Translate this page into:
de novo pyrimidine synthesis pathway inhibition reduces motility virulence of Pseudomonas aeruginosa despite complementation
⁎Corresponding author at: Department of Oral Medicine and Diagnostic Sciences, College of Dentistry, King Saud University, Riyadh, Saudi Arabia. aaniazy@ksu.edu.sa (Abdurahman A. Niazy),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objectives
Finding alternative methods to overcome antibiotic resistance in the medically important bacterium Pseudomonas aeruginosa has become increasingly vital as antibiotic resistance becomes more difficult to overcome. DNA synthesis inhibition is a strategy that has been used to inhibit cancer cells from proliferating by using pyrimidine analogues. Gateway site-directed mutagenesis is an effective way to create precise knockouts which creates good models to understand the relationship between biochemical pathways and the physiology of the organism. This article demonstrates the effect of pyrimidine synthesis inhibition on the motility virulence and physiology of P. aeruginosa.
Methods
Wild type Pseudomonas aeruginosa (PA01) was genetically modified using gateway cloning technology to inhibit the pyrimidine synthesis pathway through the complete knockout of the pyrE gene. The mutant strain was compared to the wild type PA01 in terms of its twitching, swarming and swimming motility forms and biofilm formation abilities. In addition to that, the production of the bacterial surfactant, rhamnolipids, was also measured.
Results and conclusions
The inhibition of the pyrimidine synthesis pathway resulted in significant reduction in motility and biofilm capabilities, despite complementing the mutation with uracil to bypass the mutation. Inhibition of the pyrimidine synthesis pathway is thus an effective way to reduce the virulence of Pseudomonas aeruginosa. Even though the bacterium is able to acquire pyrimidines from its surrounding, it was not sufficient enough to overcome the effects of the mutation.
Keywords
Pseudomonas aeruginosa
pyrimidine synthesis pathway
motility
virulence factors
biofilm formation
pyrE
mutation
- ΔpyrE
-
pyrE mutant
- ΔpyrE+U
-
ΔpyrE supplemented with uracil
- 5-FU
-
fluorouracil
- CTAB
-
cetyltrimethylammonium bromide
- CF
-
cystic fibrosis
- CTP
-
cytosine triphosphate
- DHODase
-
dihydroorotate dehydrogenase
- LB
-
Luria broth
- OMP
-
orotate monophosphate
- PA01
-
Pseudomonas aeruginosa wild type
- PMNs
-
polymorphonuclear neutrophils
- PRPP
-
phosphoribosyl pyrophosphate
- TS
-
thymidylate synthase
- UMP
-
uracil monophosphate
- UTP
-
uracil triphosphate
- WT
-
wild type
Abbreviations
1 Introduction
Pseudomonas aeruginosa (P. aeruginosa), a Gram-negative opportunistic bacterium, can cause acute and chronic infections in patients with suppressed immune systems, undergoing cancer chemotherapy, and with cystic fibrosis (Horcajada et al., 2019; Zifodya and Crothers, 2019). P. aeruginosa is an important cause of mortality in patients suffering from cystic fibrosis (CF), a devastating genetic disease commonly prevalent among Caucasians (Parkins et al., 2018). Although characterized by chronic lung disease, electrolyte abnormalities and pancreatic insufficiency; the high associated mortality rates of CF is primarily attributed to the formation of mucoid-alginate and biofilms by P. aeruginosa (Malhotra et al., 2019). Its pathogenicity is dictated by a variety of virulence factors including synthesis of extracellular toxins, proteases, hemolysins and exopolysaccharides (Jurado-Martín et al., 2021); in addition to its motility and biofilm production capacity (Bains et al., 2012; Kazmierczak et al., 2015; Redfern et al., 2021). The three major methods by which P. aeruginosa exhibits motility are swimming, twitching, and swarming (Burrows, 2012). While the polar flagellum aids in its swimming-type motility through liquid and extremely soft media, type IV pilus enables twitching (Kazmierczak et al., 2015). On the contrary, swarming on semi-solid surfaces is achieved through utilization of both type IV pili and flagella (Kühn et al., 2021; Talà et al., 2019). The motility of P. aeruginosa combined with its ability to produce proteins and polysaccharides, enables it to attach to and colonize surfaces forming biofilms, which are considered a key factor in its pathogenicity (Kazmierczak et al., 2015).
The alginate exopolysaccharide exclusively synthesized by P. aeruginosa, in addition to enhancing the ability of the organism to propel and colonize lung surfaces, helps it to establish a quorum sensing microbial environment, which evades host defenses and antimicrobial therapy (Chadha et al., 2021; Niazy, 2021). Novel therapies suggested for the treatment of CF include mechanisms of inhibiting biofilm formation and alginate synthesis by P. aeruginosa through the use of non-toxic drugs, regulatory proteins, and altering bio-synthetic pathways (Behzadi et al., 2021; Srinivasan et al., 2021). One reportedly achievable strategy is by means of regulating pyrimidine biosynthesis in microorganisms, which conventionally progresses through the de novo or salvage pathway (O'Donovan and Neuhard, 1970; Ralli et al., 2007).
The de novo pyrimidine synthesis pathway has been studied thoroughly in P. aeruginosa (Beckwith et al., 1962; O'Donovan and Neuhard, 1970; O'toole and Kolter, 1998) and has shown to be an important aspect in its production of virulence factors (Hosseinidoust et al., 2013). Moreover, mutations at different sites within this pathway could lead to different outcomes in terms of virulence factor production by P. aeruginosa (Niazy and Hughes, 2015; Ralli et al., 2007). Pyrimidine regulation could be achieved either at the enzymatic or transcriptional level, with the former being a feedback mechanism wherein increased cytosine triphosphate (CTP) and uracil triphosphate (UTP) lead to the decrease of pyrimidine enzyme activities and the latter targeting genes involved in the pathway (Niazy and Hughes, 2015). The first three steps in the pyrimidine pathway involves genes namely pyrB encoding aspartate transcarbamylase, pyrC encoding dihydroorotase, and pyrD encoding dihydroorotate dehydrogenase, which together catalyzes the production of orotate from carbamoyl phosphate and aspartate (Womack and O'Donovan, 1978). Secondly, the orotate phosphoribosyltransferase catalysis, a crucial step of the pathway, encoded by the pyrE gene resulting to the orotate monophosphate (OMP) synthesis from orotate and phosphoribosyl pyrophosphate (PRPP). This reaction is followed by the final step in the de novo pyrimidine pathway, wherein OMP decarboxylase encoded by pyrF results in decarboxylation of OMP to form uracil monophosphate (UMP) (Womack and O'Donovan, 1978). While mutations in pyrE and pyrF could lead to secretion and accumulation of orotate in bacteria (Womack and O'Donovan, 1978), a similar phenotype was also reported in humans exhibiting orotic aciduria (Winkler and Suttle, 1988).
Even though previous studies have reported a decrease in virulence factors as a result of orotate accumulation following mutation in pyrE (Garavaglia et al., 2012; Niazy and Hughes, 2015; Womack and O'Donovan, 1978;), the specific mutant phenotype in P. aeruginosa has not been exhaustively evaluated since. Therefore, this study aimed to determine the effect of disrupting the de novo pyrimidine synthesis pathway at pyrE on the pathogenicity of P. aeruginosa, in terms of motility and biofilm formation.
2 Materials and methods
2.1 Construction of mutant
2.1.1 Synthesis of mutant pyrE fragment
The pyrE mutant (ΔpyrE) was constructed utilizing Gateway® cloning technology (Invitrogen, CA, USA) according to the protocol previously described (Choi and Schweizer, 2005) and has been detailed in our previous work (Niazy and Hughes, 2015). Wild type (WT) P. aeruginosa strain PA01 (prototroph; Brichta et al, 2004) was used in all experimental analyses (Table1). Briefly, the 74 bps fragment from pyrE (642 bps) was replaced by the aacC1 gentamicin-resistant gene. PCR amplification of the modified pyrE was configured to: 95 °C for 2 min, 95 °C for 30 s, 50 °C for 30 s, 68 °C for 90 s for 30 cycles and 68 °C for 7 min using the Q5® High Fidelity 2X Master Mix (New England Biolabs (NEB), Ipswich, MA). After PCR amplification and addition of attB overhangs on the 5′ end, running in tandem was the amplification of the gentamicin-resistant gene from the pPS856 plasmid. Using primers that had overhangs that were complementary to the aacC1 gene in addition to the attB overhangs, a PCR was done to attach all three fragments together resulting in a mutant form of pyrE-GM with an insertion of a gentamicin-resistant gene replacing bases 337 to 411 with the following conditions: 95 °C for 2 min, 95 °C for 30 s, 50 °C for 30 s, 68 °C for 90 s for 30 cycles and finally 68 °C for 7 min using the Phusion® Hot Start Flex DNA Polymerase kit (NEB, MA, USA) (Niazy and Hughes, 2015). All amplicons were analyzed by agarose electrophoresis and corresponding bands were cut out of the gel and purified using PureLink® Quick Gel Extraction Kit (Invitrogen, MA, USA) (Table 2).
Strain or plasmid
Genotype of strain or plasmid
Source/Reference
P. aeruginosa PA01 PA0pyrE-
Prototroph
PA01 pyrE Δ
Brichta et al., 2004
E. coli DH5α
recA1 hsdR17 endA1 supE44 thi-1 relA1 gyrA96 Δ (argF-lacZYA)U169 φ 80lacZ Δ M15
Bethesda Research Laboratories, 1986
pDONR221
KanR, ccdB suicide gene, attP cloning sites
Invitrogen
pEX18ApGW
APR (Cb)R, sacB+, gateway vector, attR cloning sites, ccdB suicide gene
Dr. H. Schweizer
pDONRpyrEGM
GMR, pyrE-GM 1700 bp insert donor plasmid
This study
pEX18pyrEGM
GMR, pyr-GM 1700 bp insert gateway plasmid
This study
pUCP20
APR(CbR) PA01 replicative control plasmid
Dr. H. Schweizer
pPS856
GMR for extraction of GMR gene
Dr. H. Schweizer
Primer name
Sequence
Amplification target
pyrEUSF
TACAAAAAAGCAGGCTATGCAGGCGTATCA
Forward primer of the upstream region of pyrE
pyrEUSR
TCAGAGCGCTTTTGAAGCTAATTCGCGTGGTCCTTGGCTTCCTTGCGGTTGAAGCA
Reverse primers of the upstream region of pyrE
pyrEDSF
AGGAACTTCAAGATCCCCAATTCGGCAGATCATCGACGCCCAGGGCGCCA
Forward primers of the downstream region of pyrE
pyrEDSR
TACAAGAAAGCTGGGTTCAGATACCGTATT
Reverse primer of the downstream region of pyrE
Gm-F
CGAATTAGCTTCAAAAGCGCTCTGA
Forward primer for aacC1 gentamicin resistance cassette from pPS856
Gm-F
CGAATTGGGGATCTTGAAGTTCCT
Reverse primer for aacC1 gentamicin cassette from pPS856
GW-attB1
GGGGACAAGTTTGTACAAAAAAGCAGGCT
Forward primer attaching US-GM-DS fragments to create the pyrE-GM fragment in second PCR step
GW-attB2
GGGGACCACTTTGTACAAGAAAGCTGGGT
Reverse primer attaching US-GM-DS fragments to create the pyrE-GM fragment in second PCR step
2.1.2 Construction of donor and gateway plasmids
The connected fragment was then mixed with the enzyme Gateway® BP clonase® and pDONR221 plasmid from Invitrogen (Carlsbad, CA, USA) utilizing manufacturer’s instructions. After recombination, the recombinant plasmid was transformed into E. coli (DH5α) and plated on Luria Broth (LB) agar plates containing 60 µg/ml kanamycin. The kanamycin-resistant colonies were selected for plasmid extraction using the Invitrogen ChargeSwitch®-Pro Plasmid Miniprep kit (Carlsbad, CA, USA) according to the manufacturer’s instructions. Analysis was done using XbaI restriction enzyme (NEB, MA, USA), which cuts at the flanking region of aacC1; resulted into three plasmid fragments which were analyzed with 1.5% agarose electrophoresis. The gateway plasmid containing the proper recombination was designated as pDONR-pyrE-GM. The pEX18ApGW plasmid was then mixed with pDONR-pyrE-GM and the Gateway® LR clonase enzyme (Invitrogen, CA, USA) at 50 ng/µl each following manufacturer’s instructions. The plasmid was then transformed into E. coli (DH5α) and plated on LB agar plates containing 200 µg/ml ampicillin for selection of resistant colonies. A single resistant colony was then grown in LB broth with 200 µg/ml ampicillin and plasmid extraction was done.
2.1.3 Transformation and recombination
The constructed gateway plasmid was then transformed into P. aeruginosa by electroporation using a Gene Pulser Xcell™ Electroporation System (Bio-Rad Laboratories, CA, USA) utilizing the protocol adapted from Choi and Schweizer (2005). P. aeruginosa cells were incubated at 42 °C for 24 h; and centrifuged at 10,053 g for 2 min. The collected pellet was resuspended in 300 mM sucrose solution, wherein the cloned pEX18ApGW plasmid was added; and exposed to electroporation in a 2 mm gap electroporation cuvette. After 1 h incubation at 37 °C with shaking, the cell suspension was centrifuged; and the cell pellet was resuspended and plated on LB agar containing 30 µg/ml gentamicin. Resistant colonies were selected and grown on LB containing 5% sucrose to eliminate any merodiploids. Mutant pyrE was amplified and sent for sequencing at GENEWIZ Sequencing (South Plainfield, NJ, USA). The results were then run on NCBI Blast software for alignment results.
2.2 Analysis of growth curves
The growth curves of WT P. aeruginosa and ΔpyrE was measured according to the method described by Liu et al. (2020). Cells were grown until mid-log phase on OD600nm of 0.6 A in LB broth. A 500 µl of fresh inoculum was added to LB broth in a conical flask from the mid-log phase cells. For complementation with uracil, media containing 40 µg/ml uracil was added to the specific flask. Flasks were incubated at 37 °C with shaking at 220 rpm. Spectrophotometric readings of OD600nm were recorded hourly for 11 h on BioTek Synergy HT Microplate reader (Winooski, VA, USA).
2.3 Assessment of P. aeruginosa virulence and biofilm
2.3.1 Motility assays
Motility assays for twitching, swimming and swarming were studied. Twitching assay was done utilizing the procedure previously described (Rashid et al., 2000). Thin 3 mm layers of LB medium solidified with 1.5% agar plates were prepared. For uracil-supplemented plates, 40 µg/ml uracil was added to the medium. A sterile needle was used to touch a fresh colony and stabbed into the agar to reach the sub-surface area. Plates were then incubated at 37 °C for 24 h. The twitching diameter was measured and plates were photographed using a Nikon D3100 DSLR camera.
Swimming assay was measured on plates containing 1% tryptone, 0.5% NaCl and 0.2% agar as previously described (Li et al., 2020). Using a sterile needle, a fresh colony was touched with the tip of the needle and stabbed halfway through the agar. Plates for complementation test were supplemented with 40 µg/ml uracil. All plates were incubated for 20 h at 30 °C. The diameter of the swimming zone was measured and the plates were imaged as mentioned previously.
Swarming assay was performed on semi-solid LB agar plates containing 0.5% agar and 5% glucose as previously described with slight modifications (Radulović et al., 2018). A fresh colony was transferred to 10 ml LB broth and grown for 24 h. Cells were then standardized to a final OD600nm of 0.6. A 1 ml broth was transferred to an Eppendorf tube and cells were centrifuged at 10,053 g for 2 min. Cells were resuspended in 50 µl fresh LB broth and 5 µl was transferred to the surface of the agar plate. Plates were incubated at 30 °C for 10 h; then left at room temperature for 20 h. Uracil-treated plates were supplemented with 40 µg/ml uracil. The zone of swarming was measured and the images of the plates were obtained.
2.3.2 Rhamnolipid production assay
Rhamnolipid production was measured using prepared LB plates supplemented with 0.2 g/L cetyltrimethylammonium bromide (CTAB) and 0.005 g/L methylene blue as previously described with slight modifications (Siegmund and Wagner, 1991). A hole was stabbed with a toothpick into the center of the agar surface and 5 µl of the cell supernatant in LB was added to the punched hole. The plates were left at room temperature for 5 days and the diameter of the observed halo represented the amount of rhamnolipid produced. The halo measurements were divided by the corresponding OD600nm readings to account for cell density.
2.3.3 Biofilm formation
Cells were grown to mid-log phase in to a final OD600nm of 0.6 A; and 1 µl of bacterial cells were added to fresh 100 µl LB in 96-well round bottom plates. The inoculated plates were incubated at 37 °C for 48 h; afterwards, the well content was aspirated, and washed thrice with sterile distilled water to remove unattached cells. The wells were subsequently dried and stained with 150 µl of 0.5% (w/v) crystal violet for 10 min. After repeated washing, the air-dried wells were filled with 200 μl of 100% ethanol. The plate was incubated at room temperature for 15 min and 490 nm readings were measured on BioTek Synergy HT Microplate reader (Winooski, VA, USA) as previously described (O'toole and Kolter, 1998).
2.4 RT-qPCR analysis
Cells were grown in LB broth at 37 °C with shaking at 220 rpm for 24 h. A 1 ml inoculated broth was transferred to an Eppendorf tube and centrifuged at 10,053 g for 2 min. The collected cell pellet was used for total RNA extraction using HiGene™ Total RNA Prep Kit (BioFACT Co., Daejeon, South Korea) according to manufacturer’s instructions. RNA concentration and quality was determined using BioPhotometer Plus (Eppendorf, Germany). cDNA was synthesized using the FIREScript RT cDNA synthesis Kit (Solis BioDyne, Tartu, Estonia, https://www.sbd.ee) as per manufacturer’s instructions. Real-Time PCR was done on ABI 7500 Real-Time PCR System (Applied Biosystems, Darmstadt, Germany) using 5 × HOT FIREpol® EvaGreen® qPCR Supermix (Solis BioDyne, Tartu, Estonia, https://www.sbd.ee). 16S rRNA was used as the control. Genes selected in this study (Table 3) were determined to be associated with the motility of P. aeruginosa, as previously described (Hosseinidoust et al., 2013). Relative microRNA expression profiles were determined and analyzed in Microsoft Excel spreadsheet.
Gene
Primer sequence
RhlA
F- GGC GAT CGG CCA TCT G
R- AGC GAA GCC ATG TGC TGA T
PilA
F- TGC GCG TTC GGA AGG T
R- CGA CTC TTC AAC AGT GGT CTT CA
PilI
F- GCA CTG CAA CCC TTC ATT CAT
R- CGC ATG CGG GCT GAA C
RhlB
F- GCC TGT CGG CGT TTC ATG
R- CAT CCC CCT CCC TAT GAC AA
FliC
F- CAG TGC CAA GGA CGA TGC T
R- AAC GTT CAG ACC GCT GAT CTG
FliD
F- TGG CTG GCA CCT ACC AGA TC
R- GGC CTG GAG GGC AAT CTT
AlgR
F- CCT CGG CCA GCA ATG G
R- CGG ATA TCC AGC AGG ACG AT
2.5 Statistical analysis
All experiments were done in triplicate, and the data were presented as means ± standard deviations. Statistical comparison was done using Two-Way ANOVA followed by Tukey's post-hoc test (GraphPad Prism software version 6.00 for Windows, La Jolla California, USA; https://www.graphpad.com). Results were statistically significant when a p-value ≤ 0.05 was obtained.
3 Results
3.1 Construction of the mutant
PCR amplification results revealed a fragment approximately 1700 bps in size resulting from the insertion of the aacC1 gentamicin-resistant gene for the ΔpyrE and approximately 700 bps fragment resembling the wild type PA01 (Fig. 1A). Sequencing results revealed proper insertion of the aacC1 and confirmed the deletion of the 74 bps fragment. Results for alignment of the DNA sequence on NCBI Blast software are demonstrated in Fig. 1B.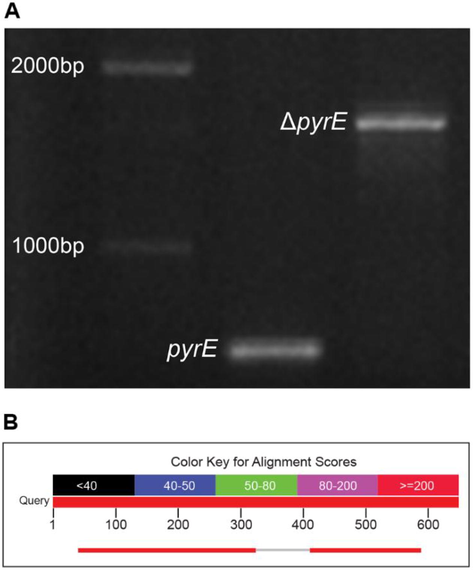
(A) Agarose gel electrophoresis (1.5%) of wild type pyrE and ΔpyrE after PCR amplification. (B) Alignment results of ΔpyrE compared to WT pyrE demonstrating the deletion.
3.2 Growth curves
Growth curves for wild type and ΔpyrE revealed that there were no differences in growth on enriched media as both the bacterial strains reached the stationary phase simultaneously. Whereas supplementation of the media with 40 µg/ml uracil did not have any considerable effect on the growth curves (Fig. 2).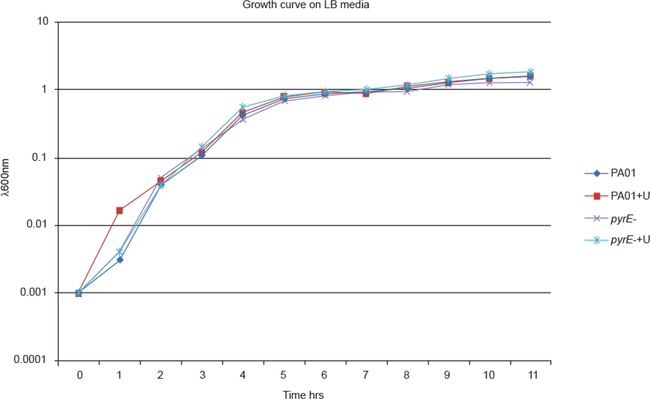
Growth curves of wild type P. aeruginosa and mutant strains in LB with and without 40 µg/ml uracil supplementation.
3.3 Motility
All three types of motility observed were reduced as a result of the mutation (Fig. 3). Upon supplementation of uracil in the growth media of ΔpyrE (ΔpyrE+U), the motility was restored to some extent but not comparable to the levels of the wild type strain. PA01 had mean diameters of 18 mm for twitching, 35 mm for swimming, and 12 mm for swarming. The ΔpyrE+U strain exhibited mean diameters of 10 mm for subsurface twitching, 15 mm for swimming and 8 mm for swarming. For the ΔpyrE strain, there was complete loss of twitching motility; and swimming and swarming motilities were severely diminished with the mean diameters being 3 mm and 4 mm respectively.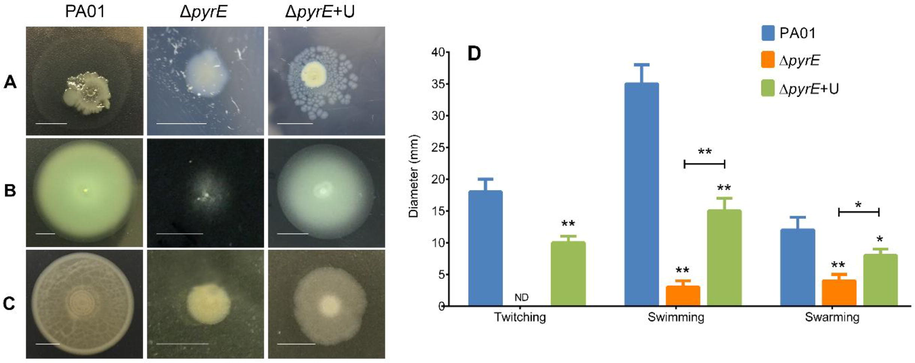
Representative images of the effect of pyrE knockout on (A) twitching, (B) swimming and (C) swarming capabilities of P. aeruginosa PA01, ΔpyrE and ΔpyrE+U on LB agar plates. The intensive growth in the middle is the initial bacterial colony while the halo around the colony represents the twitching distance. (D) Mean twitching, swimming and swarming motilities of P. aeruginosa PA01, ΔpyrE and ΔpyrE+U. Data are presented as means ± SD, n = 6 (**p≤0.01 and *p≤0.1).
3.4 Rhamnolipid production
Rhamnolipid production was decreased in the ΔpyrE strain when compared to PA01 (Fig. 4A). The wild type exhibited a mean diameter of 16 mm for the rhamnolipid halo, while the ΔpyrE strain showed a 75% drop in production and had a mean diameter of only 6 mm for the rhamnolipid halo. On the contrary, ΔpyrE+U revealed a mean diameter of 10 mm for amount of rhamnolipid produced. Dividing the results by the OD600nm, the results were as follows, 13.3 for the wild type, 9.16 for ΔpyrE+U and 5 for the ΔpyrE strain (Fig. 4B).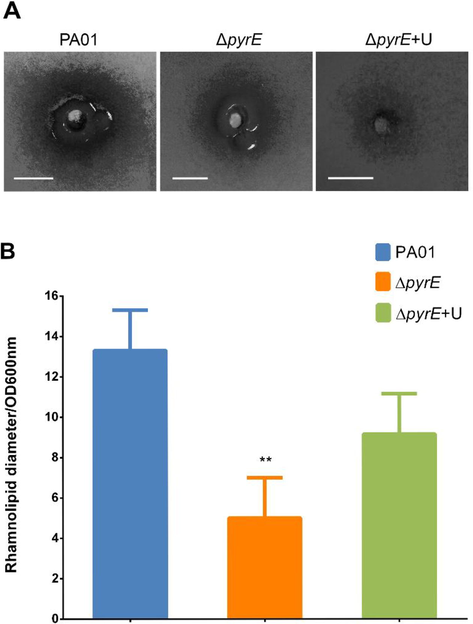
(A) Representative images of the effect of pyrE knockout on rhamnolipid production for the halo produced by P. aeruginosa PA01 wild type, mutant and uracil-supplemented strains on mineral media. (B) Rhamnolipid production assay in wild type, ΔpyrE and the ΔpyrE+U strains after dividing OD600nm readings. Data are presented as means ± SD, n = 6 (**p≤0.01).
3.5 Biofilm formation
Biofilm formation was reduced in the ΔpyrE strain when compared to PA01 by more than 50%. Supplementation with uracil increased biofilm formation by about 30% but did not restore biofilm formation to the levels of the wild type strain (Fig. 5).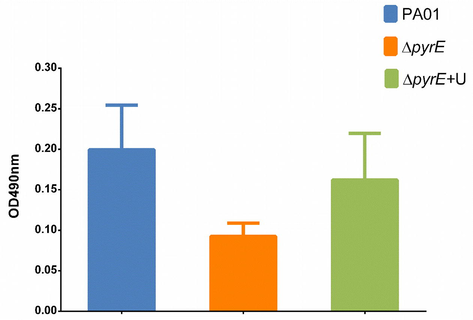
Effect of pyrE knockout on biofilm production capabilities of the PA01, ΔpyrE and complemented ΔpyrE+U at crystal violet readings of OD490nm.
3.6 RT-qPCR
Real-time PCR results showed a statistically significant drop in gene expression for rhamnolipid, pili and flagella-producing genes, except for fliD and algR which are involved in biofilm formation, in the mutant when compared to the wild type. fliD showed a marked increase in expression when compared to the wild type and uracil-supplemented mutant strain. The ΔpyrE+U showed no change in the fliD expression after supplementation with uracil. It was also determined that after supplementation with uracil, the ΔpyrE+U showed even further decreased gene expression levels compared to ΔpyrE; except for the fliD gene where there was no statistically significant change in gene expression (Fig. 6).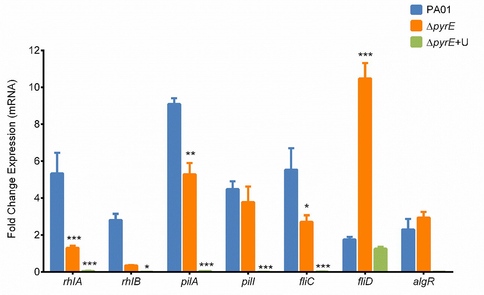
Gene expression levels of selected genes involved in motility and biofilm formation of P. aeruginosa in wild type, mutant and uracil-supplemented strains. Data are presented as mean fold change ± SD, n = 6 (***p≤0.001, **p≤0.01, and *p≤0.05).
4 Discussion
The present study observed the inhibitory effect of the disruption of de novo pyrimidine synthesis pathway upon motility and altered biofilm phenotype in P. aeruginosa (PA01) and ΔpyrE mutant. This was achieved through site-directed mutagenesis in the pyrimidine pathway, thereby leading to a complete halt in the pyrimidine biosynthesis in the mutant microorganism. Isaac and Holloway (1968) reported that the mutagenesis of the pyrimidine pathway lead to several mutant strains of P. aeruginosa and also studied their characteristics pertaining to decreased virulence. While modified biofilm production by different bacteria including P. aeruginosa through multiple genetic screens have been reported; construction of a site-directed mutagenesis is reportedly challenging (Hosseinidoust et al., 2013; Morici et al., 2007). This study focused on site-directed mutagenesis as an essential element of protein engineering, wherein proteins suitable for structural characterization of the bacteria where selectively synthesized. The preference for studying pyrE mutation in P. aeruginosa PA01 were based on promising variations in the virulence of the mutant bacterium (Hosseinidoust et al., 2013; Morici et al., 2007; Ralli et al., 2007). The mutation in this study by way of halting pyrimidine synthesis, forced the bacterium to salvage its pyrimidine from enriched media (Fig. 7). The net result was a decreased motility and biofilm formation as evidenced from the results, thereby rendering the bacterium less pathogenic.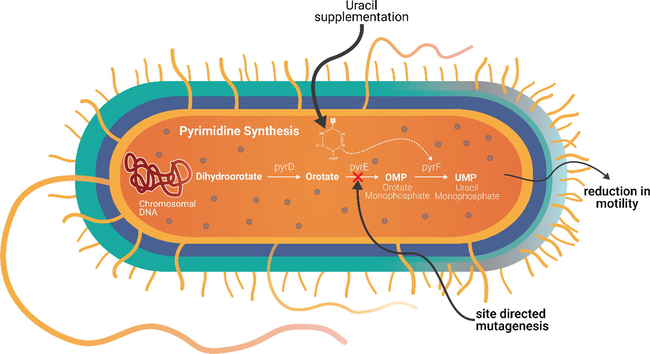
Proposed mechanism of uracil complementation in pyrE mutation of P. aeruginosa.
Based on the analysis of the growth curves in this study, it was found that enriched media had no effect on the patterns of growth in both the wild and mutant types of P. aeruginosa. However, upon supplementing the media with 40 µg/ml uracil, the wild types had a shorter lag phase in comparison to an extended lag phase combined with faster log phase in the mutant bacteria, which were pyrimidine auxotrophs. Nevertheless, the mutant bacteria were unable to grow in minimal media without the addition of uracil (data not shown). The supplementation with uracil enabled the mutant bacteria to synthesize pyrimidines using the salvage pathway rather than the slower de novo pyrimidine pathway leading to hastened nucleotide synthesis and faster growth (Ralli et al., 2007). On the contrary, the PA01 exhibited slower growth owing to their functional de novo pyrimidine pathway.
One of the most important virulence factor of P. aeruginosa to cause life-threatening infections is its motility and ability to attach and colonize surfaces (Burrows, 2012; Hosseinidoust et al., 2013; O’toole and Kolter, 1998). As a result, the mutant had marked reduction in swimming and swarming motilities, with a total loss of twitching, in comparison to PA01. However, all three forms of motility were restored in the mutant strain upon addition of uracil to the medium; indicating a correlation between motility and pyrimidine synthesis through the salvage pathway. Similarly, a decrease in rhamnolipid production and biofilm formation was also observed in the mutant bacteria in comparison to PA01. Nevertheless, uracil supplementation restored both rhamnolipid production and biofilm formation in the mutant strain to a considerable extent, which were not observed in the wild type PA01. The reduced motility of the ΔpyrE mutant, in spite of uracil supplementation, could be attributed to decreased rhamnolipid synthesis. More importantly, reduction in motility and biofilm formation in the mutant bacteria were a result of internal interference such as inhibition of quorum sensing, as there was no observable change in the growth capabilities of the wild type and mutant strains in enriched media (Bains et al., 2012; Morici et al., 2007; Ralli et al., 2007). This was further confirmed by qPCR findings of the present study which revealed a decrease in the genes expression of rhamnolipid, pili, and flagella markers in ΔpyrE.
In this study, the growth rates of wild type and mutant strains were similar in enriched media, which were comparable to what was reported by Brichta (2003). Nevertheless, motility, rhamnolipid production, and biofilm formation were reduced in the mutant type when compared to the wild type PA01. Similar results were reported by Ralli et al. (2007), following a pyrD knockout mutation in P. aeruginosa. While reporting diminished motility, rhamnolipid production and biofilm formation, they also reported reduced expression of exoproducts such as casein protease, elastase, and hemolysins and decreased iron-binding capacity. They suggested a role in inducing mutagenesis in the pyrimidine pathway as a mechanism for eliminating P. aeruginosa infection in immunosuppressed individuals. The pyrimidine auxotrophy exhibited by the ΔpyrE mutant enhances its motility, rhamnolipid production and biofilm formation upon addition of uracil to the growth media. This is in accordance to earlier findings corroborating pyrimidine auxotrophy with reduction in virulence factors of P. aeruginosa such as production of hemolysin, casein protease, elastase, pyoverdin, and pyocyanin (Ralli, 2005).
Although further gene expression studies regarding rhamnolipid, pili, and flagella function need to be further evaluated, the complementation of uracil in this study failed to restore normal gene expression in the mutant pyrE strain. Nonetheless, the decrease in the rhamnolipid gene expression observed in the present study has significant clinical implications, as rhamnolipids contribute to the ability of P. aeruginosa to resist antibacterial action of polymorphonuclear neutrophils (PMNs) in a biofilm environment (Donlan and Costerton, 2002). Based on these findings, there is potential use of DNA synthesis inhibitors specifically pyrimidine inhibitors as an alternative therapy to inhibit bacterial motility and biofilm formation in P. aeruginosa strains.
Fluorouracil (5-FU) is an anti-neoplastic drug that acts by blocking pyrimidine synthesis through inhibition of the enzyme thymidylate synthase (TS) (Attila et al., 2009). Fluorouracil has been known to cause antibacterial effects against oral microorganisms including P. aeruginosa at very high concentrations (>100 µM) (Vanlancker et al., 2016). Based on a study on P. aeruginosa PA14 strain by Ueda et al. (2009), 5-FU has the ability to suppress quorum sensing and biofilm formation through uracil-related mutations, namely carA, carB, pyrB, pyrC, pyrD, pyrE, and pyrF. Similar results were reported by Attila et al. (2009) with respect to the ability of 5-FU to inhibit biofilm formation and reduce virulence in E.coli. Using gold nanoparticle probes, Selvaraj and Alagar (2007) demonstrated the antibacterial capabilities of 5-FU against P. aeruginosa.
While bacteria efficiently develop resistance to inhibitors of bacterial growth such as antibiotics, the alternative approach of developing antibacterial drugs which inhibit virulence factors is gaining prominence (Attila et al., 2009; Guo et al., 2016). Guo et al. (2016) have reported the development of a novel compound against P. aeruginosa, with an ultimate aim of suppressing its virulence and antibiotic resistance. This novel treatment targets the pyrD, encoding for the PyrD protein. The PyrD protein is a dihydroorotate dehydrogenase (DHODase) involved in pyrimidine biosynthesis, similar to the PyrE protein investigated in the present study (Niazy and Hughes, 2015; Ralli et al., 2007). Donlan and Costerton (2002) also recommended a paradigm shift from the conventional strategies to eradicate biofilm-forming bacteria towards a futuristic approach based on inhibition of genes responsible for virulence factors such as cell adhesion, motility and rhamnolipid production.
5 Conclusions
The results of the present study revealed that pyrE mutants of P. aeruginosa PA01 exhibited a decrease in motility and biofilm formation capabilities as a result of the disruption of de novo pyrimidine synthesis. These findings demonstrate the importance of therapies targeting disruption of pyrimidine pathways, such as the pyrE, in order to treat recalcitrant P. aeruginosa infections similar to cystic fibrosis patients. Nevertheless, such possible therapies need to be validated further through molecular and clinical studies.
Conflict of interest
No conflict of interest declared.
Acknowledgements
The authors are grateful to Dr. Herbert Schweizer (Colorado State University, USA) for providing the gateway plasmids and Terrence Suministrado Sumague for assistance in preparing the figures and scientific illustrations. The authors would also like to thank Hadeel A. AlTulihe and Lorvijan G. Avila for their technical support.
Disclosure of funding
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group No (RG-1439-026). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 5-Fluorouracil reduces biofilm formation in Escherichia coli K-12 through global regulator AriR as an antivirulence compound. Appl. Microbiol. Biotechnol.. 2009;82:525-533.
- [CrossRef] [Google Scholar]
- Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol.. 2012;78:6762-6768.
- [CrossRef] [Google Scholar]
- Coordination of the synthesis of the enzymes in the pyrimidine pathway of E. coli. J. Mol. Biol.. 1962;5:618-634.
- [CrossRef] [Google Scholar]
- It's not easy being green: a narrative review on the microbiology, virulence and therapeutic prospects of multidrug-resistant Pseudomonas aeruginosa. Antibiotics (Basel, Switzerland).. 2021;10:42.
- [CrossRef] [Google Scholar]
- Construction of a Pseudomonas aeruginosa Dihydroorotase Mutant and the Discovery of a Novel Link Between Pyrimidine Biosynthetic Intermediates and the Ability to Produce Virulence Factors. University of North Texas; 2003.
- Pseudomonas aeruginosa dihydroorotases: a tale of three pyrCs. Arch. Microbiol.. 2004;182(1):7-17.
- [Google Scholar]
- Pseudomonas aeruginosa twitching motility: Type IV Pili in action. Annu. Rev. Microbiol.. 2012;66:493-520.
- [CrossRef] [Google Scholar]
- Revisiting the virulence hallmarks of Pseudomonas aeruginosa: a chronicle through the perspective of quorum sensing. Environ. Microbiol. 2021
- [CrossRef] [Google Scholar]
- Choi, K.-H., Schweizer, H. P., 2005. An improved method for rapid generation of unmarked Pseudomonas aeruginosa deletion mutants. BMC Microbiol. 5, 30-30. https://doi.org/10.1186/1471-2180-5-30.
- Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev.. 2002;15:167-193.
- [CrossRef] [Google Scholar]
- The pyrimidine nucleotide biosynthetic pathway modulates production of biofilm determinants in Escherichia coli. PLoS ONE. 2012;7(2)
- [Google Scholar]
- Identification of a small molecule that simultaneously suppresses virulence and antibiotic resistance of Pseudomonas aeruginosa. Sci. Rep.. 2016;6:19141.
- [CrossRef] [Google Scholar]
- Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev.. 2019;32:e00031-e119.
- [CrossRef] [Google Scholar]
- Predation in homogeneous and heterogeneous phage environments affects virulence determinants of Pseudomonas aeruginosa. Appl. Environ. Microbiol.. 2013;79:2862-2871.
- [CrossRef] [Google Scholar]
- Control of pyrimidine biosynthesis in Pseudomonas aeruginosa. J. Bacteriol.. 1968;96:1732-1741.
- [CrossRef] [Google Scholar]
- Pseudomonas aeruginosa: an audacious pathogen with an adaptable arsenal of virulence factors. Int. J. Mol. Sci.. 2021;22:3128.
- [CrossRef] [Google Scholar]
- Cross-regulation of Pseudomonas motility systems: the intimate relationship between flagella, pili and virulence. Curr. Opin. Microbiol.. 2015;28:78-82.
- [CrossRef] [Google Scholar]
- Mechanotaxis directs Pseudomonas aeruginosa twitching motility. PNAS. 2021;118
- [CrossRef] [Google Scholar]
- PA5001 gene involves in swimming motility and biofilm formation in Pseudomonas aeruginosa. Microb. Pathog.. 2020;144
- [CrossRef] [Google Scholar]
- Antibacterial activity and mechanism of linalool against Pseudomonas aeruginosa. Microb. Pathog.. 2020;141
- [Google Scholar]
- Cystic Fibrosis and Pseudomonas aeruginosa: the Host-Microbe Interface. Clin. Microbiol. Rev.. 2019;32:e00138-e218.
- [CrossRef] [Google Scholar]
- Pseudomonas aeruginosa AlgR represses the Rhl quorum-sensing system in a biofilm-specific manner. J. Bacteriol.. 2007;189(21):7752-7764.
- [Google Scholar]
- Accumulation of pyrimidine intermediate orotate decreases virulence factor production in Pseudomonas aeruginosa. Curr. Microbiol.. 2015;71:229-234.
- [CrossRef] [Google Scholar]
- LuxS quorum sensing system and biofilm formation of oral microflora: a short review article. Saudi Dental J.. 2021;33:116-123.
- [CrossRef] [Google Scholar]
- Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol.. 1998;30:295-304.
- [CrossRef] [Google Scholar]
- Epidemiology, biology, and impact of clonal Pseudomonas aeruginosa infections in cystic fibrosis. Clin. Microbiol. Rev.. 2018;31:e00019-e118.
- [CrossRef] [Google Scholar]
- Water-soluble gold(III) complexes with N-donor ligands as potential immunomodulatory and antibiofilm agents. Polyhedron. 2018;141:164-180.
- [CrossRef] [Google Scholar]
- Regulation of the pyrimidine biosynthetic pathway in apyrD knockout mutant of Pseudomonas aeruginosa. J. Basic Microbiol.. 2007;47:165-173.
- [CrossRef] [Google Scholar]
- Impaired Virulence factor Production in a Dihydroorotate Dehydrogenase Mutant (pyrD) of Pseudomonas aeruginosa. University of North Texas; 2005.
- Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol.. 2000;182:225-227.
- [CrossRef] [Google Scholar]
- Biofilm associated genotypes of multiple antibiotic resistant Pseudomonas aeruginosa. BMC Genomics. 2021;22(1):1-16.
- [Google Scholar]
- Analytical detection and biological assay of antileukemic drug 5-fluorouracil using gold nanoparticles as probe. Int. J. Pharm.. 2007;337:275-281.
- [CrossRef] [Google Scholar]
- New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech.. 1991;5:265-268.
- [CrossRef] [Google Scholar]
- Bacterial biofilm inhibition: a focused review on recent therapeutic strategies for combating the biofilm mediated infections. Front. Microbiol.. 2021;12:676458.
- [CrossRef] [Google Scholar]
- Pseudomonas aeruginosa orchestrates twitching motility by sequential control of type IV pili movements. Nat. Microbiol.. 2019;4:774-780.
- [CrossRef] [Google Scholar]
- Uracil influences quorum sensing and biofilm formation in Pseudomonas aeruginosa and fluorouracil is an antagonist. Microb. Biotechnol.. 2009;2:62-74.
- [CrossRef] [Google Scholar]
- 5-Fluorouracil sensitivity varies among oral micro-organisms. J. Med. Microbiol.. 2016;65:775-783.
- [CrossRef] [Google Scholar]
- Analysis of UMP synthase gene and mRNA structure in hereditary orotic aciduria fibroblasts. Am. J. Hum. Genet.. 1988;43:86.
- [Google Scholar]
- Orotic acid excretion in some wild-type strains of Escherichia coli K-12. J. Bacteriol.. 1978;136:825-827.
- [CrossRef] [Google Scholar]
- Treating bacterial pneumonia in people living with HIV. Expert Rev. Respir. Med.. 2019;13(8):771-786.
- [Google Scholar]







