Translate this page into:
Current status, spatiotemporal dynamics and genetic recombination analysis of cucurbit aphid-borne yellows virus (CABYV) infecting cucurbits in Punjab, Pakistan
⁎Corresponding author. mashfaq1642@gmail.com (Muhammad Ashfaq) mashfaq@mnsuam.edu.pk (Muhammad Ashfaq)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Cucurbits are important vegetable crops of Pakistan which are severely affected by various diseases. Keeping in view the importance of viral diseases, the present study was aimed to conduct systematic surveys of the tunnel and open field cucurbit crops in Punjab, Pakistan during 2019 and 2020. The cucurbits leaf samples with virus and virus-like symptoms were collected from the farmers fields and tunnels with no prior knowledge about sanitation and subjected to reverse transcription-polymerase chain reaction (RT-PCR) for poleroviruses incidence. The mean highest disease incidence was recorded in samples collected from district Rawalpindi (47.42%) and the mean lowest disease incidence was observed in samples of district Faisalabad (30.95%) with an overall disease incidence of 40.48% observed during 2019 and 2020. Newly detected Pakistani CABYV isolates shared 99.6 % and 94.7-96.2% nucleotide identities among themselves and with isolates retrieved from NCBI GenBank. Phylogenetic analysis showed that current study isolates i.e., OL828566 and OL828567 clustered in separate clades with isolates JQ700305, JF939814 and X7693 from Taiwan, Spain and France, respectively while isolates from Thailand, China, and Korea were grouped separately. It was evident with restriction fragment length polymorphism (RFLP) analysis that the present study CABYV isolates were grouped into two types, which seemed to be genetically similar to those identified during 2011–2015 and 2015–2021. Recombination detection analysis showed that newly detected CABYV isolates are likely to be recombinant of Spanish (JF939814) and South Korean (KR231950) isolates having the recombination breakpoint between nucleotide position number 272 and 830. Comparison and recombination detection analysis of the local isolates might help in devising a breeding program to identify resistant sources against recombinant isolates.
Keywords
Cucurbits
Recombination
In silico RFLP
RT-PCR
1 Introduction
Cucurbit crops include several economically significant vegetables and fruit crops that are being grown in various regions of Pakistan (Ahsan et al., 2020). Due to the advantage of favourable climatic conditions in Punjab, most of the cucurbits are grown throughout the year as summer and winter crops (Khokhar, 2014). In Punjab, the main cultivated cucurbit species include melon (Cucumis melo L.), cucumber (C. sativus L.), squash (Cucurbita sp.), gourd (Luffa sp.), pumpkin (Cucurbita moschata), and watermelon (Citrullus lanatus L.). The average yield of cucurbits in Pakistan is quite low as compared to other countries because of several biotic and abiotic factors but the diseases caused by RNA viruses are severely affecting cucurbits in the region (Asad et al., 2022). Among these RNA viruses, Zucchini yellow mosaic virus (ZYMV; Potyvirus), Papaya ringspot virus (PRSV; Potyvirus), Watermelon mosaic virus (WMV; Potyvirus), Cucurbit aphid-borne yellows virus (CABYV; Polerovirus) Cucumber mosaic virus (CMV; Cucumovirus) are the significant ones which have been reported to infect cucurbits in Pakistan (Ahsan et al., 2020a,b, Asad et al., 2019, 2022, Ashfaq et al., 2015, 2017, 2021; Ashfaq and Ahsan, 2017).
The Cucurbit aphid-borne yellows virus (Polerovirus: family Luteoviridae) was identified for the very first time in France in 1982 (Knierim et al., 2010). CABYV is considered to be the first known polerovirs infecting cucurbit crops (Lecoq et al., 1992). It was reported from Pakistan in 2020 (Ahsan et al, 2020). Poleroviruses are ssRNA(+) viruses with six open reading frames (ORFs) in an approximately 5.7 kb genome (d’Arcy et al., 2005). The three 5′ proximal ORFs (ORF 0–2) were separated from ORF 3–5 (three 3′ proximal ORFs) by 200 nucleotides (nt) non-coding intergenic region (IR) (Krueger et al., 2013; d’Arcy et al., 2005).
Poleroviruses are reported to have effects on the yield of different crops but they remain unnoticed due to several reasons, including the yellowing of leaves that look like a deficiency of nutrients (Desbiez et al., 2016). CABYV infections are often mixed with infections of other viruses affecting cucurbits (Mnari-Hattab et al., 2009). The initial symptoms due to CABYV on cucumber, watermelon and squash are yellow mottling and patches of interveinal chlorosis which results in coalesce, yellowing, thickening and brittleness of leaves (Lecoq and Desbiez, 2017). Commonly, signs of diseases are retained in the early leaves but significant changes have been seen from cultivar to cultivar. In some cultivars, only mild symptoms developed on a few leaves while others showed bright yellowing at the complete plant (Lecoq et al., 1992). Furthermore, a reduction in the number of fruits and dropping of blossoms are often observed (Menzel et al., 2020). The severity of the symptoms can change with time, may vary according to the season and are more prominent in the hot season than in cold (Chung et al., 2015; Relevante et al., 2012). Thickening with yellowing appears on older leaves (Knierim et al., 2010). On young leaves, interveinal chlorosis occurs and dull green veins appear on the earlier leaves. The leaves of infected plants become thicker while the leaf edges roll upwards (Romay et al., 2014). The transmission of CABYV is possible with the help of particular aphid species. The viral transmission is reported to be done through circulation, persistent and non-propagation manner by specific aphid vectors and there is no possibility of transmission by mechanical ways (Jeger, 2020; Khanal and Ali, 2019). It is demonstrated that transmission of CABYV is possible through Myzus persicae and Aphis gossypii only (Lecoq et al., 1992).
The data regarding full length and partial sequences of CABYV isolates is already being deposited in GenBank from several parts and different geographical regions of the world (Caciagli, 2009; Lecoq and Desbiez, 2012; Romay et al., 2014; Ahsan et al., 2020). Several Asian countries including Thailand, Philippines, and Taiwan have described another cucurbit infecting polerovirus that emerged as recombinant isolates between melon aphid-borne yellows virus and CABYV (Knierim et al., 2010, 2014). Whereas, the scientists in Brazil recently described a recombinant isolate of CABYV and undetermined polerovirus (Costa et al., 2019). Two major molecular groups; basal Asian and Mediterranean have been defined among non-recombinant isolates and Eastern Asia represented minor highly divergent groups (Kassem et al., 2013; Kwak et al., 2018). In Pakistan, limited research on CABYV has been done previously and scarce information about its molecular diversity is available. Therefore, this study aimed to characterize the variability of Pakistani CABYV isolates using movement protein (MP), RdRp, and partial coat protein (CP) sequences followed by determination of its relationship with other reported isolates.
2 Materials and methods
2.1 Field surveys for polerovirus in Punjab
Surveys were carried out in Punjab (Rawalpindi, Chakwal, Sialkot, Faisalabad, Multan, Vehari, Bahawalpur, Muzaffargarh, Khanewal and Lodhran) during 2019 and 2020 for the detection of Poleroviruses infecting cucurbits (Fig. 1). In each season, randomly selected cucurbits open fields and tunnels were visited and 650–670 samples depicting suspected Poleroviruses symptoms notably interveinal chlorosis which results in coalesce, yellowness, and thickness and brittleness of leaves (Ahsan et al., 2020) were collected (Fig. 2). The coordinates of surveyed sites were recorded using Global Positioning System (GPS) and visited again for sample collection. Data regarding poleroviruses, varietal response, vector and its control was collected from surveyed farmers using a brief questionnaire.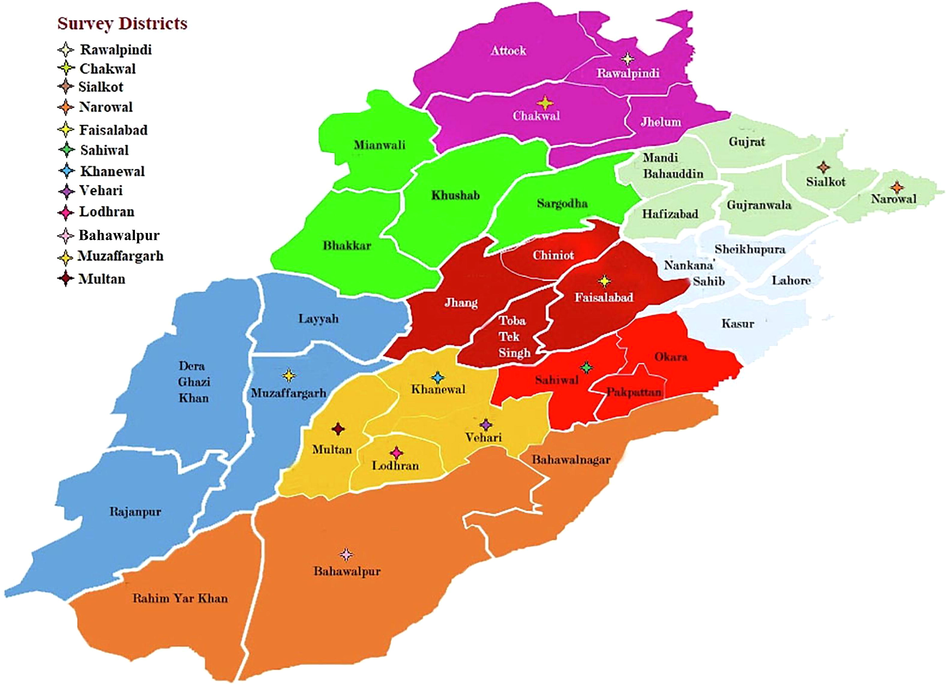
Map of Survey Districts.
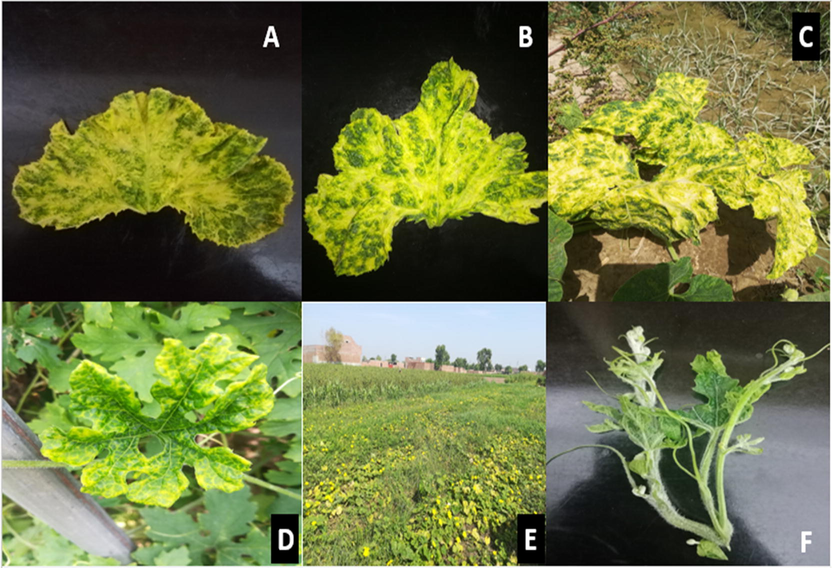
Typical symptoms of CABYV infecting cucurbits; A (Round gourd), B (Pumpkin) C (Squash), D (Bitter gourd), E (Ridge gourd) and F (Watermelon).
2.2 Screening of polerovirus using group-specific degenerate primers
In a preliminary screening for the prevalence of Poleroviruses, TRIzol® Reagent (Cat# 15596, Life Technologies, Carlsbad, USA) was used to extract total RNA followed by quantification using Nanodrop (Thermo Scientific Co. USA) as per the instruction manual. Nuclease-free water was used to make a 500 ng/μL RNA working dilution followed by synthesis of first-strand cDNA using Revert Aid Reverse Transcription Kit (K1691, Thermo Fischer Scientific, USA) and PococpR-140 (Xiang et al., 2008) as reverse primer. PCR amplification was done from the resultant cDNA through poleroviruses specific degenerate primers PococpR-140 and PococpF-139 as described by Xiang et al. (2008). A pre-stained 1% (w/v) agarose gel was used to examine the PCR product using electrophoresis followed by visualization under UV trans-illuminator (Vilber Lourmat, S. No. 6532). A sample for poleroviruses was considered positive if a band of 1.4 kb was detected and the below-given formula was used to calculate the disease incidence (Rao et al., 2002).
2.3 Cloning and sequencing
GeneJET PCR Purification Kit (Thermoscientific, USA) was used to purify the positive amplicons of 1.4 kb and propagated through Escherichia coli XL1-Blue as the host of pTZ57R/T TA cloning vector (K1214, Thermo Scientific, USA). To purify recombinant plasmid DNA, GeneJET Plasmid Miniprep Kit (K0503, Thermo Fisher Scientific, USA) was used as per manufacturer directions. The existence of an insert in transformants was validated using restriction digestion with ECoR1 enzyme and positive colonies were sent to Macrogen (South Korea) for sequencing in both directions. NCBI BLAST tool was used to compare the obtained sequences with polerovirus sequences available in GenBank.
2.4 Phylogenetic analysis of polerovirus
Polerovirus sequences of the present study and sequences submitted from other parts of the world were retrieved from GenBank and aligned with ClustalW embedded in MegaX software (Kumar et al., 2018). After alignment in MegaX software, the Maximum Likelihood method with 1000 bootstrap replicates was used to detect their ancestral lineage and phylogenetic relationship. BioEdit v7.2.6.1 having the Sequence Identity Matrix option was used to determine the nucleotide and amino acid sequences identities (Hall, 1999).
2.5 In silico RFLP and recombination analysis
In silico restriction fragment length polymorphism (RFLP) of CP gene of our isolate from this present study isolate and 30 other CABYV isolates retrieved from GenBank, was performed using BccI restriction enzyme in CLC Main Workbench 20 (https://www.qiagenbioinformatics.com/). The resulting virtual gel was analysed in PyElph v1.4 (Pavel and Vasile, 2012) for serotyping of isolates and construction of phylograms. Recombinant events in the two new Pakistani and 10 other CABYV isolates were analyzed with RDP4 (Martinl et al., 2017) using all the available methods with default settings viz. MaxChi, BootScan, RDP, 3SEQ, Siscan, GENECONV, PhylPro and Chimaera.
3 Results
3.1 Polerovirus screening
During 2019–2020, the incidence of Poleroviruses was tested in 671 symptomatic samples of cucurbits through RT-PCR with group-specific degenerate primers PococpR-140 and PococpF-139 (Xiang et al., 2008). In 2019, 123 samples out of 308 produced 1.4 kb RT-PCR product of expected size while 149 out of 363 samples produced the expected product in 2020. The production of the expected 1.4 kb RT-PCR product implies the poleroviruses incidence. Based on RT-PCR results, the disease incidence percentage was calculated as shown in Table 1. Overall 40.48% of disease incidence was recorded in Punjab, Pakistan during 2019 and 2020. In 2019, higher disease incidence (45.45%) was recorded in Rawalpindi, followed by Khanewal (44.44), Multan and Lodhran (42.85%), Sialkot (40%), Vehari (37.77%), Muzaffargarh (37.50%), Bahawalpur (35.71%), Chakwal (33.33%) and lowest in Faisalabad (28.57%). A relatively higher disease incidence was recorded in 2020 with the same trend as observed previously in Rawalpindi (50%) as highest and Faisalabad as lowest (33.33%). This higher incidence may be attributed to the already establishment of aphids as a vector of poleroviruses that are actively transmitted by aphids during the growing season.
Sampling Sites
2019
2020
S. No.
RT-PCR
RT-PCR
+Ve/Total
%D.I
+Ve/Total
%D.I
1
Rawalpindi
15/33
45.45
15/30
50
2
Chakwal
10/30
33.33
15/40
37.50
3
Faisalabad
2/7
28.57
5/15
33.33
4
Vehari
17/45
37.77
19/45
42.22
5
Muzaffargharh
15/40
37.50
20/45
44.44
6
Multan
15/35
42.85
15/33
45.45
7
Lodhran
15/35
42.85
18/40
45
8
Bahwalpur
10/28
35.71
20/45
44.44
9
Khanewal
20/45
44.44
25/55
45.45
10
Sialkot
4/10
40
7/15
46.66
D.I in Punjab
123/308
39.93
149/363
41.04
3.2 Molecular characterization and phylogenetic analysis
Genus specific primers PoconF-139/PococpR-140 were used to characterize poleroviruses based on RdRp and CP gene and the amplified fragment of 1.4 kb of RdRp-CP gene was obtained from maximum number of cucurbit samples (Xiang et al., 2008). BLASTn revealed that the sequence consisted of partial RdRp, entire CP, and overlapping MP genes. Furthermore, all Pakistani Polerovirus isolates showed a conserved sequence of eight nucleotides ACAAAAGA immediately upstream of the intergenic NCR, which is identical to the first eight residues of the 5′terminus NCR and has been acknowledged as the transcription beginning site for polerovirus subgenomic RNA1. After careful analysis, two isolates of CABYV have been submitted to GenBank with accession numbers given in Table 2.
Accession No.
Location
Host
Year
NT identity % of Pakistani isolates
AA identity % of Pakistani isolates
OL828566
OL828567
OL828566
OL828567
JF939814
Spain
Cucurbita pepo
2013
95.5
95.3
96.6
96.9
X76931
France
unknown
1994
95
95.2
94.4
94.7
KF791040
Thailand
Cucumis sativus
2015
94.6
93.9
94.7
94.9
KR231947
South Korea
Cucumis melo
2015
94.2
94.5
96
96.2
EU636992
China
C. melo var. cantalupensis
2010
93
94
95.6
95.8
KR231950
South Korea
Cucumis melo
2015
94.2
94.3
94.9
95.1
KF815682
Thailand
Cucumis sativus
2015
94.3
94.4
95.1
95.3
EU000535
China
C. argyrosperma
2008
94
94
94.9
95.1
JQ700305
Taiwan
Momordica charantia
2012
96.2
95
94.2
94.4
GQ221223
China
Cucurbita sp.
2012
93.8
93.5
95.1
95.3
BLASTn analysis revealed that each sequence has 597 nucleotides that encode 198 amino acids (aa) of incomplete RdRp gene, 199 nucleotides of intergenic-NCR, and 600 nucleotides that encode 299 CP gene, and 576 nucleotides that encode 192 aa of MP that overlaps CP gene. All Pakistani CABYV isolates had a conserved sequence of ACAAAAGA immediately upstream of the intergenic NCR and a CP conserved motif “GILKAYHE” at position 94–101.
Sequence Identity Matrix results showed that the current study Pakistani CABYV isolates shared 99.6% nucleotide identity among themselves while 94.7–96.2% identity with previously reported CABYV isolates. In case of amino acids, they shared 99.7% similarity with each other and 94.2–96.9% similarity with other isolates reported from other parts of the world and used in this study for analysis. Isolate OL828566 of this study shared 96.2% similarity with JQ700305 from Taiwan and 95 and 95.5% similarity with X76931 and JF939814 from France and Spain, respectively. Similarly. OL828567 isolate shared 95.3% similarity with JF939814 from Spain and 95–95.2% with JQ700305 and X76931 from Taiwan and France, respectively. A similarity of 94.2–96.9% was evident between amino acid based sequences of present study isolates and isolates reported from elsewhere (Table 2). Phylogenetic analysis showed that current study isolates form separate clades with isolates JQ700305, JF939814 and X7693 from Taiwan, Spain and France, respectively while other isolates from China, Thailand and South Korea form separate clad (Fig. 3). Phylogenetic analysis revealed that CABYV Pakistani isolates didn’t follow the geographic lineage i.e. Asiatic and Mediterranean.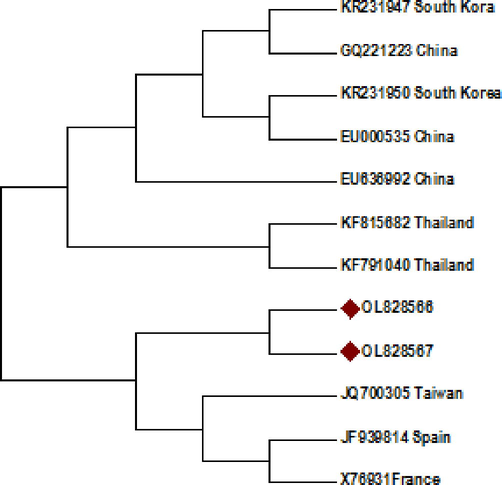
Evolutionary relationships based on all the understudied genes of nucleotides sequences of Pakistani CABYV isolates with already reported sequences.
3.3 In silico RFLP and spatiotemporal analysis
In silico RFLP simulation of CP gene of CABYV by using BccI delineates 32 isolates into two different patterns (Pattern I and Pattern II). Pattern I and pattern 2 consisted of approximately 120 bp and 206 bp fragments, respectively (Fig. 4). Of all isolates studied, 9 and 17 isolates corresponded to the pattern I and pattern II, respectively while the other six are considered as others. Interestingly, it is observed that both the genetic groups were present in both time periods (2011–2015 and 2015–2021) but all the accession of the CP gene were not equally distributed into two genetic groups, however, a few accessions were not found in either group, categorized as other. Interestingly, it is observed during the study that in group 1 (Pattern I) only accessions from Asia were present but in group 2 (pattern II) isolates from Asia, Europe and America were present. The spatial and temporal study revealed that genetic variation and geographical distribution of CABYV was increased after 2015 than before 2015.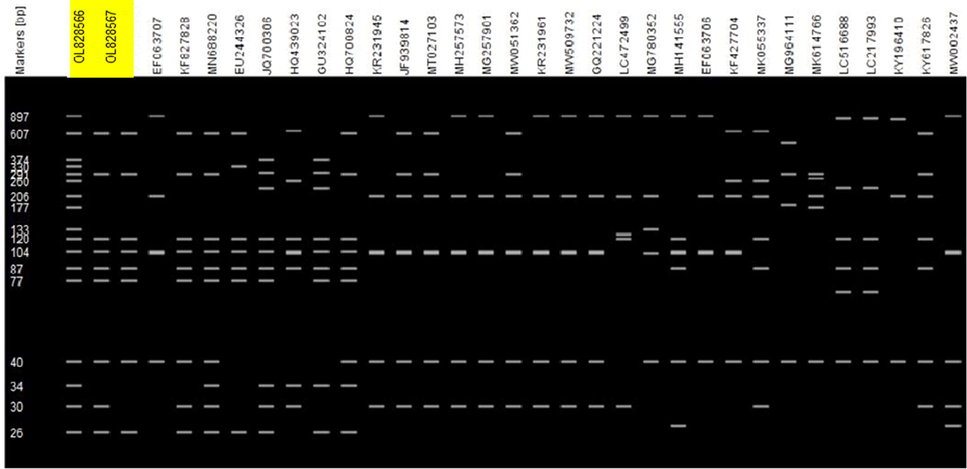
In silico RFLP analysis and genotyping of 32 isolates of CABYV.
3.4 Genetic recombination analysis
For genetic recombination analysis, all of the methods provided in RDP4 with default settings were used to evaluate recombination breakpoint events in the understudied gene of CABYV isolates presented in Table 3, Fig. 5. The analysis detected two recombination events, one of which (Event 2) was significant because it was recognized by more than three methods out of nine methods with a significant p-value, while the other was not. Event no 2 revealed that Pakistani isolates OL828566 and OL828567 of CABY virus might be originated from possible recombination between KR231950 as a major parent with 96.9% similarity and an unknown minor parent might be JF939814. This event was significantly detected with nine methods i.e. GENECONV, MaxChi, BootScan, RDP, Chimaera, 3Seq, LARD, Phylprp and SiScan. The recombination breakpoint was ranged between 269 and 832, 272–830, 272–830, 271–828, 271–830, 272–830, 271–832, 272–828 and 269–828 nucleotide in GENCONVE, Chimaera, MaxChi, LARD, Phylpro, SiScan, 3Seq, BootScan and RDP, respectively. The P-value for each method is presented in Table 3.
Recombinant Isolate
Breaking point
Major
ParentMinor
ParentP-Value
Starting
Ending
RDP
GENECONV
Bootscan
MaxChi
Chimaera
SiScan
3SEQ
LARD
PhylPro
OL828566
272
830
KR231950
JF939814
NS
9.850 × 10−2
NS
1.505 × 10−1
7.725 × 10−2
NS
1.889 × 10−2
NS
NS
OL828567
272
830
KR231950
JF939814
NS
9.850 × 10−2
NS
1.505 × 10−1
7.725 ×
10−2
NS
1.889 × 10−2
NS
NS
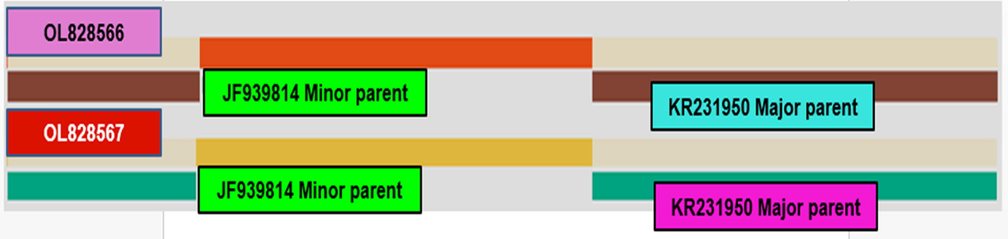
Recombination events identified in Pakistani CABYV isolates employing available methods in RDP4 with major and minor parents.
4 Discussion
Pakistan is a major contributor to cucurbits production in Asia with an area of 67360 ha under cucurbit cultivation with 1 million tonnes of yield (Statistics of Pakistan 2019–2020). The cucurbit production is hampered by many biotic and abiotic factors generally and viral diseases particularly. According to our assessment of the current scenario of CABYV in cucurbits samples showing yellowing symptoms with an overall 40.48% disease incidence is emerging as a major threat to cucurbits production in core cucurbits producing areas of Punjab, Pakistan as well as all over the world (Ahsan et al., 2020a,b; Juarez et al., 2013; Juárez et al., 2019; Lecoq, 1999). CABY virus reproted to be the most frequent virus damaging cucurbits in temperate, Mediterranean, and subtropical climates (Lecoq et al., 1992). Two aphid species; M. persicae and A. gossypii were found to be involved in the transmission of CABYV in a persistent and circulative manner. Therefore, the cultivated or wild plant species could be linked with the high prevalence and occurrence of CABYV while the earlier may serve as a source of inoculum for its spread. (Kassem et al., 2013; Khanal and Ali, 2019). Furthermore, CABYV-infected plants have been shown to alter aphid feeding behaviour in such a way that it helps in virus acquisition while healthy plants were preferably used by viruliferous aphids to settle (Carmo-Sousa et al., 2016). This altered feeding behavior of aphid along with overlapping system of cucurbit and existence of wild plants near cultivation fields may lead to the high prevalence of CABYV in those cucurbit crops lacking CABYV-resistant sources. BLASTn revealed that sequence consisted of partial RdRp, entire CP, and overlapping MP gene. Moreover, an intergenic non translated region (NTR) of 199–203 nucleotide region was also observed between RdRp and CP which is in accordance with the finding of Kassem et al. (2013), Knierim et al. (2014), Xiang et al. (2008). Detailed sequence compassion revealed that all Pakistani Polerovirus isolates showed a conserved sequence of eight nucleotides ACAAAAGA immediately upstream of the intergenic NCR, which is identical to the first eight residues of the 5′terminus NCR and has been acknowledged as the transcription beginning site for polerovirus subgenomic RNA1 (Beuve et al., 2008; Chen et al., 2016; Guilley et al., 1994; Krueger et al., 2013; Pazhouhandeh, 2007) and a CP conserved motif “GILKAYHE” (Guilley et al., 1994). Sequence Identity Matrix results showed that the Pakistani isolates from the current study shared 99.6% of nucleotide identity among themselves while 94.7–96.2% identity with previously reported CABYV isolates. Phylogenetic analysis revealed that the current study isolates form separate clades with isolates JQ700305, JF939814 and X7693 from Taiwan, Spain and France, respectively while other isolates from China, Thailand and South Korea form separate clad. Phylogenetic analysis revealed that CABYV Pakistani isolates didn’t follow the geographic lineage i.e. Asiatic and Mediterranean as observed by Kassem et al. (2013), Kwak et al. (2018) and Shang et al. (2009). The key sources of genetic variability in RNA viruses are mutation, reassortment, and recombination (Akinyemi et al., 2016; Holmes, 2006). This could lead to the encapsulation of discrete sequence components, as well as the repetition, interchange, or obliteration of existing viral elements. All of the methods provided in RDP4 with default settings were used to evaluate recombination breakpoint events. (Martin et al., 2015) for estimation of recombinant events in the CP + MP + RdRp gene of CABYV isolates. The analysis found that two recombination events were detected, one of which (Event 2) was significant because it was recognized by more than three methods out of nine methods with a significant p-value, while the other was not. Currently, various recombinant polerovirus isolates including CABY virus isolates, have been identified as significant (Knierim et al., 2010; Costa et al., 2020), which necessitates extensive studies on the genetic variability of CABYV infecting cucurbit crops. RFLP analysis with BccI suggested that the simulated CABYV isolates may be alienated into three genetic groups i.e. group 1 (Pattern I) and group 2 (Pattern II) comprised of nine and seventeen isolates, respectively while remaining six isolates with no definite pattern were considered as others. Interestingly, it is observed that both the genetic groups were present in both time periods (2011–2015 and 2015–2021). Similar patterns were also observed by (Garcia-Arenal et al., 2001; Kassem et al., 2007). The presence of recombinant isolates and their uneven spatiotemporal dynamics threatened efficient crop production which should be resolved by appropriate viral diagnostic and management strategies.
5 Conclusion
The present study revealed that CABYV is an emerging threat to cucurbit crops, in Punjab, Pakistan though the extent of damages posed by yellows disease in cucurbits is unknown. It is important to estimate epidemics caused by plant viruses as they are frequently emerging and transmitted by diverse species of plants as well as varies according to the different agro-ecological zone that can affect viral disease epidemiology. This spatial/temporal flow of viral infections between overlapping crops could play a significant role, as it may obstruct early disease diagnosis. Therefore, the response of cultivated and wild plant species against genetically diverse CABYV population and its distribution study is need of the hour that would help in the measures related to disease control.
Acknowledgement
This study was funded by Higher Education Commission (HEC) of Pakistan under NRPU project No. 8162 to Muhammad Ashfaq. The authors also extend their appreciation to the support of the Research Center for Advanced Materials Science (RCAMS) at King Khalid University, Abha, Kingdom of Saudi Arabia through a project number RCAMS/KKU/G003-21.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- First report of Cucurbit aphid borne yellows virus (CABYV) infecting melon in Pakistan. J. Plant. Pathol.. 2020;102:563-564.
- [CrossRef] [Google Scholar]
- Current status and genetic variability of cucumber mosaic cucumovirus (CMV) isolates infecting major cucurbits and solanaceous vegetables in Pothwar region of Pakistan. Pak. J. Agri. Sci.. 2020;57(5):1353-1361.
- [Google Scholar]
- Ecogenomic survey of plant viruses infecting Tobacco by Next generation sequencing. Virol. J.. 2016;13(1):181.
- [Google Scholar]
- Genetic diversity of cucumber green mottle mosaic virus (CGMMV) infecting cucurbits. Saudi J. Biol. Sci.. 2022;29:3577-3585.
- [CrossRef] [Google Scholar]
- Incidence and distribution of Zucchini yellow mosaic virus (ZYMV) infecting Cucumber (Cucumis sativus L.) crop in Pothowar. Pakistan. Pur. Appl. Biol.. 2019;8:2036-2043.
- [Google Scholar]
- Current status and molecular characterization of Zucchini yellow mosaic virus (ZYMV) infecting Ridge gourd (luffa acutangula L) in different regions of punjab, pakistan. Pak. J. Bot. 2022;54(2):467-474.
- [Google Scholar]
- First Report of Zucchini yellow mosaic virus in Round Gourd (Praecitrullus fistulosus) in Pakistan. Plant. Dis.. 2017;101(1):265.
- [Google Scholar]
- First report of Zucchini yellow mosaic virus in ridge gourd in Pakistan. Plant. Dis.. 2015;99(12):1870.
- [Google Scholar]
- Natural Occurrence and Host Range Studies of Cucumber Mosaic Virus (CMV) Infecting Ornamental Species in Rawalpindi-Islamabad Area of Pakistan. Philipp. Agric. Sci.,. 2017;100(1):55-61.
- [Google Scholar]
- Molecular characterization and identification of economically important Potyviruses in Cucurbitaceae family from Gujranwala division of Punjab, Pakistan. J. King Saud Univ. Sci.. 2021;33(8):101642.
- [Google Scholar]
- Biological and molecular characterization of an American sugar beet-infecting Beet western yellows virus isolate. Plant. Dis.. 2008;92(1):51-60.
- [Google Scholar]
- Caciagli, P., 2009. Vegetable viruses. In Desk encyclopedia of plant and fungal virology, 1st ed.; Mahy BW, Van-Regenmortel MH. Eds; Elsevier Academic press: London.
- Cucurbit aphid-borne yellows virus (CABYV) modifies the alighting, settling and probing behaviour of its vector Aphis gossypii favouring its own spread. Ann. Appl. Biol.. 2016;169(2):284-297.
- [Google Scholar]
- Characterization of a novel polerovirus infecting maize in China. Viruses. 2016;8(5):120.
- [Google Scholar]
- Effects of temperature on systemic infection and symptom expression of Turnip mosaic virus in Chinese cabbage (Brassica campestris) Plant Pathol. J.. 2015;31(4):363.
- [Google Scholar]
- Cucurbit aphid-borne yellows virus from melon plants in Brazil is an interspecific recombinant. Arch. Virol.. 2019;164(1):249-254.
- [CrossRef] [Google Scholar]
- The recombinant isolate of cucurbit aphid-borne yellows virus from Brazil is a polerovirus transmitted by whiteflies. Plant Pathol.. 2020;69(6):1042-1050.
- [Google Scholar]
- d’Arcy, C., Domier, L., Mayo, A., 2005. Family luteoviridae. Virus Taxonomy. VIIIth Report of International Committee on Taxonomy of Viruses.
- Desbiez, C., Millot, P., Wipf-Scheibel, C., Blancard, D., Chesneau, T., Lecoq, H., 2016. First report of Pepo aphid-borne yellows virus in cucurbits in Tanzania and Mayotte. New Dis. Rep. 33, 20.
- Variability and genetic structure of plant virus populations. Annu. Rev. Phytopathol. 2001;39(1):157-186.
- [Google Scholar]
- Nucleotide sequence of cucurbit aphid-borne yellows luteovirus. Virology. 1994;202(2):1012-1017.
- [Google Scholar]
- BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic acids symposium series. 1999;c1979–c2000:95-98.
- The evolution of viral emergence. Proc. Natl. Acad. Sci.. 2006;103(13):4803-4804.
- [CrossRef] [Google Scholar]
- The epidemiology of plant virus disease: Towards a new synthesis. Plants. 2020;9:1768.
- [Google Scholar]
- Relative incidence, spatial distribution and genetic diversity of cucurbit viruses in eastern Spain. Ann. Appl. Biol.. 2013;162(3):362-370.
- [Google Scholar]
- Natural hosts and genetic diversity of the emerging tomato leaf curl New Delhi virus in Spain. Front. Microbiol.. 2019;10:140.
- [Google Scholar]
- Cucurbit aphid-borne yellows virus is prevalent in field-grown cucurbit crops of southeastern Spain. Plant Dis.. 2007;91(3):232-238.
- [Google Scholar]
- Genetic diversity and potential vectors and reservoirs of Cucurbit aphid-borne yellows virus in Southeastern Spain. Phytopathol.. 2013;103:1188-1197.
- [CrossRef] [Google Scholar]
- Complete genome sequence of a zucchini yellow mosaic virus isolated from pumpkin in Oklahoma. Microbiol. Resour. Announce.. 2019;8(2):e01583-e01618.
- [Google Scholar]
- Production status of major vegetables in Pakistan, their problems and suggestions. Agric. Corner. 2014;9
- [CrossRef] [Google Scholar]
- Molecular identification of three distinct Polerovirus species and a recombinant Cucurbit aphid-borne yellows virus strain infecting cucurbit crops in Taiwan. Plant. Pathol.. 2010;59:991-1002.
- [CrossRef] [Google Scholar]
- Molecular diversity of poleroviruses infecting cucurbit crops in four countries reveals the presence of members of six distinct species. Arch. Virol.. 2014;159:1459-1465.
- [CrossRef] [Google Scholar]
- The complete nucleotide sequence of the genome of Barley yellow dwarf virus-RMV reveals it to be a new Polerovirus distantly related to other yellow dwarf viruses. Front. Microbiol.. 2013;4:205.
- [Google Scholar]
- MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.. 2018;35(6):1547-1549.
- [Google Scholar]
- Complete genome sequences and evolutionary analysis of Cucurbit aphid-borne yellows virus isolates from melon in Korea. Plant Pathol. J.. 2018;34(6):532-543.
- [Google Scholar]
- A new yellowing disease of cucurbits caused by a luteovirus, Cucurbit aphid-borne yellows virus. Plant Pathol. J.. 1992;41(6):749-761.
- [Google Scholar]
- Viruses of cucurbit crops in the Mediterranean region: an ever-changing picture. Adv. Virus. Res. Elsevier.. 2012;84:67-126.
- [Google Scholar]
- Moroccan watermelon mosaic. newly reported on zucchini squash in France. Plant. Pathol.. 2017;57(4)
- [CrossRef] [Google Scholar]
- Lecoq, H., 1999. Epidemiology of Cucurbit aphid-borne yellows virus. 243-248 in: The Luteoviridae. H. G. Smith and H. Baker, eds. CAB International, Walingford, UK.
- RDP4: Detection and analysis of recombination patterns in virus genomes. Virus. Evolution. 2015;1(1)
- [CrossRef] [Google Scholar]
- Martinl, D.P, Murrel, B, Khoosal, A., Muhire, B., 2017. Detecting and analyzing genetic recombination using RDP4. In Bioinformatics; Springer. 433-460.
- First report of Cucurbit aphid-borne yellows virus infecting cucurbits in Germany. New Dis. Rep.. 2020;41(1)
- [Google Scholar]
- Biological and mo-lecular characterization of the Cucurbit aphid-borne yellows virus affecting cucurbits in Tunisia. Plant. Dis.. 2009;93(10):1065-1072.
- [Google Scholar]
- PyElph-a software tool for gel images analysis and phylogenetics. BMC. Bioinform.. 2012;13:1-6.
- [Google Scholar]
- The mechanism of action of Polerovirus P0 in RNA silencing suppression. (PhD Doctoral Thesis), University of Strasbourg, France. plant virus populations. Annu. Rev. Phytopathol.. 2007;39:157-186.
- [Google Scholar]
- Occurrence and incidence of banana bunchy top disease in southern part of Sindh. Plant Pathol. J.. 2002;1(2):74-75.
- [Google Scholar]
- Emerging new poleroviruses and tospoviruses affecting vegetables in Asia and breeding for resistance. Extension Bull.-Food Fertilizer Technol. Center. 2012;657:1-12.
- [Google Scholar]
- Current status of cucurbit viruses in V enezuela and characterization of V enezuelan isolates of Z ucchini yellow mosaic virus. Plant. Pathol.. 2014;63:78-87.
- [Google Scholar]
- Distribution and molecular diversity of three cucurbit-infecting poleroviruses in China. Virus. Res.. 2009;145(2):341-346.
- [Google Scholar]
- Complete sequence analysis reveals two distinct poleroviruses infecting cucurbits in China. Arch. Virol.. 2008;153(6):1155-1160.
- [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2022.102255.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







