Translate this page into:
Delivery of siRNAs against MERS-CoV in Vero and HEK-293 cells: A comparative evaluation of transfection reagents
⁎Corresponding author at: King Fahd Medical Research Center, King Abdulaziz University, Post Box No- 80216, Jeddah 21589, Saudi Arabia. ssohrab@kau.edu.sa (Sayed Sartaj Sohrab)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Background
A new coronavirus was identified in Jeddah, Saudi Arabia in 2012 and designated as Middle East Respiratory Syndrome Coronavirus (MERS-CoV). To date, this virus has been reported in 27 countries. The virus transmission to humans has already been reported from camels. Currently, there is no vaccine or antiviral therapy available against this virus.
Methods
The siRNAs were in silico predicted, designed, and chemically synthesized by using the MERS-CoV-orf1ab region as a target. The antiviral activity was experimentally evaluated by delivering the siRNAs with Lipofectamine™ 2000 and JetPRIMER as transfection reagents in both Vero cell and HEK-293-T cell lines at two different concentrations (10.0 nM and 5.0 nM). The Ct value of quantitative Real-Time PCR (qRT-PCR) was used to calculate and determine the reduction of viral RNA level in both cell supernatant and cell lysate isolated from both cell lines.
Results
The sequence alignment resulted in the selection of highly conserved regions. The orf1ab region was used to predict and design the siRNAs and a total of twenty-one siRNAs were finally selected from four hundred and twenty-six siRNAs generated by online software. Inhibition of viral replication and significant reduction of viral RNA was observed against selected siRNAs in both cell lines at both concentrations. Based on the Ct value, the siRNAs # 11, 12, 18, and 20 were observed to be the best performing in both cell lines at both concentrations.
Conclusion
Based on the results and data analysis, it is concluded that the use of two different transfection reagents was significantly effective. But the Lipofectamine™ 2000 was found to be a better transfection reagent than the JetPRIMER for the delivery of siRNAs in both cell lines.
Keywords
In silico prediction
MERS-CoV
siRNAs
Vero Cells
HEK-293-T cells
Saudi Arabia
- MERS-CoV
-
Middle East Respiratory Syndrome Coronavirus
- qRT-PCR
-
quantitative Real-Time PCR
- siRNAs
-
short interfering RNAs
Abbreviations
1 Introduction
Coronaviruses are well-known for respiratory illness in both humans and animals. A novel Coronavirus, known as Middle East Respiratory Syndrome Coronavirus (MERS-CoV) was identified in 2012 from a hospitalized patient from Jeddah, Saudi Arabia. This is the sixth human pathogenic coronavirus that had significant genomic sequence similarity with SARS-CoV to cause disease in humans and animals. The infected patient developed severe pneumonia symptoms followed by death after 11 days of hospitalization (Zaki et al. 2012). The infected persons develop variable symptoms like shortness of breath, fever, and in severe cases, multiorgan failure (Assiri et al. 2013; Yin and Wunderink 2018). Currently, this virus has spread to 27 countries with 2591 confirmed cases and 894 deaths, and a mortality rate up to greater than 35 % including WHO; (last Accessed on 10.9.2022; https://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html) and became a global threat to the human population (Chafekar and Fielding 2018; WHO 2022; Zaki et al. 2012). The dromedary camels are known as the main source of virus spread and their role in infection to humans have been reported (Azhar et al. 2014; Lee and Wong 2015; Memish et al. 2014; Oboho et al. 2015). However, camel workers are also known as an intermediary source of the virus spread to humans. The source of infection remains uncertain as some of the infected patients had no history of close contact with camel (Alshukairi et al. 2018). The MERS-CoV belongs to the family Coronaviridae and the genome of coronaviruses is single-stranded positive sense RNA with an approximately 25–32 kb genome size. The virus has been divided into Alpha, Beta, Gamma, and Delta coronaviruses groups and MERS-CoV belongs to the lineage - C Betacoronavirus (βCoVs). As it has been reported that coronaviruses have a very high rate of mutation, recombination, and sequence diversity which favors the new virus strain and isolates emergence with novel features and characteristics (Al-Omari et al. 2019).
Currently, no vaccines or antiviral therapy is available for MERS-CoV but many therapeutic compounds and vaccines are under various stages of an investigation, and few have reached an advanced stage with promising results. The role of RNA interference (RNAi) has shown significant antiviral activity against many viruses as well as other pathogens and diseases by using short interfering RNAs (siRNAs) and micro-RNA (miRNAs). Long noncoding RNAs (lncRNAs) against cancers (Mahmoodi et al. 2019; Hattab et al. 2021), bacterial infections (Menanteau-Ledouble et al. 2020) fungal infections (Bruch et al., 2022; Wang et al. 2022), parasitic infections (Somarathne et al. 2018; Portet 2021), viral infections (Levanova et al. 2018). Several potential RNA interference-based (RNAi) drugs have been recently reported (Setten et al.2019). Additionally, the clustered regularly interspaced short palindromic repeats (CRISPR-Cas) system was identified in 2005 as an adaptive immune system against viral and plasmid infections and currently it has been divided into two classes, class 1 and class 2 (Escalona-Noguero et al. 2021). The details and effective use of CRISPR have been recently proposed as a potential therapeutic tool for the treatment of viral diseases (Baddeley et al., 2021; Kong et al. 2021; Lin et al. 2021; Najafi et al.2022).
The genome-wide molecular screening and bioinformatics approaches have provided a platform to predict, design, and filter the potential siRNAs, shRNA, and miRNAs against various diseases (Levanova and Poranen 2018; Setten et al. 2019). Many siRNAs and miRNAs have been designed in silico and evaluated experimentally in more than 20 clinical trials against viral diseases like HIV, Flock house virus (FHV), DENV, HBV, HCV, HPV, Influenza, SARS-CoV, SARS-CoV-2, & MERS-CoV and shown promising results (Fakhr et al. 2016; Hasan et al. 2014; Huang et al. 2017; Idrees and Ashfaq 2013; Kumar et al. 2013; Liu et al. 2017; Nur et al. 2015; Shahid et al. 2017; Sohrab et al. 2021; Sohrab, Aly El-Kafrawy, et al. 2020a, 2020b; Sohrab et al. 2018; Taning et al. 2018; Tsai et al. 2018; Wang et al. 2016; Zeng et al. 2017; Zhang and Lu 2020). As per the status and information, we designed this study to conduct the in-silico prediction, designing, and experimental evaluation of potential siRNAs at 5 and 10 nM concentrations by using two different transfection reagents in Vero and HEK-293-T cell lines.
2 Materials and methods
2.1 Sequence retrieval and analysis
The MERS-CoV (Human/Camels) genome was retrieved from NCBI-PubMed. The analysis of the genome sequence was performed by using the online software BioEdit (Version 7.2). The multiple sequence alignment was done using ClustalW. As it has been reported that the orf1ab region plays an important role in virus replication. Based on the multiple sequence alignment and homology, the orf1ab region was selected as the target for siRNAs design.
2.2 Designing, filtration, and chemical synthesis of siRNAs
The multiple sequence alignment homology provided the selection of orf1ab as a target for the prediction, designing, and filtration of probable potential siRNAs. We have used an online integrated bioinformatics approach for the prediction, designing, and filtration of potential siRNAs as per the guidelines for the strict criteria of selection and filtration (ElHefnawi et al. 2016; Fakhr et al. 2016; Hasan et al. 2014; Naito and Ui-Tei 2012; Nur et al. 2015; Sohrab et al. 2021; Sohrab, Aly El-Kafrawy, et al. 2020a, 2020b; Sohrab et al. 2018). By applying the criteria for selection, we have filtered only twenty-one siRNAs for their in-vitro evaluation study. Integrated DNA Technologies (IDT-USA) was used for the chemical synthesis of selected siRNAs.
2.3 Cytotoxicity assay
The cytotoxicity of designed and synthesized siRNAs was evaluated and determined in both cells by using the Invitrogen™ CyQUANT™ MTT Cell Viability Assay following the manufacturer’s instruction. The absorbance was measured at 570 nm using a SpectraMax i3x imaging cytometer and the mean OD value was used for cytotoxicity calculation using the standard formula.
2.4 Experimental evaluation of siRNAs against MERS-CoV
The experimental evaluation of chemically synthesized siRNA was performed in triplicates at two different concentrations (10.0 nM and 5.0 nM) in selected cell lines (Vero cells and HEK-293-T cells) by using two different transfection reagents. The transfection reagents were selected based on their transfection efficiency. The first one was Lipofectamine™ 2000 (ThermoFisher Scientific, USA). According to its manufacturer, Lipofectamine reagents have become the most referenced transfection reagents since their launch in 1993. They are therefore considered the goldstandard of transfection reagents and are used as a basis of comparison for efficiencies of other transfection methods. Lipofectamine™ 2000 is a versatile transfection reagent that has been shown to effectively transfect the widest variety of adherent and suspension cell lines. This is being used and works effectively with common cell lines and many challenging ones. The second one was JetPRIMER ((Polyplus, France), which is a non-liposomal, polymer-based transfection reagent. This is cost-effective and is widely used for many siRNA and DNA delivery in many cell lines with better cell viability and higher transfection efficiency. The virus replication inhibition and reduction of viral RNA were determined by the Ct value of qRT-PCR with proper negative and positive control.
2.5 Delivery of siRNAs by Lipofectamine 2000 and Jet prime and virus inoculation
The siRNAs were delivered through reverse transfection method by using Lipofectamine™ 2000 and JetPRIMER transfection reagent into grown Vero and HEK-293-T cells (60–80 % confluency (1x104) at various concentrations (10.0 nM and 5.0 nM). Briefly, 50 μM siRNAs stocks were diluted to various concentrations (10.0 and 5.0 nM) in 100 μl Opti-MEM medium by adding Lipofectamine™ 2000 as well as JetPRIMER following incubation at Room temperature for 30 min. The siRNA-lipid complex (1 μl) was added to the grown cells at various siRNA concentrations (10.0 nM-5.0 nM) and mixed gently and incubated at 37 °C for 72 h. The transfected cells were grown for 24 h at 37 °C and then MERS-CoV at 0.01 MOI was used for inoculation following the published protocol from our lab (Azhar et al. 2014) and cells were incubated for 1 h. The siRNA-transfected and virus-inoculated cells were replenished with fresh DMEM and further grown for seventy-two hours. All the experiments were performed in triplicates with proper negative and positive control. The virus-infected cells were treated as a positive control, while the non-infected cells were treated as a negative control. The cytopathic effect (CPE) in both cells was observed daily for 72 hrs. and after full CPE, the cells were harvested, and the viral RNA was purified from both cell lysate and supernatant using the commercial QIAmp Viral RNA Mini Kit (Qiagen, USA) as per kit instructions.
2.6 Confirmation of virus inhibition by quantitative-Real-Time PCR
The antiviral potency of siRNAs and inhibition of virus replication and reduction of viral RNA level was determined by quantitative-Real-Time PCR (qRT-PCR) using the MERS-CoV primers as described earlier (Azhar et al. 2014). The Ct value of qRT-PCR was used to analyze the inhibitory effect of each siRNAs in Vero and HEK-293-T in cell supernatant as well as cell lysate at selected concentrations (10.0 nM-5.0 nM).
3 Results
3.1 Sequence analysis
The multiple sequence alignment results of the MERS-CoV full genome showed significant similarities at various locations. Based on the role of orf1ab in the virus replication process and high sequence homology, we selected this region as a target for in silico prediction, designing, and filtration of potential siRNAs. Fig. 1 shows the sequence similarity with the MERS-CoV-orf1ab region of human and camel isolates.
Multiple sequence alignmnet of MERS-CoV-orf1ab gene from Human and camels isolates.
3.2 In silico prediction and chemical synthesis of siRNAs
The software generated a total of four hundred and sixty-two siRNAs from the or1ab gene of MERS-CoV, but we have selected only twenty-one siRNAs based on their strict criteria for selection and filtration with no off-target, and no match with human mRNA sequences (Fakhr et al. 2016; Naito and Ui-Tei 2012; Sohrab, Aly El-Kafrawy, et al. 2020a, 2020b; Sohrab et al. 2018). The selected siRNAs were chemically synthesized by Integrated DNA Technologies (IDT), USA, and used for experimental evaluation in selected cells. The predicted siRNAs were expected to be highly specific and potent against the orf1ab gene of MERS-CoV. The predicted siRNAs have been listed in Table 1.
S.N.
Target sequence
Predicted RNA oligo sequences
(5′→3′)
1
AGCAATCTATTTTTACTATTAAT
UAAUAGUAAAAAUAGAUUGCU
CAAUCUAUUUUUACUAUUAAU
2
ATGGATAATGCTATTAATGTTGG
AACAUUAAUAGCAUUAUCCAU
GGAUAAUGCUAUUAAUGUUGG
3
GCGACTTTATGTCTACAATTATT
UAAUUGUAGACAUAAAGUCGC
GACUUUAUGUCUACAAUUAUU
4
GACACTTTAGATGATATCTTACA
UAAGAUAUCAUCUAAAGUGUC
CACUUUAGAUGAUAUCUUACA
5
ATGCTATTAGTTTGAGTTTTAAT
UAAAACUCAAACUAAUAGCAU
GCUAUUAGUUUGAGUUUUAAU
6
TGCTATTAGTTTGAGTTTTAATA
UUAAAACUCAAACUAAUAGCA
CUAUUAGUUUGAGUUUUAAUA
7
GAGCTAGTTTGCGTCAAATTTTT
AAAUUUGACGCAAACUAGCUC
GCUAGUUUGCGUCAAAUUUUU
8
CTCTAATATCTTTGTTATTAACA
UUAAUAACAAAGAUAUUAGAG
CUAAUAUCUUUGUUAUUAACA
9
CTCTTAGAAACTCTTTAACTAAT
UAGUUAAAGAGUUUCUAAGAG
CUUAGAAACUCUUUAACUAAU
10
TGGTTTGATTTTGTTGAAAATCC
AUUUUCAACAAAAUCAAACCA
GUUUGAUUUUGUUGAAAAUCC
11
ACGCAAATTGCGTTAATTGTACT
UACAAUUAACGCAAUUUGCGU
GCAAAUUGCGUUAAUUGUACU
12
TGGTATCTAAAGGTTTCTTTAAG
UAAAGAAACCUUUAGAUACCA
GUAUCUAAAGGUUUCUUUAAG
13
GTCTTGTATTCGGCTTATACAAG
UGUAUAAGCCGAAUACAAGAC
CUUGUAUUCGGCUUAUACAAG
14
TCCTTCTATAGTTGAATTTAATA
UUAAAUUCAACUAUAGAAGGA
CUUCUAUAGUUGAAUUUAAUA
15
GTCTACAATAATAAATTGTTAGC
UAACAAUUUAUUAUUGUAGAC
CUACAAUAAUAAAUUGUUAGC
16
AACAACATTAACAGATTTAATGT
AUUAAAUCUGUUAAUGUUGUU
CAACAUUAACAGAUUUAAUGU
17
CTCTACAATTAGGATTTTCAACT
UUGAAAAUCCUAAUUGUAGAG
CUACAAUUAGGAUUUUCAACU
18
TTGTATAAGAAAGTCAATAATGA
AUUAUUGACUUUCUUAUACAA
GUAUAAGAAAGUCAAUAAUGA
19
CTCAACTATTCATAACTATTTTA
AAAUAGUUAUGAAUAGUUGAG
CAACUAUUCAUAACUAUUUUA
20
TGCCAATATGCGTGTTATACATT
UGUAUAACACGCAUAUUGGCA
CCAAUAUGCGUGUUAUACAUU
21
GGGTACTATTAAAGAAAATATAG
AUAUUUUCUUUAAUAGUACCC
GUACUAUUAAAGAAAAUAUAG
3.3 Cytotoxicity Assay
The cytotoxicity of selected siRNAs in both cells was determined by using both Lipofectamine™ 2000 and JetPRIMER transfection reagent. Based on the results obtained in this study, no cytotoxicity was observed for any siRNAs in both cells at tested concentrations.
3.4 Experimental evaluation of siRNAs against MERS-CoV
The in-vitro evaluation of siRNAs was performed by using Lipofectamine™ 2000 and JetPRIMER as transfection reagents to Vero and HEK-293 cell lines. The inhibition of virus replication and the reduction of viral RNA were determined by the Ct value of qRT-PCR performed by using the cell supernatant and lysate for all selected siRNAs. The better inhibition of virus replication and reduction of viral RNA level in cell supernatant as well as lysate was observed in Vero cells by Lipofectamine™ 2000 at both concentrations (10.0 nM and 5.0 nM) of siRNAs tested than the JetPRIMER transfection reagent.
By using the JetPRIMER as a transfection reagent, the Ct value of qRT-PCR was observed to be variable in both cells. The Ct value has been presented in Table 2 and Fig. 2. The inhibition of MERS-CoV replication was observed to be comparable with the dose-dependent in Vero cells as well as HEK-293-T cells at both concentrations of siRNAs. In Vero cells supernatant, the highest Ct value of qRT-PCR was observed with siRNAs#12 (22.98/22.70 (at 10.0 nM /5.0 nM)) followed by siRNA#18 (19.96/20.99) and siRNA#20 (17.22/17.35). The Ct value in cell lysate also varied significantly as compared to the control group. The highest Ct value was observed in siRNA#11 (22.93/22.66) followed by siRNA#20 (22.91/22.65) and siRNA#18 (21.73/22.55). Interestingly, the Ct value of most of the siRNAs was significantly better in cell lysate than the cell supernatant at both concentrations as well as the higher Ct value was observed at 5.0 nM, which indicates that the lower concentration is more effective than the higher concentration of siRNAs tested (Table 2).
siRNAs combinations
Vero cells
HEK-293-T cells
(Cell Supernatant)
(nM)(Cell Lysate)
(nM)(Cell Supernatant)
(nM)(Cell Lysate)
(nM)
10.0
5.0
10.0
5.0
10.0
5.0
10.0
5.0
siRNA1
16.00
16.95
15.81
15.75
34.49
33.31
36.50
36.13
siRNA 2
16.57
16.95
15.95
15.71
34.14
34.24
35.85
35.99
siRNA3
14.54
14.81
15.21
15.35
34.93
33.89
35.48
36.59
siRNA4
16.61
16.84
16.49
15.98
33.25
33.76
33.87
35.53
siRNA5
16.09
16.75
16.11
15.98
33.75
33.99
34.32
33.74
siRNA6
15.21
14.12
16.64
16.28
33.79
33.80
34.78
35.75
siRNA7
15.97
16.78
15.35
15.87
35.94
36.85
35.97
34.76
siRNA8
17.46
17.95
15.94
15.79
34.65
33.84
35.89
35.23
siRNA9
17.89
17.91
16.23
16.87
34.46
34.14
34.96
34.11
siRNA10
17.51
17.96
16.95
16.90
33.85
33.10
35.47
35.21
siRNA11
17.53
19.67
22.93
22.66
35.93
35.71
35.32
35.61
siRNA 12
22.98
22.70
20.21
21.13
35.78
35.97
37.60
37.78
siRNA 13
14.22
14.59
20.64
20.06
34.74
35.63
36.82
36.48
siRNA14
15.29
19.86
20.55
21.45
34.86
36.61
35.72
35.76
siRNA15
15.00
14.10
22.85
21.43
34.55
36.06
35.27
36.50
siRNA16
15.05
16.45
21.8
20.82
33.76
35.91
35.84
36.48
siRNA17
15.67
15.11
22.46
21.49
33.08
35.67
34.19
35.81
siRNA18
19.96
20.99
21.73
22.55
35.88
35.81
37.34
37.99
siRNA19
16.21
16.53
21.66
21.80
39.61
39.68
36.73
36.87
siRNA20
17.22
17.35
22.91
22.65
35.64
35.97
35.97
35.70
siRNA 21
15.49
16.97
20.06
21.90
35.82
36.71
34.87
34.61
Positive Control
15.99
16.95
15.81
15.75
33.32
33.12
34.72
33.24
Negative Control
80.90
80.78
80.75
80.71
90.10
90.15
90.18
90.16
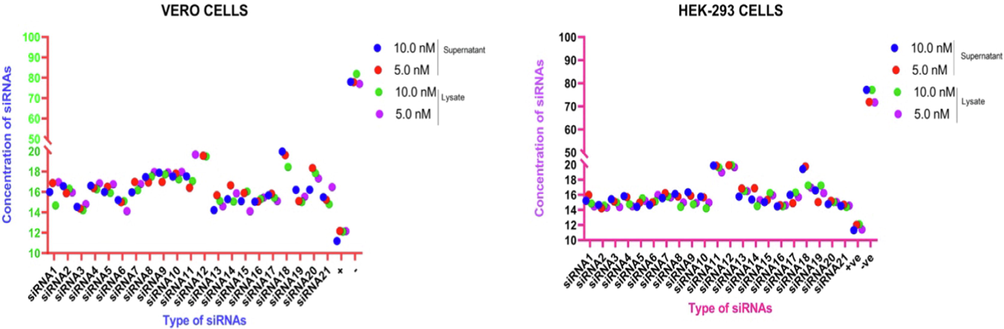
Graphical representation of Ct value of qRT-PCR of siRNAs delivered by JetPRIMER in Vero cells and HEK-293-T cells.
In HEK-293-T cells transfected with Jet prime transfection reagent, the Ct value of qRT-PCR for each siRNAs was variable in cell supernatant as well as cell lysate at both concentrations. In cell supernatant, the highest Ct value was observed with siRNA#19 (39.61/ 39.68 (at 10.0 nM /5.0 nM)) followed by siRNA#7 (35.94/36.85) and siRNA#21 (35.82/36.71). The cell lysate showed the variable Ct value for all the tested siRNAs. The highest Ct value was observed with siRNA#12 (37.60/37.78) followed by siRNA#18 (37.34/37.99), siRNA#13 (36.82/36.48), and siRNA#19 (36.73/36.87) at both concentrations (Table 2).
In the case of Lipofectamine™ 2000 as a transfection reagent, the qRT-PCR results were variable in both Vero and HEK-293-T cells. The results of the Ct value for each siRNAs have been presented in Table 3 and Fig. 3. The level of viral RNA was reduced as indicated by the Ct value which indicates the inhibition of virus replication in both cells as compared to the control. The cell supernatant isolated from Vero cells showed the high Ct value of many siRNAs at both concentrations. The highest Ct value in cell supernatant was observed to be 20.18 for siRNA#12 at 10.0 nM and 19.56 at 5.0 nM followed by 19.96 and 19.62 for siRNA#18 and 17.89 and 16.98 for siRNA#9. While the Ct value was higher (>15) for siRNAs#1, siRNAs#2, siRNAs#4–12, and siRNAs#14–21. The cell lysate of Vero cells also showed a better Ct value as compared to the positive control. The highest Ct value of siRNA#12 was 19.49/20.70 (at 10.0 nM /5.0 nM) followed by siRNA#18, 18.44/20.19, 17,83/17.33 for siRNA#20. The siRNA#8–10 showed almost similar Ct values ranging from 17.23 to 17.96 at both concentrations.
siRNAs combination
Vero cells
HEK-293-T cells
(Cell Supernatant)
(nM)(Cell Lysate)
(nM)
(Cell Supernatant)
(nM)
(Cell Lysate)
(nM)
10.0
5.0
10.0
5.0
10.0
5.0
10.0
5.0
siRNA1
15.99
16.87
15.97
16.95
15.21
15.98
14.83
14.48
siRNA 2
16.57
16.98
16.31
15.95
14.65
14.21
14.56
14.31
siRNA3
14.54
14.38
15.21
15.81
15.41
15.12
14.99
14.38
siRNA4
16.31
16.89
16.26
16.84
15.82
15.71
14.75
14.50
siRNA5
15.99
16.95
15.90
16.75
14.43
14.94
15.52
15.21
siRNA6
15.21
14.99
15.10
14.12
14.65
14.98
15.13
15.98
siRNA7
15.97
16.98
16.17
16.78
15.56
16.21
15.78
15.67
siRNA8
17.46
16.93
17.50
17.95
16.12
15.78
14.91
14.99
siRNA9
17.89
16.98
17.71
17.91
16.34
15.89
14.75
14.91
siRNA10
17.51
17.81
17.23
17.96
15.75
15.67
14.21
14.97
siRNA11
17.53
16.41
19.06
19.67
19.89
19.86
19.63
18.98
siRNA 12
20.18
19.56
19.49
20.70
20.61
19.92
19.93
19.66
siRNA 13
14.22
15.67
15.12
14.59
15.78
16.83
16.50
16.49
siRNA14
15.29
16.65
15.06
15.86
15.37
16.87
14.52
15.29
siRNA15
15.10
15.89
16.04
14.10
15.03
15.30
16.22
15.93
siRNA16
15.05
15.06
15.30
15.45
14.47
14.62
14.49
14.63
siRNA17
15.67
15.82
15.42
15.11
15.99
14.89
16.30
15.71
siRNA18
19.96
19.62
18.44
20.19
19.41
19.75
17.25
17.96
siRNA19
16.21
15.10
15.01
15.53
16.59
15.03
17.24
16.22
siRNA20
16.22
18.34
17.83
17.33
15.76
15.20
15.03
15.06
siRNA 21
15.49
15.23
14.79
16.47
14.54
14.69
14.38
14.57
Positive Control
11.19
11.17
12.10
12.15
11.32
12.00
12.09
12.41
Negative Control
78.01
77.82
81.92
77.01
77.11
71.90
77.12
71.72
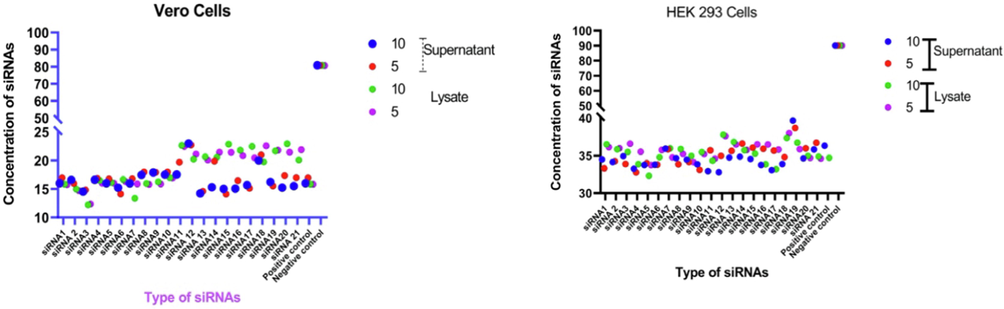
Graphical representation of Ct value of qRT-PCR of siRNAs delivered by Lipofectamine™ 2000 in Vero cells and HEK-293-T Cells.
The HEK-293-T cells also showed the variations in Ct value of many siRNAs in both cell supernatant and cell lysate. The highest Ct value in cell supernatant was observed to be 20.61/19.92 (at 10.0 nM /5.0 nM) for siRNA#12 followed by 19.89/19.86 for siRNA#11 and 19.41/19.65 for siRNA#18. A higher Ct value, ≤15 was observed in many siRNAs tested in cell supernatant. The cell lysate also showed the variation in Ct value in many siRNAs at both concentrations. The highest Ct value was 19.93/19.66 for siRNA#12 followed by 19.63/18.98 for siRNA#12 and 17.25/17.96 for siRNA#18. Only 3 siRNAs that showed a higher Ct value of more than 16 and the remaining were more than 14 Ct values as compared to the positive control group. The significant variation of Ct value was observed in many siRNAs at a lower concentration as compared to positive control which indicates that the better inhibition of virus replication resulted in the lower level of viral RNA in both cell supernatant and cell lysate in both cells.
4 Discussion
The new virus was identified in 2012 from a hospitalized patient in Jeddah, Saudi Arabia, and based on the novel characters, properties, and sequence homology with another known coronavirus, it was finally designated as MERS-CoV. Since 2012, this virus has been reported from 27 countries with over 2591 confirmed cases and 894 deaths (last Accessed on 10.9.2022 https://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html). Due to the status of the virus spread and reports from various locations globally, with tremendous efforts, significant progress has been made with valuable information published about the MERS-CoV. But still, there is no USFDA-approved vaccine or antiviral therapy available for MERS-CoV. Many vaccines and antiviral therapies are under the various stage of investigation and some of them have reached an advanced stage including oligonucleotide-based therapy (siRNAs/miRNAs) based therapy (Folegatti et al. 2020; Hashem et al. 2019; Li et al. 2020; Mubarak et al. 2019; Xu et al. 2019; Zhou et al. 2019). This RNAi-based approach has emerged as an alternative therapy against many deadly diseases including viral-mediated (Carneiro et al. 2015; Chakraborty et al. 2017; Moon et al. 2016). The oligonucleotide-based therapy includes the use of siRNA/miRNA/shRNAs and the ALNRSV01 was the first siRNA that was documented for human use (Levanova and Poranen 2018). In silico guided experimental evaluation against MERS-CoV has been recently described with promising results which identified the potential siRNAs (ElHefnawi et al. 2016; Fakhr et al. 2016; Hasan et al. 2014; Nur et al. 2015; Sohrab et al. 2021; Sohrab, Aly El-Kafrawy, et al. 2020a, 2020b; Sohrab et al. 2018). Additionally, a similar strategy was used to identify, design, and evaluate the siRNAs against the newly emerged SARS-CoV-2, and some of them were found to be potentially effective (Sohrab et al. 2021).
Similar RNAi technology can be used to design, and filter by integrated bioinformatics approach, and experimentally evaluated against MERS-CoV. The replication of MERS-CoV is mediated by the orf1ab gene and the attachment with the host cell is mediated by Spike (S) protein gene. The inhibition of virus replication can be inhibited in many alternative ways including the use of RNAi technology applying the use of siRNAs. The orf1ab region includes two-thirds of the Coronavirus genome and encodes non-structural proteins. Very few siRNAs have been designed by using in-silico software but none of them have been evaluated in cell lines (Hasan et al. 2014; Nur et al. 2015). A few studies have been conducted on the in silico designing and experimental evaluation of siRNAs against HCV and MERS-CoV and some siRNAs were observed to be potentially effective and inhibited the virus replication resulting in the reduction of viral RNA level in cell lysate and supernatant. The reduction of viral RNA level was determined by the Ct value of qRT-PCR (El Hefnawi et al. 2016; Sohrab et al. 2021; Sohrab, Aly El-Kafrawy, et al. 2020a, 2020b).
In this study, we have discussed the in-silico prediction, designing, and experimental evaluation of siRNAs against MERS-CoV delivered by two different transfection reagents, namely, Lipofectamine™ 2000 and JetPRIMER in Vero cells and HEK-293-T cell lines. A total of four hundred and sixty-two siRNAs from the orf 1ab genome were generated by online software (Fakhr et al. 2016; Sohrab et al. 2018) but only twenty-one siRNAs were selected and chemically synthesized and further used. The synthesized siRNAs were delivered by Lipofectamine™ 2000 and JetPRIMER for the experimental evaluation of the reduction of viral RNA by using the two different concentrations in both Vero cells and HEK-293-T cell lines. The results obtained from this work provided a significant reduction of viral RNA as determined by the Ct value of qRT-PCR performed by using both cell supernatant and lysate of both Vero cells and HEK-293-T cells. The use of two different transfection reagents for the delivery of siRNA in two different cells was almost similar at both tested concentrations. But based on the Ct value of each siRNAs as compared to the control group, the Lipofectamine™ 2000 was better as compared to JetPRIMER and Vero cells were better than HEK-293-T cells for the in-vitro evaluation of siRNAs against MERS-CoV. This variation could be due to better growth and multiplication of viruses in Vero cells. During data analysis, we observed that some siRNAs (siRNA# 11, 12, 18 and, 20) delivered by either Lipofectamine™ 2000 or JetPRIMER showed the best Ct value and were common in both cell supernatant and lysate collected from both cell lines at both concentrations of siRNAs.
The siRNA#9 and siRNA#12 were observed to be the best-performing siRNAs at 10.0 nM and 5.0 nM concentrations in both cell lysate and the supernatant collected from the Vero cells. While siRNA# 12 and siRNA#19 were observed to be the best for HEK-293-T cells delivered by JetPRIMER transfection reagent. Based on the Ct value, the siRNAs (#18 and 20) were better performing siRNAs in Vero cell lines, while siRNA#13, and siRNA#16 were better in HEK-293-T cells at both concentrations in both cell supernatant and cell lysate. The siRNA#11,12,18, and 20 are the best performing siRNAs in both cell lines delivered by JetPRIMER transfection reagent.
The data analysis of siRNAs delivered by Lipofectamine™ 2000 as a transfection reagent, revealed that the siRNA# 8, 11, 12, 18, and 20, were the best performing siRNAs at 10.0 nM and 5.0 nM concentration as per their Ct value in both cell supernatant and cell lysates collected from Vero cell lines while the siRNA# 8, 11, 12, 13, 15, 18 and 20 were best in HEK-293-T cell lines. The siRNAs # 8, 11, 12 18 and, 20 were common for both and the best-performing siRNAs in both cell lines at both concentrations and in cell supernatant and cell lysate. The better performing siRNAs# 1, 2, 4, and 5 in the Vero cell line while siRNAs# 7, 19, and 20 were the better performing siRNAs in HEK-293-T cell lines in both cell supernatant and cell lysate at both 10.0 nM and 5.0 nM concentrations. Additionally, it was also observed that there were many siRNAs that showed higher Ct values as compared to the positive control group delivered by both Lipofectamine™ 2000 and JetPRIMER transfection reagent at both concentrations in both cell supernatant and cell lysate isolated from both cell lines. The siRNAs#2,8,10, 11, 12 13, 14, 15,16,18 19 and, 20 were common and showed higher Ct values than the positive control in both cell lines at both concentrations (10.0 nM and 5.0 nM) delivered by Jet Prime while the siRNAs# 1,2,4,5, 8, 9, 10,11,12, 18 and 20 were showed higher Ct value than positive control in Vero cells while, only siRNAs# 8,11, 12,13,15, 18 and 20 were with higher Ct value than the positive control group. Based on the above findings, we observed that the many siRNA was found to be potentially active to inhibit the replication/multiplication of the virus that indicated low RNA level in cell lines which resulted in higher Ct value than the positive control at both concentrations of siRNAs in qRT-PCR analysis in both cell supernatant and cell lysate isolated from Vero and HEK-293-T cell lines. However, better inhibition of virus replication was observed in Vero cells as compared to HEK-293-T cell lines in both siRNAs’ transfection reagents tested.
The findings of this study are supported by other recent publications (El-Kafrawy et al. 2021; Sohrab et al. 2021; El-Kafrawy, et al. 2020). In a study, it was observed that siRNA#1 and 4 were found to be potentially effective to inhibit the MERS-CoV replication in Vero cells (Sohrab 2021; Aly El-Kafrawy, et al. 2020). While in another study conducted on HEK-293 cells, the siRNAs# 1, 2, 4, 6, and 9 were found to be effective against MERS-CoV replication inhibition at various concentrations, delivered by Lipofectamine™ 2000 as transfection reagent (Sohrab et al. 2021). Additionally, in another study conducted on the Huh-7 cells line, the siRNAs# 2,6,16 and 19 were the best-performing siRNAs at various concentrations tested in both cell supernatant and cell lysate (El-Kafrawy et al. 2021). The data generated after the result analysis from this work encouraged us to evaluate these siRNAs in multiple cell lines against other coronaviruses. The evaluation of these siRNA alone or in combinations will provide a clear understanding of the potential use of siRNA as oligonucleotide-based antiviral therapeutics not only against MERS-CoV but other coronaviruses. The limitation of this study was that the evaluation of the siRNAs was conducted in only selected cell lines because the MERS-CoV does not grow and multiply in most other cell lines. This study requires a long time of work in the BSL3 lab only. The findings of this study should be further evaluated on mice and other human primates which is lacking here in our facility.
5 Conclusion
The results and data analysis from this study provided a clear observation that the use of two different transfection reagents significantly affected the delivery of siRNAs in two different cell lines which resulted in the reduction of viral RNA level as determined by the Ct value of qRT-PCR. A better reduction of viral RNA was observed in Vero cell lines than the HEK-293-T cell lines by Lipofectamine™ 2000 as compared to JetPRIMER at both concentrations of siRNAs.
Acknowledgments
This study was financially supported by King Abdulaziz City for Science and Technology (KACST), Riyadh, Saudi Arabia, by providing the special grant on MERS-CoV (Project # 39-2). Therefore, all authors are grateful for the financial support from KACST, Riyadh, Saudi Arabia. The authors would also like to gratefully acknowledge the research facility provided by Special Infectious Agents Unit (SIAU), King Fahd Medical Research Centre (KFMRC), King Abdulaziz University, Jeddah, Saudi Arabia.
Author contributions
SSS, SAE designed the experiments, ZM, SSS performed bioinformatics study and analysis. SSS, AMH and FA executed the experiments. SSS, SAE, ZM wrote and edited the manuscript. EIA: Contributed to designing of experiments and reviewed the manuscript. All authors provided critical feedback and analysis of manuscript. All authors reviewed the MS and approved.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- MERS coronavirus outbreak: implications for emerging viral infections. Diagn. Microbiol. Infect. Dis.. 2019;93:265-285.
- [Google Scholar]
- High Prevalence of MERS-CoV infection in camel workers in Saudi Arabia. MBio 2018:9.
- [Google Scholar]
- Epidemiological, demographic, and clinical characteristics of 47 cases of Middle East respiratory syndrome coronavirus disease from Saudi Arabia: a descriptive study. Lancet Infect. Dis.. 2013;13:752-761.
- [Google Scholar]
- Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med.. 2014;370:2499-2505.
- [Google Scholar]
- The application of CRISPR/Cas systems for antiviral therapy. Front. Genome Editing. 2021;3
- [CrossRef] [Google Scholar]
- RNA-based therapeutics to treat human fungal infections. Trends Microbiol.. 2022;30:411-420.
- [CrossRef] [Google Scholar]
- Evaluation of canonical siRNA and Dicer substrate RNA for inhibition of hepatitis C virus genome replication–a comparative study. PLoS One. 2015;10:e0117742.
- [Google Scholar]
- Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids. 2017;8:132-143.
- [Google Scholar]
- Small interfering RNAs (siRNAs) in cancer therapy: a nano-based approach. Int. J. Nanomed.. 2019;14:3111-3128.
- [Google Scholar]
- In Silico Design and Experimental Validation of siRNAs Targeting Conserved Regions of Multiple Hepatitis C Virus Genotypes. PLoS One. 2016;11:e0159211.
- [Google Scholar]
- In vitro inhibitory analysis of rationally designed siRNAs against MERS-CoV replication in Huh7 cells. Molecules. 2021;26(9):2610.
- [Google Scholar]
- CRISPR/Cas technology as a promising weapon to combat viral infections. Bioessays. 2021;43:e2000315.
- [Google Scholar]
- Precise and efficient siRNA design: a key point in competent gene silencing. Cancer Gene Ther.. 2016;23:73-82.
- [Google Scholar]
- Safety and immunogenicity of a candidate Middle East respiratory syndrome coronavirus viral-vectored vaccine: a dose-escalation, open-label, non-randomised, uncontrolled, phase 1 trial. Lancet Infect. Dis.. 2020;20:816-826.
- [Google Scholar]
- A Computational Approach for Predicting Role of Human MicroRNAs in MERS-CoV Genome. Adv Bioinformatics. 2014;2014:967946
- [Google Scholar]
- A highly immunogenic, protective, and safe adenovirus-based vaccine expressing Middle East respiratory syndrome coronavirus S1-CD40L fusion protein in a transgenic human dipeptidyl peptidase 4 mouse model. J. Infect Dis.. 2019;220:1558-1567.
- [Google Scholar]
- Clinical advances of siRNA-Based nanotherapeutics for cancer treatment. Pharmaceutics. 2021;13
- [CrossRef] [Google Scholar]
- In vivo inhibition of influenza A virus replication by RNA interference targeting the PB2 subunit via intratracheal delivery. PLoS One. 2017;12:e0174523.
- [Google Scholar]
- RNAi: antiviral therapy against dengue virus. Asian Pac. J. Trop. Biomed.. 2013;3:232-236.
- [Google Scholar]
- Advanced Nanotheranostics of CRISPR/Cas for Viral Hepatitis and Hepatocellular Carcinoma. Adv. Sci. (Weinheim Baden-Wurttemberg, Germany). 2021;8:e2102051.
- [CrossRef] [Google Scholar]
- Anti-SARS coronavirus agents: a patent review (2008 - present) Expert Opin. Ther. Pat.. 2013;23:1337-1348.
- [Google Scholar]
- Probable transmission chains of Middle East respiratory syndrome coronavirus and the multiple generations of secondary infection in South Korea. Int. J. Infect. Dis.. 2015;38:65-67.
- [Google Scholar]
- RNA interference as a prospective tool for the control of human viral infections. Front. Microbiol.. 2018;9:2151.
- [Google Scholar]
- Coronavirus vaccine development: from SARS and MERS to COVID-19. J. Biomed. Sci.. 2020;27:1-23.
- [Google Scholar]
- The Use of CRISPR/Cas9 as a Tool to Study Human Infectious Viruses. Front. Cell. Infect. Microbiol.. 2021;11
- [CrossRef] [Google Scholar]
- Efficacy Analysis of Combinatorial siRNAs against HIV Derived from One Double Hairpin RNA Precursor. Front. Microbiol.. 2017;8:1651.
- [Google Scholar]
- Prevalence of MERS-CoV nasal carriage and compliance with the Saudi health recommendations among pilgrims attending the 2013 Hajj. J Infect Dis. 2014;210:1067-1072.
- [Google Scholar]
- Effects of siRNA silencing on the susceptibility of the fish cell line CHSE-214 to Yersinia ruckeri. Vet. Res.. 2020;51:45.
- [Google Scholar]
- Inhibition of Hepatitis C Virus in Mice by a Small Interfering RNA Targeting a Highly Conserved Sequence in Viral IRES Pseudoknot. PLoS One. 2016;11:e0146710.
- [Google Scholar]
- Middle East respiratory syndrome coronavirus (MERS-CoV): infection, immunological response, and vaccine development. J. Immunol. Res.. 2019;7(2019):6491738.
- [Google Scholar]
- siRNA design software for a target gene-specific RNA interference. Front. Genet.. 2012;3:102.
- [Google Scholar]
- Therapeutic potentials of CRISPR-Cas genome editing technology in human viral infections. Biomed. Pharmacother.. 2022;148:112743
- [Google Scholar]
- Design of Potential RNAi (miRNA and siRNA) Molecules for Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Gene Silencing by Computational Method. Interdiscip. Sci.. 2015;7:257-265.
- [Google Scholar]
- 2014 MERS-CoV outbreak in Jeddah–a link to health care facilities. N. Engl. J. Med.. 2015;372:846-854.
- [Google Scholar]
- Hemocyte siRNA uptake is increased by 5' cholesterol-TEG addition in Biomphalaria glabrata, snail vector of schistosome. Peer J. 2021;9:e10895.
- [Google Scholar]
- The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov.. 2019;18:421-446.
- [Google Scholar]
- In vitro inhibitory analysis of consensus siRNAs against NS3 gene of hepatitis C virus 1a genotype. Asian Pac. J. Trop. Med.. 2017;10:701-779.
- [Google Scholar]
- Design and Delivery of Therapeutic siRNAs: Application to MERS-Coronavirus. Curr. Pharm. Des.. 2018;24:62-77.
- [Google Scholar]
- In silico prediction and experimental validation of siRNAs targeting ORF1ab of MERS-CoV in Vero cell line. Saudi J. Biol. Sci.. 2020;28(2):1348-1355.
- [Google Scholar]
- Designing and evaluation of MERS-CoV siRNAs in HEK-293 Cell line. J. Infect. Public Health. 2020;14(2):238-243.
- [Google Scholar]
- In silico prediction and designing of potential siRNAs to be used as antivirals against SARS-CoV-2. Curr. Pharm. Des.. 2021;27(32):3490-3500.
- [Google Scholar]
- Development of siRNA mediated RNA interference and functional analysis of novel parasitic nematode-specific protein of Setaria digitata. Exp. Parasitol.. 2018;186:42-49.
- [CrossRef] [Google Scholar]
- Engineered Flock House Virus for Targeted Gene Suppression Through RNAi in Fruit Flies (Drosophila melanogaster) in Vitro and in Vivo. Front. Physiol.. 2018;9:805.
- [Google Scholar]
- Influenza A virus-derived siRNAs increase in the absence of NS1 yet fail to inhibit virus replication. RNA. 2018;24:1172-1182.
- [Google Scholar]
- LncRNA: A Potential Target for Host-Directed Therapy of Candida Infection. Pharmaceutics. 2022;14
- [CrossRef] [Google Scholar]
- Recent developments in antivirals against hepatitis B virus. Virus Res.. 2016;213:205-213.
- [Google Scholar]
- WHO, World Health Organization (WHO). 2022. 'Middle East respiratory syndrome coronavirus (MERS-CoV). Monthly Summary November 2019. Available from: https://www.who.int/health-topics/middle-east-respiratory-syndromecoronavirus-mers#tab=tab_1. [Last Accessed on 10 September 2022].
- Antibodies and vaccines against Middle East respiratory syndrome coronavirus. Emerging Microbes Infect.. 2019;8:841-856.
- [Google Scholar]
- MERS, SARS and other coronaviruses as causes of pneumonia. Respirology. 2018;23:130-217.
- [Google Scholar]
- Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med.. 2012;367:1814-1820.
- [Google Scholar]
- Effects of siRNA-mediated suppression of HPV-11 L1 expression on the proliferation and apoptosis of vaginal epithelial cells. Exp. Ther. Med.. 2017;13:1561-2155.
- [Google Scholar]
- In Silico Design of siRNAs Targeting Existing and Future Respiratory Viruses with VirusSi. bioRxiv. 14 2020.08.13.250076 2020:..
- [Google Scholar]
- Advances in MERS-CoV vaccines and therapeutics based on the receptor-binding domain. Viruses. 2019;11:60.
- [Google Scholar]







