oxytrichidae into two subfamilies, Oxytrichinae and Stylonychinae, on the basis that the former had more “flexible” bodies and the latter more ‘‘rigid’’ ones (Berger, 1999; Berger and Foissner, 1997). Sequence analysis of species from Oxytrichidae during the early years of the 21st century has confirmed this subdivision, albeit with relatively stronger support for the Stylonychinae subfamily than the Oxytrichinae (Bernhard et al., 2001; Hewitt et al., 2003; Schmidt et al., 2007). Analyses based on molecular data have also strongly supported the monophyly of the subfamily Stylonychinae (Bernhard et al., 2001; Chen et al., 2013; Foissner et al., 2004; Hu et al., 2011; Schmidt et al., 2007). The subfamily, Stylonychinae is characterised by the rigid body mentioned above, a lack of cortical granules and an adoral zone of membranelles that is usually more than 40% of the body length (Berger and Foissner, 1997) and also includes the so-called “18-cirri oxytrichids” (Berger, 1999).
The genus Tetmemena, first designated by Eigner (1999), is classified as a member of the subfamily Stylonychinae, family Oxytrichidae, suborder stichotrichida, order hypotrichia within the class Spirotrichea (Abraham et al., 2019; Adl et al., 2019; Lynn, 2008).
Species of the genus Tetmemena are characterised by elliptical to ovoid shape, comparatively rigid pellicle; adoral zone of membranelles formed like a question mark; endoral and paroral membranes in a Stylonychia-pattern; transverse cirri arranged in a J-shape; one right and one left marginal row; six dorsal rows formed in an Oxytricha pattern; three caudal cirri, often distinctly elongated (Eigner, 1997, 1999; Shao et al., 2013, 2015). Within the genus, Tetmemena pustulata has been well characterised based on silver staining, morphogenesis and SSU rRNA gene, ITS1-5.8S-ITS2 sequences.
During an investigation of the diversity of fresh-water ciliates within Okhla Bird Sanctuary in September 2013, an oxytrichid ciliate was isolated. Observations of its morphology both in vivo and following protargol impregnation demonstrate that the isolate represents a novel species within the genus Tetmemena. In this study, we describe its morphology and morphogenesis during cell division. Moreover, the SSU rRNA gene and the ITS1-5.8S-ITS2 regions of the new isolate were sequenced and analyzed, moreover, the secondary structures of the ITS1 and ITS2 regions were compared to investigate its systematic status and to assess its phylogenetic position.
2 Materials and methods
Water samples were collected on 18th September 2013 from Okhla Bird Sanctuary (N28°32′43.5″, E77°18′41.7″), Delhi, India. Water temperature and pH at the time of collection were 23 °C and 7.2 respectively. The cells were identified and isolated in vivo using stereoscopic and phase contrast microscopes. The clonal cultures of Tetmemena saprai n. sp. were maintained at 22–23 °C in Pringhsheim's medium (Chapman-Andresen, 1958) was used for culturing. Small pieces of boiled cabbage were added to promote the growth of the bacteria which serve as the primary food source for the ciliates. At some point the addition of cabbage was halted for a week in order to starve the cells and encourage them to encyst. Cysts were identified and viewed under phase contrast microscopy to study their morphology. Protargol impregnation and scanning electron microscopy were used to visualise the surface ciliature (Foissner, 2014; Kamra and Sapra, 1990). Nuclear observations were made by using the Feulgen staining technique (Chieco and Derenzini, 1999). Biometric characterisation was done at a magnification of 400 × and 1000 × directly from a phase contrast microscope. Line diagrams were prepared using Corel Draw Graphics software. Classification follows Berger (1999) and Lynn (2008), while general terminology follows Berger (1999, 2001, 2006, 2007, 2008, 2011, 2012), Borro (1972) and Wallengren (1900).
2.1 Generation time
The time it took T. saprai n. sp. to generate was calculated by isolating a dividing cell in a separate cavity block to which a small piece of boiled cabbage was added. The time taken for each daughter cell to divide into two daughter cells was observed and noted. This was done in triplicates to obtain a mean value.
2.2 DNA extraction, amplification and sequencing
Total genomic DNA was extracted using Qiagen DNA blood and tissue kit (Qiagen, India) with the cell concentration being 50 cells / ml. PCR was performed to isolate the SSU rRNA gene and ITS1-5.8S-ITS2 region. Primers used for isolating the SSU rRNA gene were 5′-AAC CTG GTT GAT CCT GCC AGT-3′ as forward and 5′-TGA TCC TTC TGC AGG TTC ACC TAC-3′ as reverse (Lv et al., 2013; Medlin et al., 1988), along with two internal primers: 5′-CGG TAA TTC CAG CTC CAA TAG-3′ as forward and 5′-AAC TAA GAA CGG CCA TGC AC-3′ as reverse. Primers used for isolating the ITS1-5.8S-ITS2 region were 5′- GCT CCT ACC GAT TTC GAG TG-3′ as forward and 5′-TTA AGT TCA GCG GGT GAT CC-3′ as reverse. The PCR protocol employed was as follows: 95 °C for 5 min, 50 °C (for the SSU rRNA gene)/ 54 °C (for the ITS1-5.8S-ITS2 region) for 1 min, 72 °C for 1 min, followed by 30 identical amplification cycles of denaturation at 95 °C for 45 s, annealing at 50 °C/54 °C for 45 s, extension at 72 °C for 45 s and a final step of 95 °C for 45 s, 50 °C/54 °C for 45 s and a final extension at 72 °C for 10 min. The PCR product was eluted using QIAquick Gel Extraction kit (Qiagen, India). The eluted product was sequenced using the Applied Biosystems 3130xl Automated DNA sequencer.
2.3 Phylogenetic analyses
The phylogenetic position of T. saprai n. sp. on the basis of the SSU rRNA gene was determined by retrieving sequences of 39 other taxa including one outgroup from GenBank. Similarly, for the ITS1-5.8S-ITS2 region, the phylogenetic position was determined by retrieving 16 other taxa including an outgroup. In both cases, Pseudourostyla sp. (Urostylida) was selected as the outgroup taxon. The sequences were aligned using ClustalX2 (Jeanmougin et al., 1998) and both ends of the alignment were manually trimmed using BioEdit software (Hall, 1999). Phylogenetic trees were constructed using both the Maximum Likelihood (ML) method and Bayesian inference (BI). ML analyses were carried out using RAxML-HPC2 v8.2.12 on the CIPRES Science Gateway (Miller et al., 2010) and BI analyses by using MrBayes 3.2.2 software (Ronquist and Huelsenbeck, 2003). ML analysis was carried out with 1000 bootstrap replicates using the GTR + gamma model. In the BI method, the tree was constructed using the GTR + G + I model with Markov chain Monte Carlo (MCMC) simulation for 100,000 generations, with a burn-in of 250 generations. The GTR + G + I model is recognized to be the most complex model with the most parameter rich rate matrix thus, giving the highest (maximised) likelihood to the data (Foster, 2003). This was obtained by entering the command “lset nst = 6 rates = invgamma” in the MrBayes software after entering the data (Ronquist et al., 2011). This generated General Time Reversible model with a proportion of invariable sites and a gamma-shaped distribution of rates across the sites (GTR + G + I). The genetic distances between T. saprai n. sp. and its congeners were calculated by aligning them using BioEdit software.
2.4 Secondary structure prediction of the ITS region
Sequences of ITS1 and ITS2 from four different species of the genus Tetmemena (T. saprai n. sp., T. pustulata, T. vorax and T. bifaria) were obtained individually by aligning them using BioEdit software. Secondary structures of these genes were predicted using mfold web server (http://unafold.rna.albany.edu/?q=mfold/RNA-Folding-Form) (Wang et al., 2015; Zuker, 2003) with default settings. The structures obtained were then viewed and edited using RnaViz 2.0 software (Li et al., 2013; Rijk and Wachter, 1997; Wang et al., 2015).
3 Results
3.1 Tetmemena saprai n. sp. (Fig. 1A–G, 2A–C, Table 1)
3.1.1 Diagnosis
Tetmemena saprai n. sp. with body size in vivo about 125–140 × 50–60 µm; rigid oxytrichid; dorsoventrally flattened with body length to width ratio around 2:1; body lanceolate anteriorly and rounded posteriorly; two macronuclear nodules and 2–4 micronuclei; 42 adoral membranelles; 18 frontal-ventral-transverse cirri; one right (about 28 cirri) and one left marginal (about 22 cirri) cirral row; six dorsal rows with dorsal kinety 4 (DK4) shortened anteriorly; two dorsomarginal rows; three caudal cirri; generation time about eight hours; resting cysts with three layers: wavy outer layer, spiny middle layer and smooth inner layer.
3.1.2 Type locality and ecology
The Okhla Bird Sanctuary (N28°32′43.5″, E77°18′41.7″) is a bird sanctuary covering approximately 4 square kilometers at the Okhla - barrage over the Yamuna River in Delhi, India. Between Okhla - village to the west and Gautam Budh Nagar to the east the site contains a large lake, and the sanctuary is surrounded by thorny shrub, grassland and wetland. The sediment consists of organic debris and fine sand. Water hyacinths grow extensively. The samples were collected at a depth of approximately one meter.
3.1.3 Voucher material
The protargol impregnated slide containing the holotype specimen (Acc. No.: Pt.3665) has been deposited in the Zoological Survey of India, Kolkata, India.
3.1.4 GenBank submission
The nucleotide sequences of SSU rRNA gene and ITS1-5.8S-ITS2 region were deposited in GenBank with accession numbers KP336401 and KT731103, respectively.
3.1.5 Etymology
The species name acknowledges Professor G. R. Sapra’s contributions to the field of ciliate biology.
3.1.6 Description
Body 125–140 × 50–60 µm in vivo (Fig. 1A), 100 × 45 µm on average in protargol preparations (Fig. 1F, G, Table 1); rigid; dorsoventrally flattened with lanceolate anterior and rounded posterior ends; cortex colourless; cortical granules absent; two macronuclear nodules; macronuclei ellipsoidal with 19–23 × 7.5–10 µm (stained); 2–4 micronuclei (Fig. 1D); micronuclei globular of 0.1–2.6 µm in diameter (stained); contractile vacuole about 15 µm in size, located below the adoral zone of membranelles near the left body margin (Figs. 1A and 2A); voracious feeder of algae, bacteria, other small ciliates like Tetrahymena and even exhibits cannibalism; locomotion slow to moderately crawling on the bottom of the petri dish; generation time 8 ± 0.5 hrs; resting cysts about 20–23 µm across in vivo; outer surface wavy, middle layer spiny and inner layer smooth with distinct macronuclear nodule (Fig. 1E).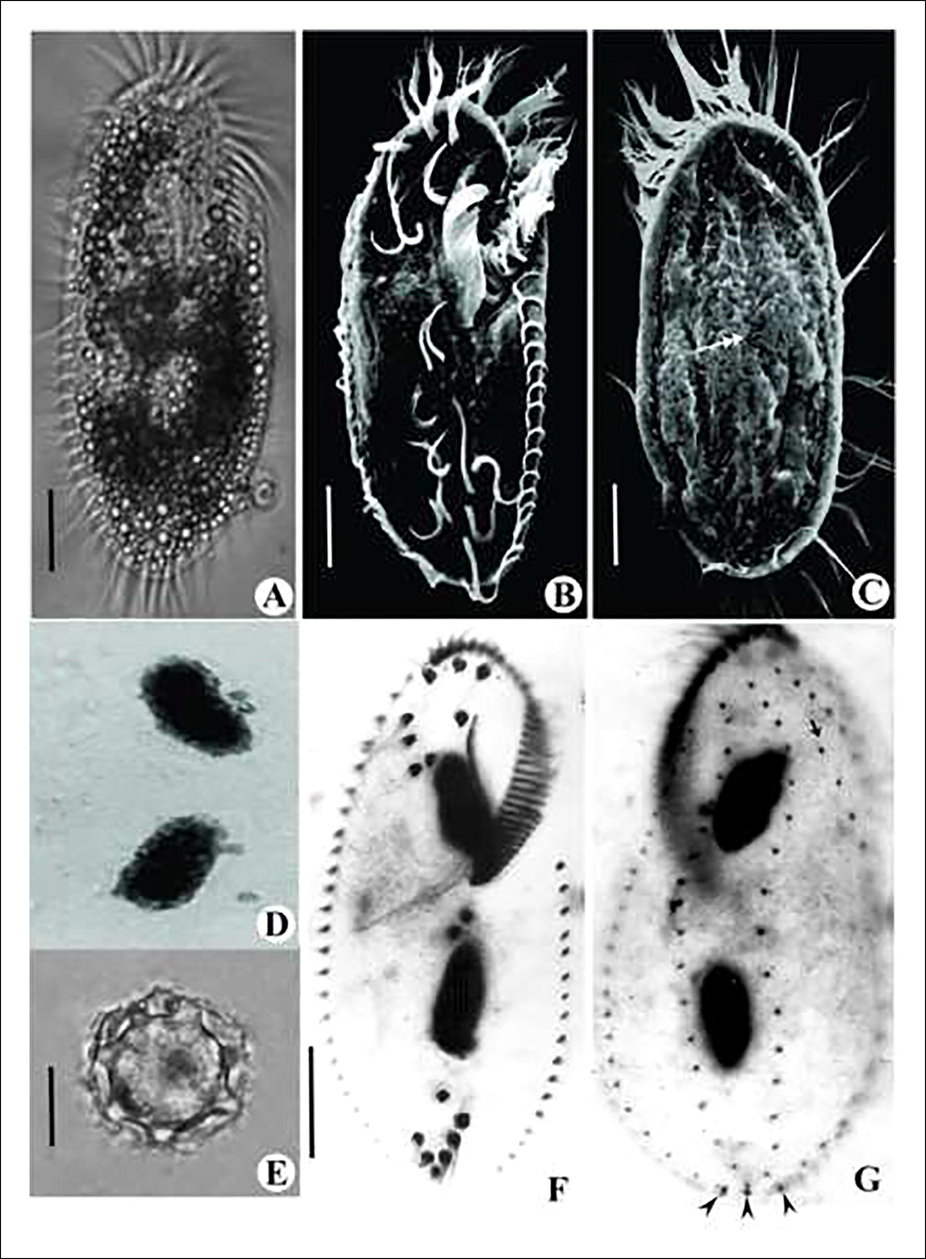
Photomicrographs of live (A), Scanning electron microscopic (B, C), Feulgen stained (D), Cyst (E) and protargol impregnated (F, G) cells of Tetmemena saprai n. sp. A. Live cell in ventral view. B. Ventral view showing ciliature. C. Dorsal view showing ciliature; DK4 shortened anteriorly (arrow); dorsal bulge (double arrow) D. Two macronuclear nodules and four micronuclei. E. Cyst showing wavy exocyst, spiny mesocyst and smooth endocyst. F. Ventral view of a vegetative cell with five transverse cirri arranged in a J-shaped row. G. Dorsal view of a vegetative cell; DK4 shortened anteriorly (arrow); caudal cirri (arrow head) Scale bars: 20 μm (A-C; F & G) and 10 μm (E).
Character
Mean
Min
Max
SD
CV
n
Body, length
98.42
88.90
109.38
6.93
7.05
10
Body, width
47.39
43.40
53.38
3.46
7.30
10
Macronuclear nodules, number
2
2
2
0
0
25
Macronuclear nodule, length
21.91
19.60
23.98
1.43
6.51
10
Macronuclear nodule, width
8.82
7.53
9.98
0.81
9.22
10
Micronuclei, number
2–4
2
4
0.80
29.93
25
Micronucleus, diameter
1.91
0.17
2.63
0.96
50.09
10
Adoral membranelles, number
41.80
37
45
2.78
6.65
10
AZM, length
43.75
37.98
50.23
4.20
9.61
10
Left marginal row, number of cirri
22.10
20
25
1.52
6.90
10
Right marginal row, number
28.80
23
32
2.74
9.52
10
Frontal cirri, number
8
8
8
0
0
25
Post oral ventral cirri, number
3
3
3
0
0
25
Pretransverse cirri, number
2
2
2
0
0
25
Transverse cirri, number
5
5
5
0
0
25
Caudal cirri, number
3
3
3
0
0
25
Dorsal kineties, number
6
6
6
0
0
25
Dikinetids in DK1
29.27
27
33
1.74
5.93
11
Dikinetids in DK2
22.09
20
24
1.22
5.53
11
Dikinetids in DK3
19.45
17
22
1.69
8.71
11
Dikinetids in DK4
21.45
19
24
1.69
7.90
11
Dikinetids in DM1
11.82
11
14
0.87
7.39
11
Dikinetids in DM2
4.27
4
5
0.47
10.93
11
Dorsal bristles no.
108.36
99
115
5.03
4.64
11
Distance between CC1 and 2
2.14
1.81
2.41
0.23
10.66
10
Distance between CC2 and 3
2.70
1.81
3.1
0.36
13.38
10
Distance between F8 (III/2) – UM
3.70
2.28
4.32
0.59
15.83
10
Distance between F1 (II/2) - F8 (III/2)
4.39
3.39
5.63
0.70
15.93
10
Distance between F2 (I/1) - F8 (III/2)
10.72
8.77
12.44
1.12
10.44
10
Distance between F3 (II/3) - F8 (III/2)
11.42
9.51
12.91
1.12
9.70
10
Distance between F4 (III/3) - F8 (III/2)
9.17
7.62
10.14
0.75
8.13
10
Distance between F5 (VI/4) - F8 (III/2)
4.97
4.57
6.08
0.44
8.86
10
Distance between F6 (VI/3) - F8 (III/2)
2.99
2.09
5.40
0.95
31.64
10
Distance between F7 (IV/3) - F8 (III/2)
2.66
2.24
3.94
0.51
19.28
10
Distance between V1 (IV/2) - V2 (V/4)
2.99
2.28
4.60
0.63
21.17
10
Distance between V2 (V/4) - V3 (V/3)
5.91
5.31
6.67
0.42
7.11
10
Distance between V3 (V/3) - V4 (V/2)
5.10
4.39
5.99
0.59
11.57
10
Distance between V4 (V/2) - V5 (VI/2)
7.09
5.95
7.93
0.65
9.15
10
Infraciliature is as shown in Figs. 1B, C, F, G and 2A–C. Adoral zone of membranelles less than 50% of the body length with 37–45 adoral membranelles, cilia about 8 µm long, Undulating membranes in a Stylonychia pattern. 18 frontal-ventral-transverse (FVT) cirri, including three frontal, one buccal, four frontoventral, three postoral ventral, two pretransverse ventral and five transverse cirri. Frontal cirri slightly larger than buccal and frontoventral cirri; the posterior most ventral VI/2 lies adjacent to the transverse VI/1. Of the five transverse cirri, four are characteristically arranged in an oblique row adjoining the fifth cirrus. Consistently, one left marginal row with 20–25 cirri and one right marginal row with 23–32 cirri, posterior ends of the marginal rows are not confluent.
Dorsal ciliature composed of four kineties and two dorsomarginal rows, dorsal cilia about 4 µm long, dorsal rows 1–6 composed of 27–33, 20–24, 17–22, 19–24, 11–14, 4–5 bristles, respectively; first three dorsal kineties 1–3 almost bipolar, DK4 shortened anteriorly at the level of the 4th or 5th bristle of DK3 (Figs. 1C, G and 2C). Caudal cirri invariably three and not equidistant, about 11 µm long, located at the ends of dorsal kineties 1, 2 and 4 (Fig. 2C).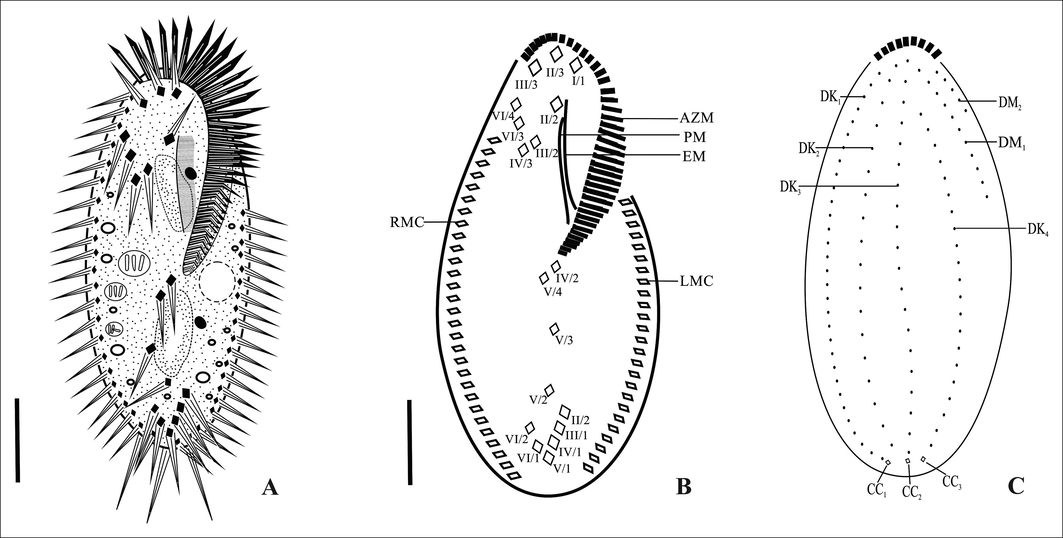
Line diagrams showing protargol impregnated vegetative cells of Tetmemena saprai n. sp. A. From life B. Ventral surface C. Dorsal surface. AZM, adoral zone of membranelles; CC1–3, caudal cirri; DK1–4, dorsal kineties; DM1–2, dorsomarginal kineties; EM, endoral membrane; LMC, left marginal cirri; PM, paroral membrane; RMC, right marginal cirri; II/2, buccal cirri; I/1, II/3, III/3, frontal cirri; VI/4, VI/3, IV/3, III/2, frontoventral cirri; IV/2, V/4, V/3, postoral ventral cirri; V/2 and VI/2, pretransverse ventral cirri; II/1, III/1, IV/1, V/1, VI/1, transverse cirri. Scale bars: 20 μm.
3.2 Morphogenesis:
3.2.1 Oral apparatus and ventral ciliature (Fig. 3A–H; Fig. 4 A-H)
The oral primordium (OP) arises as a few kinetosomes near the leftmost transverse cirri II/1. After the first 8 or 9 kinetosomes have been formed the OP moves anteriorly, away from II/1. The primordium extends to the level of the posterior end of the adoral zone of membranelles (Figs. 3A, B, C and 4A, B). Membranelles differentiate from the anterior-right portion of the OP to form the adoral membranelles of the opisthe and splits into three primordial streaks I-III (Figs. 3D and 4C). Simultaneously, the posterior-most frontoventral cirrus IV/3 differentiates and forms a common primordium for streaks IV-VI of the proter (Figs. 3D and 4C). Postoral ventral cirrus V/4 differentiates into primordia for streak V and VI and cirrus IV/2 differentiates to form primordial streak IV for the opisthe (Figs. 3D, E and 4C, D). Frontoventral cirri, II/2 and III/2 differentiate into primordial streaks II and III respectively for the proter (Figs. 3E and 4E). The left frontal cirrus of the opisthe, I/1 develops from the leftmost anlage. In the proter, the parental undulating membranes are reorganised and form the left frontal cirrus, I/1. The newly formed FVT cirri migrate to their final positions (Figs. 3F, G, H and 4F, G, H).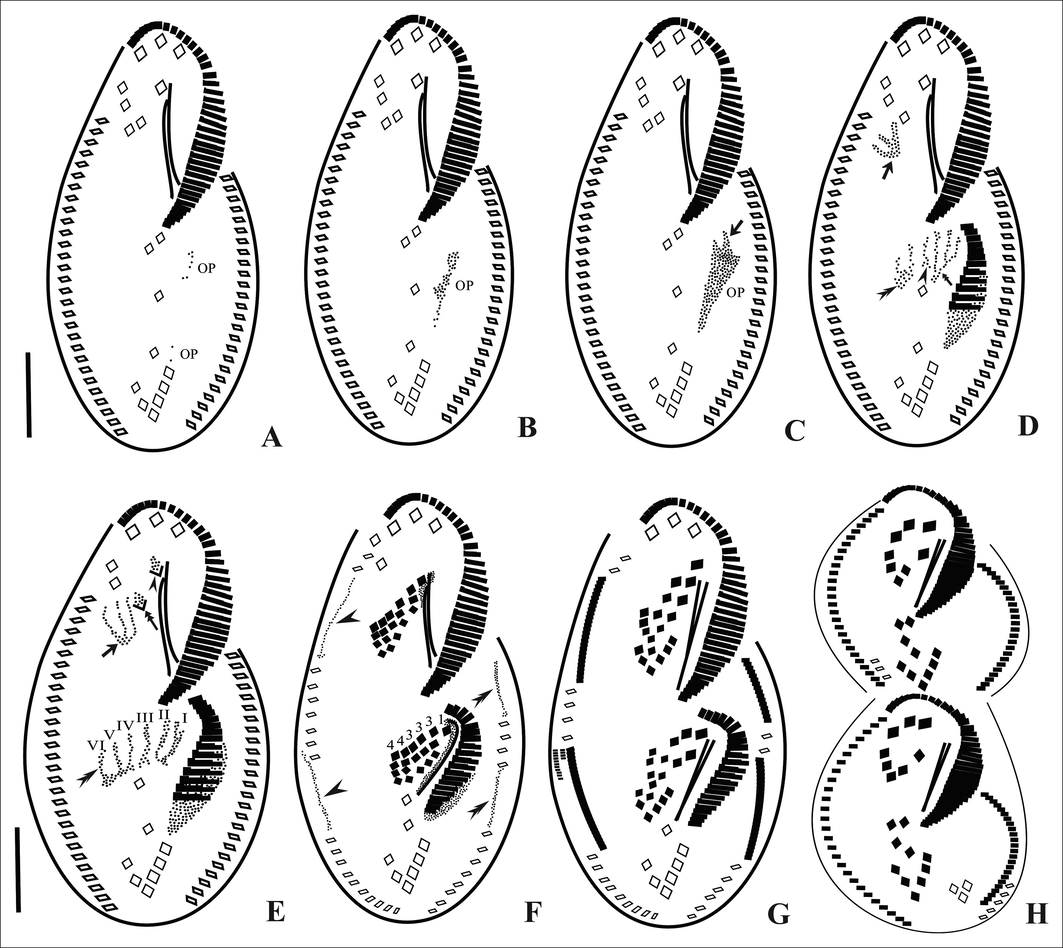
Line diagrams of the ventral surface of Tetmemena saprai n. sp. showing the morphogenetic stages (based on protargol impregnated cells). A, B. Origin of OP as a few kinetosomes near the leftmost transverse cirrus (II/1). C. Kinetosomes proliferate, OP moves anteriorly and loses its connection with (II/1), origin of streak I of opisthe from OP (arrow). D. Disaggregation of IV/3 to form streak IV-VI for proter (arrow), disaggregation of V/4 to form streaks V and VI for opisthe (double arrowhead), formation of streaks I–III anteriorly (double arrow) and disaggregation of IV/2 to form streak IV for opisthe (arrowhead) E. Primordial streaks IV–VI of proter formed from IV/3 (arrow), disaggregation of frontal cirri III/2 (double arrow) and II/2 (arrowhead), disaggregation of IV/2 forming complete set (I-VI) of primordial streaks for opisthe (double arrowhead). F. Differentiation of cirri in a 1, 3, 3, 3, 4, 4 pattern, within-row formation of marginal primordia for RMC (arrowheads) and LMC (double arrowheads). G, H. Late divider. OP, oral primordium. Scale bars: 20 μm.
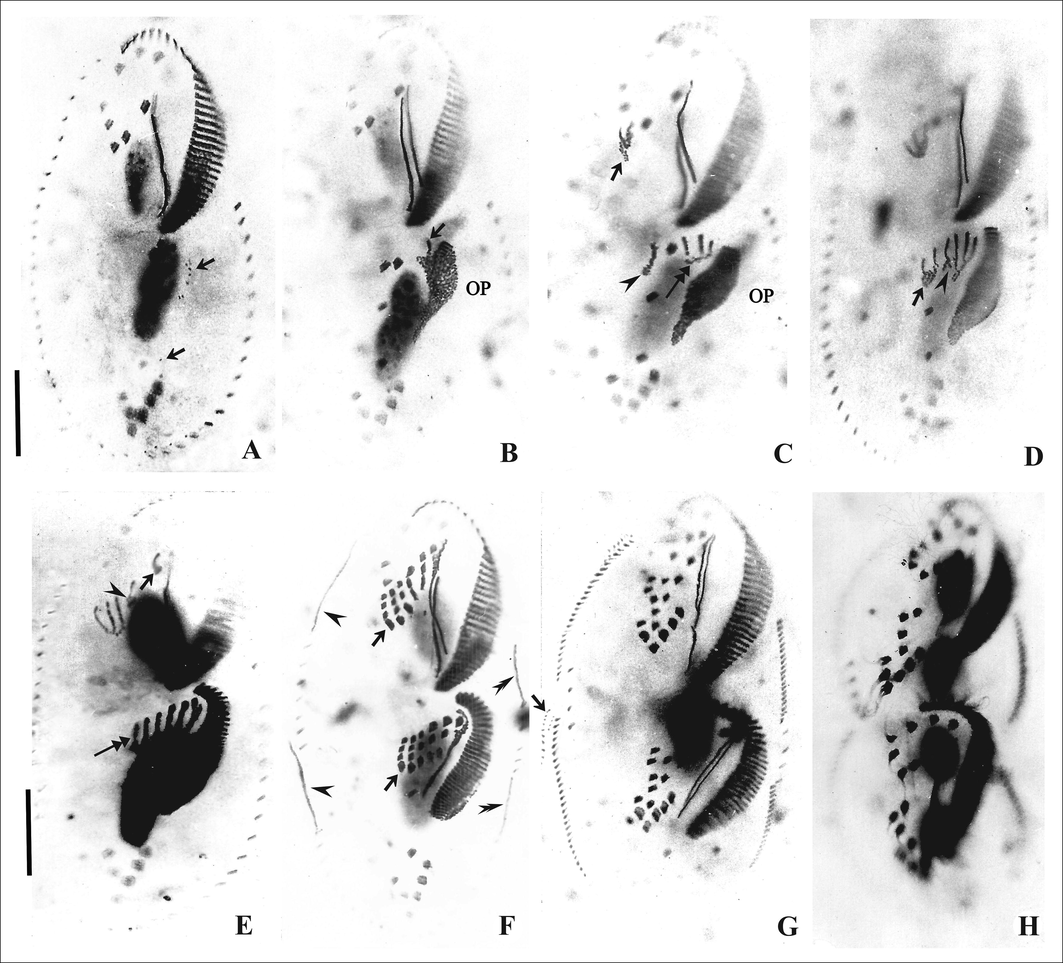
Photomicrographs of the ventral surface of Tetmemena saprai n. sp. showing the morphogenetic stages (based on protargol impregnated cells). A, B. Origin of OP as a few kinetosomes near the leftmost transverse cirrus (II/1) (arrow), origin of streak I from OP (arrow). C. Disaggregation of IV/3 to form streaks IV-VI for proter (arrow), formation of streaks I–III (double arrow) from OP for opisthe, and disaggregation of V/4 (arrowhead). D. Disaggregation of V/4 (arrow) to form streaks V and VI and disaggregation of IV/2 to form streak IV (arrowhead) for opisthe E. Disaggregation of frontoventral cirrus III/2 to form streak III of proter (arrowhead), and of II/2 (arrow) to form streak II; complete set of streaks formed for opisthe (double arrow). F. Differentiation of cirri in a 1, 3, 3, 3, 4, 4 pattern in the proter and opisthe (arrows), within-row formation of marginal primordia for RMC (arrowhead) and LMC (double arrowhead). G. Late divider showing formation of new dorsomarginals (arrow) close to newly formed RMC. H. Cell in cytokinesis. OP, oral primordium. Scale bars: 20 μm.
In all, five parental cirri (two postoral ventral and three frontoventral cirri) are involved in the origin of the two sets of six primordial streaks each for proter and opisthe of the cell. Streaks I-VI differentiate into 1, 3, 3, 3, 4, 4 cirri, respectively (Figs. 3F and 4F).
3.2.2 Marginal and dorsal ciliature (Figs. 3F, G, 4F, G, 5A–G)
A few cirri from the anterior and middle parts of the parental marginal rows are incorporated within the row to form the marginal cirral primordia (Figs. 3F and 4F). These marginal cirral primordia extend posteriorly and differentiate into new marginal cirri replacing the parental marginal rows. Simultaneously, two sets of primordia are formed for the three dorsal kineties 1–3 which extend in two directions and the parental dorsal kineties are incorporated or resorbed (Fig. 5A, E). The third dorsal primordium splits into two parts forming primordia for dorsal kineties 3 and 4, respectively (Fig. 5B, C, F). Caudal cirri develop at the posterior ends of the dorsal kineties 1, 2 and 4 and are not equidistant (Fig. 5C, G). Dorsomarginal rows develop at/near the anterior end of the newly formed right marginal rows of the proter and opisthe and extend posteriorly (Figs. 3G and 4G).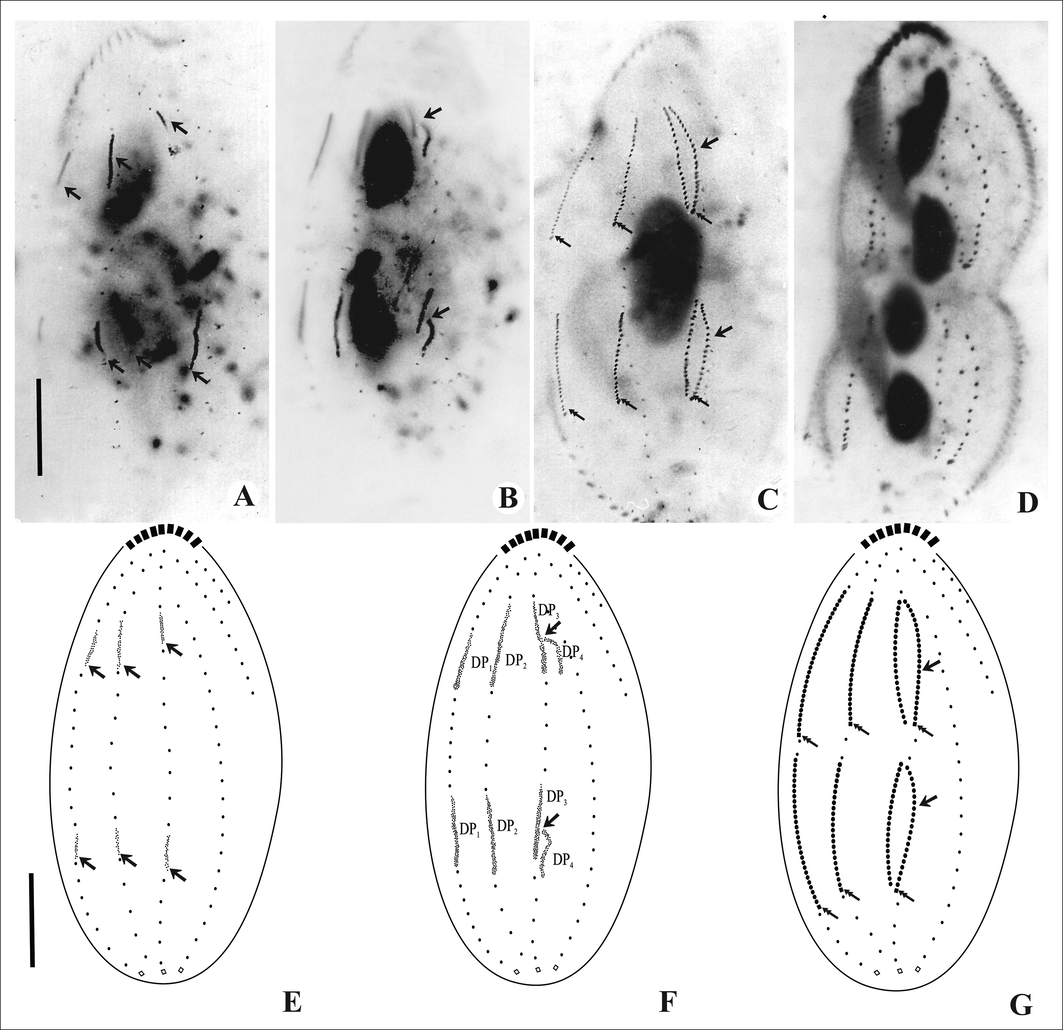
Photomicrographs and line diagrams of the dorsal surface of Tetmemena saprai n. sp. showing morphogenetic stages (based on protargol impregnated cells). A, E. Within- row formation of the dorsal primordia for the proter and opisthe (arrows); B, F. Unequal split of the third dorsal primordia (arrows). C, G. Formation of fourth dorsal row after splitting (arrows), caudal cirri formed at the ends of newly formed DK1, 2 & 4 (double arrows) for proter and opisthe. D. Cell in cytokinesis. Scale bars: 20 μm.
3.2.3 SSU rRNA gene sequence
The SSU rRNA gene sequence of T. saprai n. sp. has been submitted to GenBank with the accession number KP336401. The length of the sequence was 1695 bp. On the basis of this sequence, the genetic distance was calculated with its congeners retrieved from GenBank [sequences from two different strains of T. pustulata viz., KM222092 and X03947 and T. bifaria FM209296].
The genetic distance between T. saprai n. sp. and T. pustulata (KM222092) was 0.94% (16 nucleotide differences in 1695 bp) while with T. pustulata (S. pustulata) (X03947) it was 1.95% (33-nt differences in 1695 bp) and with T. bifaria (S. bifaria) (FM209296) it was 1.89% (32 nt differences in 1695 bp). In the phylogenetic tree T. saprai n. sp. showed monophyletic relationship with T. pustulata (KM222092) with high support (100-% bootstrap value and 1.00 BI) and was paraphyletic with T. pustulata (X03947) with moderate support (73-% bootstrap value and 0.74 BI) (Fig. 6).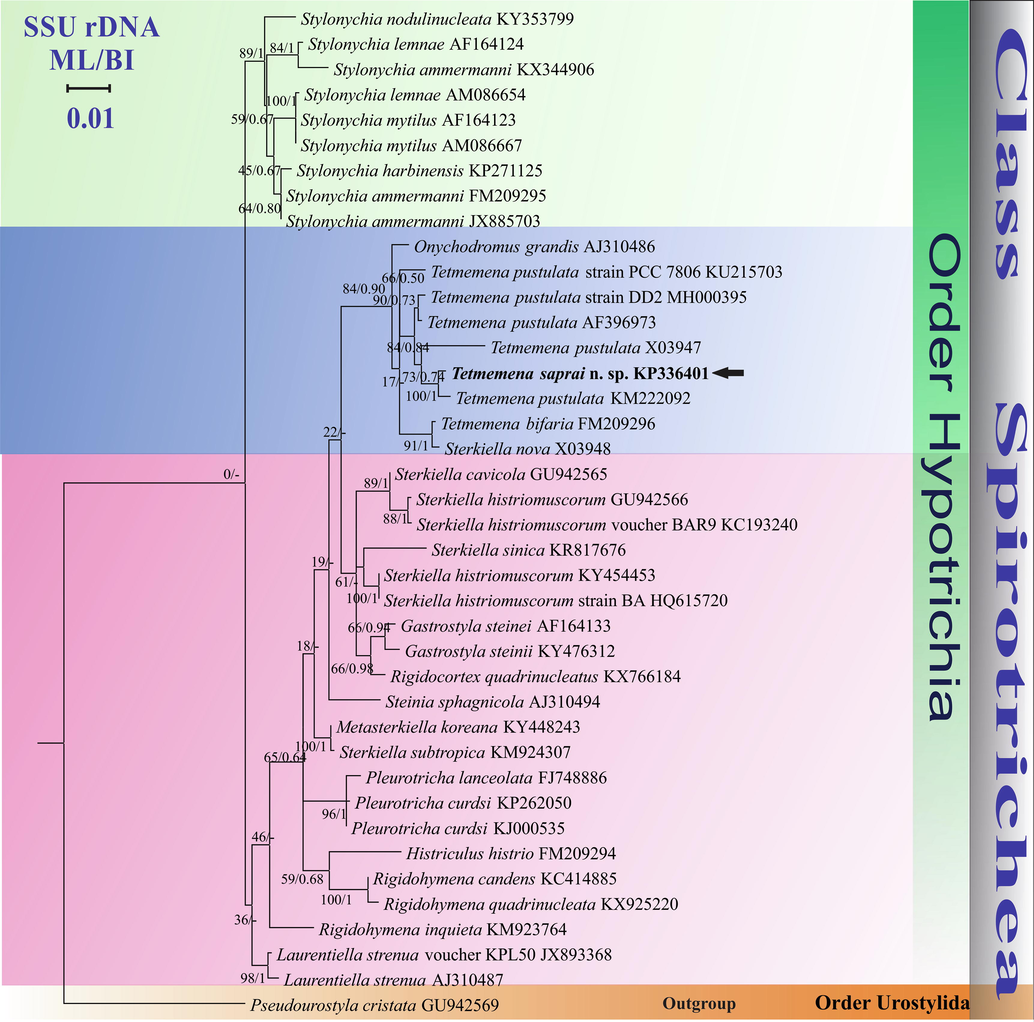
Phylogenetic tree based on SSU rDNA sequences and using GTR + I + G as the nucleotide substitution model showing the position of Tetmemena saprai n. sp. (highlighted in bold and indicated with an arrow). Each node contains bootstrap values from ML analysis while the posterior probabilities are from BI. Accession numbers are provided after species names. The symbol “–” at the nodes indicates disagreement between the two methods. The scale bar corresponds to 0.01 expected substitutions per site.
3.2.4 ITS1-5.8S-ITS2 sequence
ITS1-5.8S-ITS2 sequence of T. saprai n. sp. has also been submitted to GenBank with the accession no. KT731103. The length of the sequence was 667 bp. On the basis of this sequence, the genetic distance was calculated with its congeners retrieved from GenBank (sequences from two different strains of T. pustulata viz., KM222037 and AJ286791, T. bifaria (AJ286796) and T. vorax (AJ286795)).
The genetic distance between T. saprai n. sp. and T. pustulata (KM222037) was 1.01% (5 nt differences in 493 bp); while with T. vorax (S. vorax) (AJ286795) it was 4.24% (28 nt differences in 661 bp); with T. pustulata (S. pustulata) (AJ286791) it was 4.08% (27 nt differences in 662 bp) and with T. bifaria (AJ286796) it was 6.15% (41 nt differences in 667 bp). In the phylogenetic tree, T. saprai n. sp. showed a monophyletic relationship with T. pustulata (KM222037, Chinese population) with high support (100% bootstrap value and 0.99 BI) and is paraphyletic with T. pusilla (AJ286793) and T. vorax (AJ286795) with moderate support (72% bootstrap value, 0.99 BI and 57% bootstrap value, 0.73 BI, respectively). T. saprai also showed a paraphyletic relationship with a German population of T. pustulata (AJ286791), (88% bootstrap value and 0.84 BI) (Fig. 7).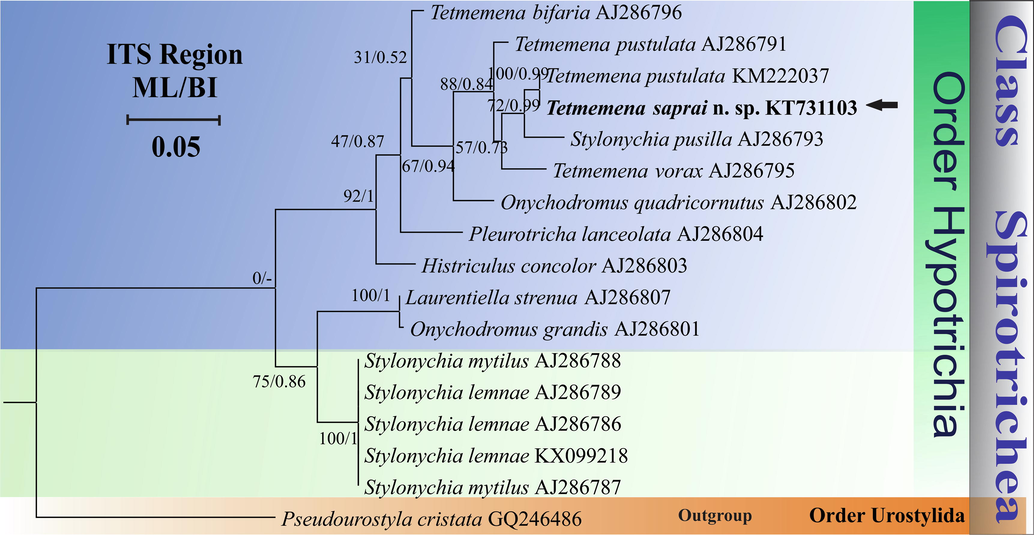
Phylogenetic tree based on ITS1-5.8S-ITS2 region sequences and using GTR + I + G as the nucleotide substitution model showing the position of Tetmemena saprai n. sp. (highlighted in bold and indicated with an arrow). Each node contains bootstrap values from ML analysis while the posterior probabilities are from BI. Accession numbers are provided after species names. The symbol “–” at the nodes indicates disagreement between the two methods. The scale bar corresponds to 0.05 expected substitutions per site.
3.2.5 Secondary structures of the ITS region in genus Tetmemena
The available sequences of the ITS1-5.8S-ITS2 region of T. pustulata (KM222037, AJ286791), T. vorax (AJ286795) and T. bifaria (AJ286796) along with T. saprai n. sp. were selected in order to predict their secondary structures. The secondary structures of ITS1 and ITS2 of these four ciliate species have one large loop and two helices (I and II; Fig. 8). The secondary structure of ITS1 was found to be more variable in T. bifaria with changes in one of helices when compared with other species of the genus Tetmemena. Some nucleotide changes were noticed in both the ITS1 (Fig. 8A–E) and ITS2 (Fig. 8F–J) region among these ciliate species, except for T. pustulata (KM222037), which appeared to be similar to T. saprai n. sp. In having few nucleotide variations in the whole length of ITS1-5.8-ITS2 region.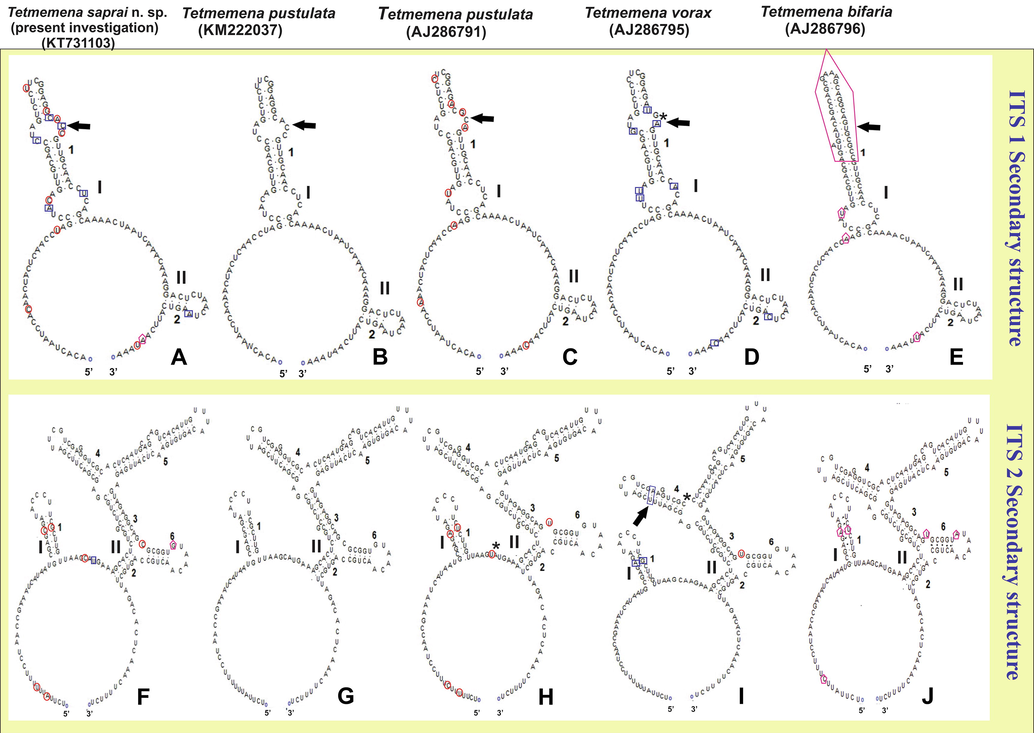
Secondary structures of the internal transcribed spacer 1 and 2 (ITS1 and ITS2) RNA transcript of four species of Tetmemena (T. saprai n. sp, T. pustulata, T. vorax and T. bifaria). Diagrams of ITS1 (A–E) and ITS2 (F–J) illustrate a large loop with two helices, labelled I and II. The red circle marks the only nucleotide variation between T. saprai n. sp. and T. pustulata (AJ286791), blue squares indicate the nucleotide variation between T. saprai n. sp. and T. vorax (AJ286795) and pink pentagons show nucleotide variations between T. saprai n. sp. and T. bifaria (AJ286796). Arrows indicate the differences in the loop between the species. The symbol ‘*’ marks the gaps in the nucleotide sequence.
4 Discussion
4.1 Comparison of Tetmemena saprai n. sp. with its congeners
T. saprai n. sp. is distinct from other species of the genus that have been described thus far in so far as it exhibits a new combination of characters. Although it appears to be closely related to T. pustulata (Shao et al., 2013; Wirnsberger et al., 1985) it is distinct in several respects, as shown in Table 3.
4.2 Justification for the establishment of a new species
T. saprai n. sp. possesses characteristic features of the genus Tetmemena. In light of the differences in morphology and morphometry, as well as morphogenetic and molecular data between the presently investigated species and other reported species of Tetmemena, it is affirmed that this species belongs to the Tetmemena-pustulata-vorax complex but is distinctly a new species (Tables 2 and 3). Morphological and ontogenetic information has helped in inferring phylogenetic relationships in many reported taxon of ciliates (Chen et al., 2017; Gupta et al., 2001; Kumar and Foissner, 2017; Shi and Li, 1993; Shi and Ammermann, 2004).
Character
Tetmemena saprai n. sp.(present investigation)
Tetmemena pustulata (Wirnsberger et al., 1985)
Tetmemena pustulata (Shao et al., 2013)
Tetmemena vorax(Wirnsberger et al., 1985)
Tetmemena bifaria (bi3 and bi4 from Kumar et al., 2016)
pu1
pu2
pu3
pu4
bi3
bi4
Body length
98.42
65
94
79
53
97.20
115
82
100.20
Body width
47.39
34
47
36
26
40.40
51.50
42
45.50
Macronuclear no.
2
2
2
2
–
2
2
2
2
Macro length
21.91
14
18
15
11.50
15.50
17
12.10
18.80
Macro width
8.82
7
9
7
6.10
10.80
12
8.70
12.20
Micronuclear no.
2–4
2
2
2
2
2
2
2.10
–
Micro diameter
1.91
4
4
4
–
–
4
2.45
–
AZM no.
41.80
35
38
34
32.10
38.70
50.50
27
29
AZM length
43.75
35
49
42
29
41.70
–
36.50
47
LMC no.
22.10
19
21
19
14.90
20.50
33
11.70
12.60
RMC no.
28.80
26
31
27
24.20
20.00
24
14.20
14.60
FC no.
8
8
8
8
8
8
8
8
8
VC no.
5
5
5
5
5
5
5
5
5
TC no.
5
5
5
5
5
5
5
5
5
CC no.
3
3
3
3
3
3
3
3
3
Dorsal rows no.
6
6
6
6
6
6
6
6
6
DK1
29.27
–
–
–
–
28
–
19.40
–
DK2
22.09
–
–
–
–
21.60
–
18.50
–
DK3
19.45
–
–
–
–
22.20
–
15.30
–
DK4
21.45
–
–
–
–
21.50
–
12.60
–
DM1
11.82
–
–
–
–
9.70
–
6.70
–
DM2
4.27
–
–
–
–
6.70
–
4.20
–
Dorsal bristles no.
108.36
–
–
–
–
109.70
–
76.7
–
Character
Tetmemena saprai n. sp. (Present Investigation)
Tetmemena pustulata (Wirnsberger et al. 1985)
Tetmemena pustulata (Shao et al. 2013)
Tetmemena vorax (Wirnsberger et al. 1985)
Shape
Anterior end lanceolate and Posterior end rounded
Elliptical and both end rounded
Elliptical or Ovoid and posterior end rounded
Anterior end rounded and posterior end tapering
Size
100 × 45 µm not much variation
48–129 × 26–83 µm, variation in related populations
75–115 × 40–60 µm
93–137 × 48–55 µm, variation in related populations
DK4
Terminates before anterior half of the body (Shortened anteriorly)
Extends anteriorly (Full complete row)
Extends anteriorly (Full complete row)
Terminates before anterior half of the body (Shortened anteriorly)
Central Dorsal Bulge
No bulge
Strongly bulged
–
No bulge
Position of Frontal Cirri
Posterior 4 frontal cirri are arranged in an oblique, hook-shaped row
F8-UM = 3.70 µmF8 with other Frontal cirri = 6.62 µmPosterior 4 frontal cirri are arranged in an oblique, hook-shaped row
–Posterior 4 frontal cirri are arranged in an oblique, hook-shaped row
F8-UM = 6.30 µmF8 with other Frontal cirri = 5.20 µmPosterior 4 frontal cirri are arranged in two pairs separated by a gap
–
Position of Ventral Cirri
Distance between V2, V3 and V4 is less and equidistantV3-V4 = 5.10 µm V4-V5 = 7.09 µm V2-V3 = 5.91 µm
Distance between V2,V3 and V4 are almost equidistant
Distance between V2,V3 and V4 is more , and almost equidistantV3-V4 = 10 µm V4-V5 = 12.5 µm V2-V3 = 9.90 µm
More gap between V2 and V3 and V3 is closer to V4
Distance between T3 and T4
Less
Less
Less
More
OP Origin
T1
De-novo
De-novo
T1 and T2
4.3 Phylogenetic position of Tetmemena saprai n. sp.
T. saprai n. sp. lies in a cluster belonging to the pustulata-vorax complex of the family Oxytrichidae. The phylogenetic analysis based on the SSU rRNA gene sequence supports the monophyly of the genus Tetmemena. T. saprai n. sp. shows a monophyletic relationship with only one of the strains of T. pustulata (KM222092) and a paraphyletic one with T. bifaria (FM209296) and one of the strains of T. pustulata (X03947) (Fig. 6). Since the SSU rRNA gene is highly conserved it is known to be most suitable for interspecific separation but not at intraspecific level (Gao et al., 2010; Yi et al., 2008). In comparison to the SSU rRNA gene, the ITS region (ITS1-5.8S-ITS2 region) shows relatively high variation in nucleotide sequences, making it more suitable for biodiversity analysis (Gao et al., 2010). This contention is strongly supported by the present study in that the genetic distance within the genus Tetmemena calculated on the basis of the SSU rRNA gene was was 1–2% whereas it was 1–6% on the basis of the ITS region. It is known from previous studies that even 1% nucleotide variation can be considered sufficient for designation as a new species (Schmidt et al., 2007). Since the nucleotide variation in T. saprai n. sp. with other known species of genus Tetmemena is more than 1%, this would suggest that T. saprai n. sp. is a distinct species, and this conclusion is in fact strongly supported by the predicted secondary structures of the ITS region, in that these show remarkable variations in the helices between T. saprai and its congeners. Since, the percentage of nucleotide variations calculated on the basis of the ITS region was more than that for SSU rRNA gene.
Remarks on the synonymy of T. saprai n. sp and S. notophora sensu Sapra and Dass 1970.
Over the last two decades the investigation of biodiversity among various fresh water bodies in India has shown the widespread presence of the species we designate as T. saprai n. sp. The morphological and morphogenetic description of S. notophora sensu Sapra and Dass (1970) from Delhi, India, corresponds well with T. saprai n. sp. In fact, Berger (1999) synonymised S. notophora sensu Sapra and Dass 1970 with T. pustulata (S. pustulata). It is agreed that S. notophora sensu Sapra and Dass (1970) is different from S. notophora described by Kahl (1932) but is also distinctly different from T. pustulata (S. pustulata). While we would agree that S. notophora sensu Sapra and Dass (1970) is different from the S. notophora described by Kahl (1932) we would argue that it is also distinctly different from T. pustulata (S. pustulata). The similarity between S. notophora sensu Sapra and Dass (1970) and T. saprai n. sp. suggests that in future all papers concerning S. notophora from India should be referring to T. saprai n. sp.
5 Conclusion
In light of the tools available to present day taxonomy, an integrated approach is required in order to arrive at confident identification of closely related species (Clamp and Lynn, 2017; Santoferrara and McManus, 2017; Sun et al., 2016; Warren et al., 2017). Comparisons are available of various species of Stichotrines based on molecular markers (Dai and Xu, 2011; Gao et al., 2016; Paiva et al., 2009; Schdmidt et al., 2007; Tamura et al., 2011), which also include species of Stylonychia and Tetmemena but in the absence of in-depth morphological and morphogenetic information, a detailed comparison with T. saprai n. sp. is not feasible. It is clear, therefore, that more data concerning the morphology, ontogenetic processes and more gene information of other species of are still required to achieve a comprehensive understanding for the whole genus Tetmemena.
Acknowledgements
This study was supported by the researchers supporting project number (RSP-2019/19) of King Saud University, Riyadh, Saudi Arabia. Also it was supported by the Junior Research Fellowships to JSA and SM from CSIR (Council of Scientific and Industrial Research), India and SS from UGC (University Grants Commission) India, respectively. Authors thank King Saud University, Saudi Arabia and the Principal, Acharya Narendra Dev College & Maitreyi College, University of Delhi for providing the necessary facilities infrastructure for the research and for the support and encouragement.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Techniques and tools for species identification in ciliates: a review. Int. J. Syst. Evol. Microbiol.. 2019;69:877-894.
- [Google Scholar]
- Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol.. 2019;65:623-649.
- [Google Scholar]
- Monograph of the Oxytrichidae (Ciliophora, Hypotrichia). Monographiae Biol.. Kluwer Academic Publishers; 1999. p. :1-722.
- Catalogue of Ciliate Names: Hypotrichs. Salzburg: Verlag Helmut Berger, Austria; 2001.
- Monograph of the Urostyloidea (Ciliophora, Hypotricha) (Monographiae Biologicae, vol. 85). Dordrecht: Springer; 2006.
- Monograph of the Urostyloidea (Ciliophora, Hypotricha). New York: Springer Science & Business Media; 2007.
- Monograph of the Amphisiellidae and Trachelostylidae (Ciliophora, Hypotricha). New York: Springer Science & Business Media; 2008.
- Monograph of the Gonostomatidae and Kahliellidae (Ciliophora, Hypotricha). New York: Springer Science & Business Media; 2011.
- Monograph of the Oxytrichidae (Ciliophora, Hypotrichia) (first ed.). New York: Springer Science & Business Media; 2012.
- Cladistic relationships and generic characterization of oxytrichid hypotrichs (Protozoa, Ciliophora) Arch. Protistenkunde.. 1997;148:125-155.
- [Google Scholar]
- Phylogenetic relationships within the class Spirotrichea (Ciliophora) inferred from small subunit rRNA gene sequences. Mol. Phylogenetics. Evol.. 2001;21:86-92.
- [Google Scholar]
- Revision of the order hypotrichida (Ciliophora, Protozoa) J. Protozool.. 1972;19:1-23.
- [Google Scholar]
- Pinocytosis of inorganic salts by Amoeba proteus (Chaos diffluens) C. R. Trav. Lab. Carlsberg. Chim.. 1958;31:77-92.
- [Google Scholar]
- Morphology, morphogenesis and small-subunit rRNAgene sequence of the novel brackish-water ciliate Strongylidium orientale sp. nov. (Ciliophora, Hypotrichia) Int. J. Syst. Evol. Microbiol.. 2013;63:1155-1164.
- [Google Scholar]
- The diverse morphogenetic patterns in spirotrichs and philasterids: researchers based on five-year-projects supported by IRCN-BC and NSFC. Eur. J. Protistol.. 2017;61:439-452.
- [Google Scholar]
- Investigating the biodiversity of ciliates in the ‘Age of Integration’. Eur. J. Protistol.. 2017;61:314-322.
- [Google Scholar]
- Taxonomy and phylogeny of Tunicothrix (Ciliophora, Stichotrichia), with description of two novel species, Tunicothrix brachystica n. sp. and Tunicothrix multinucleata n. sp., and the establishment of Parabirojimidae n. fam. Int. J. Syst. Evol. Microb.. 2011;61:1487-1496.
- [Google Scholar]
- Evolution of morphogenetic processes in the Orthoamphisiellidae n. fam., Oxytrichidae, and Parakahliellidae n. fam., and their depiction using a computer method (Ciliophora, Hypotrichida) J. Eukaryot. Microbiol.. 1997;44:553-573.
- [Google Scholar]
- Comparison of divisional morphogenesis in four morphologically different clones of the genus Gonostomum and update of the natural hypotrich system (Ciliophora, Hypotrichida) Eur. J. Protistol.. 1999;35:34-48.
- [Google Scholar]
- An update of ‘basic light and scanning electron microscopic methods for taxonomic studies of ciliated protozoa’. Int. J. Syst. Evol. Microbiol.. 2014;64:271-292.
- [Google Scholar]
- Reconciling classical and molecular phylogenies in the stichotrichines (Ciliophora, Spirotrichea), including new sequences from some rare species. Eur. J. Protistol.. 2004;40:265-281.
- [Google Scholar]
- Likelihood in Molecular Phylogenetics. London, Lausanne: The Natural History Museum; 2003.
- Phylogenetic consideration of two scuticociliate genera, Philasterides and Boveria (Protozoa, Ciliophora) based on 18S rRNA gene sequences. Parasitol. Int.. 2010;59:549-555.
- [Google Scholar]
- The all data based evolutionary hypothesis of ciliated protists with revised classification of the phylum Ciliophora (Eukaryota, Alveolata) Sci. Rep.. 2016;6:1-14.
- [Google Scholar]
- Stylonychia ammermanni sp. n., a new oxytrichid (Ciliophora: Hypotrichida) ciliate from the river Yamuna, Delhi, India. Acta Protozool.. 2001;40:75-82.
- [Google Scholar]
- BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids Symp. Ser.. 1999;41:95-98.
- [Google Scholar]
- Phylogenetic relationships among 28 spirotrichous ciliates documented by rDNA. Mol. Phylogenet. Evol.. 2003;29:258-267.
- [Google Scholar]
- Insights into the phylogeny of sporadotrichid ciliates (Protozoa, Ciliophora: Hypotricha) based on genealogical analyses of multiple molecular markers. Chin. J. Oceanol. Limnol.. 2011;29:96-102.
- [Google Scholar]
- Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria) Tierwelt Dtl.. 1932;25:399-650.
- [Google Scholar]
- Partial retention of parental ciliature during morphogenesis of the ciliate Coniculostomum monilata (Dragesco and Njiné 1971) Njiné 1978 (Oxytrichidae, Hypotrichida) Eur. J. Protistol.. 1990;25:264-278.
- [Google Scholar]
- Fine-tune investigations on three stylonychid (Ciliophora, Hypotricha) ciliates. Eur. J. Protistol.. 2016;56:200-218.
- [Google Scholar]
- Morphology and ontogenesis of Stylonychia (Metastylonychia) nodulinucleata nov. subgen. (Ciliophora, Hypotricha) from Australia. Eur. J. Protistol.. 2017;57:61-72.
- [Google Scholar]
- Multigene-based analysis of the phylogenetic evolution of Oligotrich ciliates, with consideration of the internal transcribed spacer 2 secondary structure of three systematically ambiguous genera. Eukaryot. Cell.. 2013;12:430-437.
- [Google Scholar]
- Morphogenesis and molecular phylogeny of a new freshwater ciliate, Notohymena apoaustralis n. sp. (Ciliophora: Oxytrichidae) J. Eukaryot. Microbiol.. 2013;60:455-466.
- [Google Scholar]
- The Ciliated Protozoa: Characterization, Classification, and Guide to the Literature (third ed.). New York: Springer Science & Business Media; 2008.
- The characterization of enzymatically 373 amplified eukaryotic 16 S–like rRNA– coding regions. Gene. 1988;71:491-499.
- [Google Scholar]
- Miller, M.A., Pfeiffer, W., Schwartz, T., 2010. In Proceedings of the Gateway Computing Environments Workshop (GCE). New Orleans, LA, pp. 1–8.
- Comparative phylogenetic study of stichotrichia (Alveolata: Ciliophora: Spirotrichea) based on 18S-rDNA sequences. Genet. Mol. Res.. 2009;8:223-246.
- [Google Scholar]
- RnaViz, a program for the visualization of RNA secondary structure. Nucleic Acids Res.. 1997;25:4679-4684.
- [Google Scholar]
- MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572-1574.
- [Google Scholar]
- Ronquist, F., Huelsenbeck, J., Teslenko, M., 2011. Draft MrBayes version 3.2 manual: Tutorials and model summaries, pp. 1–105.
- Integrating dimensions of biodiversity in choreotrichs and oligotrichs. Eur. J. Protistol.. 2017;61:323-330.
- [Google Scholar]
- Organization and development of the macronuclear anlage in Stylonychia notophora Stokes. J. Cell Sci.. 1970;6:351-363.
- [Google Scholar]
- Phylogeny of Stichotrichia (Ciliophora; Spirotrichea) reconstructed with nuclear small subunit rRNA gene sequences: discrepancies and accordances with morphological data. J. Eukaryot. Microbiol.. 2007;54:201-209.
- [Google Scholar]
- Shao, C., Pan, X., Jiang, J., Ma, H., Al-Rasheid, K.A.S., Warren, A., Lin, X., 2013. A redescription of the oxytrichid Tetmemena pustulata (Müller, 1786) Eigner, 1999 and notes on morphogenesis in the marine urostylid Metaurostylopsis salina Lei et al., 2005 (Ciliophora, Hypotrichia). Eur. J. Protistol. 49, 272–282.
- A general overview of the typical 18 frontal-ventral-transverse cirri Oxytrichidae s. l. Genera (Ciliophora, Hypotrichia) J. Ocean Univ. China. 2015;14:522-532.
- [Google Scholar]
- Stylonychia harbinensis sp. n., a new oxytrichid ciliate (Ciliophora, Hypotrichia) from the Heilongjiang Province, China. Protistology. 2004;3:219-222.
- [Google Scholar]
- Discovery of Stylonychia nodulinucleata sp. nov. (Ciliophora, Hypotrichida, Oxytrichidae) and comparison of its neighbouring species. Zool. Res.. 1993;14:10-14.
- [Google Scholar]
- An integrative approach to phylogeny reveals patterns of environmental distribution and novel evolutionary relationships in a major group of ciliates. Sci. Rep.. 2016;6:1-12.
- [Google Scholar]
- MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol.. 2011;28:2731-2739.
- [Google Scholar]
- Zur Kenntnis der vergleichenden Morphologie der hypotrichen Infusorien. Bih. K. Svensk Vetensk Akad. Handl.. 1900;26:1-31.
- [Google Scholar]
- A case study to estimate the applicability of secondary structures of SSU-rRNA gene in taxonomy and phylogenetic analyses of ciliates. Royal Swedish Acad. Sci.. 2015;44:574-585.
- [Google Scholar]
- Warren, A., Patterson, D.J., Dunthorn, M., Clamp, J.C., Achilles-Day, U.E.M., Aescht, E., Al-Farraj, S.A., Al-Quraishy, S., Al-Rasheid, K., Carr, M., Day, J.G., Dellinger, M., El-Serehy, H.A., Fan, Y., Gao, F., Gao, S., Gong, J., Gupta, R., Hu, X., Kamra, K., Langlois, G., Lin, X., Lip-scomb, D., Lobban, C.S., Luporini, P., Lynn, D.H., Ma, H., Macek, M., Mackenzie-Dodds, J., Makhija, S., Mansergh, R.I., Martín-Cereceda, M., McMiller, N., Montagnes, D.J.S., Niko-laeva, S., Ong’ondo, G.O., Pérez-Uz, B., Purushothaman, J., Quintela-Alonso, P., Rotterová, J., Santoferrara, L., Shao, C., Shen, Z., Shi, X., Song, W., Stoeck, T., La Terza, A., Vallesi, A., Wang, M., Weisse, T., Wiackowski, K., Wu, L., Xu, K., Yi, Z., Zufall, R., Agatha, S., 2017. Beyond the “Code”: A Guide to the Description and Documentation of Biodiversity in Ciliated Protists (Alveolata, Ciliophora). J. Eukaryot. Microbiol. 64, 539–554.
- Morphological, biometric and morphogenetic comparison of two closely related species, Stylonychia vorax and S. pustulata (Ciliophora: Oxytrichidae) J. Protozool.. 1985;32:261-268.
- [Google Scholar]
- Molecular phylogeny of Pseudokeronopsis (Protozoa, Ciliophora, Urostylida), with reconsideration of three closely related species at inter- and intra-specific levels inferred from the small subunit ribosomal RNA gene and the ITS1-5.8S-ITS2 region sequences. J. Zool.. 2008;275:268-275.
- [Google Scholar]
- Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res.. 2003;31:3406-3415.
- [Google Scholar]







