Translate this page into:
CpG methylation analysis of tumour suppressor gene and expression of Cathepsin B in renal cell carcinoma
⁎Corresponding authors. venzymes@gmail.com (P. Vijayaragavan), vkgopalakrishnan@gmail.com (P. Vijayaragavan), vkgopalakrishnan@gmail.com (V.K. Gopalakrishnan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Renal cell carcinoma (RCC) is one of the most common types of cancers, representing about 2.3% of all malignancies throughout the world. In RCC, Cathepsin B (CtsB) plays a major role in signalling pathways, processing, and intracellular protein degradation. CtsB is involved in tumour cell invasion, matrix remodelling and metastasis and this mechanism was controlled by natural inhibitors. In this study targeted methylation changes were analyzed in primary renal cancer and metastatic tissues and the expression of CtsB was analyzed in RCC and compared with normal cells. The levels of CpG methylation of Runt-trelated transcription factor 3 (RUNX3) were analyzed. The decreased level of RUNX3 was observed in metastatic tissues due to CpG methylation. The increased methylation in CpG islands increased metastasis and was associated with RUNX3. The amount of CtsB mRNA was high in the RCC tissues than in surrounding normal tissues. Increased expression of CtsB was observed in RCC. Overexpression of CtsB was confirmed by Western blotting analysis and expression of CtsB increased the proliferation of RCC. The present finding revealed that CtsB has a major role in the development and proliferation of RCC.
Keywords
Renal cell carcinoma
CpG methylation
Cathepsin B expression
Metastasis
1 Introduction
Renal cell carcinoma (RCC) is one of the common kidney neoplasia and is reported at 3 % in women and 5 % in men, respectively (Capitanio et al., 2019). In most of cases, RCCs are commonly diagnosed in 25 % of patients in the early stages at the time of clinical diagnosis. In recent years, therapeutic approaches have developed continuously to improve the health of patients associated with RCC, however, the survival of individuals with metastatic disease is very less (Patel et al., 2019). Kidney cancer is one of the most important risky solid tumours worldwide and more than 400,000 cases and about 175,000 deaths were reported in 2018 (Ferlay et al., 2019). RCC accounts for about 90 % of all kidney tumours and the clear cell (ccRCC) is an important type of kidney tumour. About 70 % of the RCC is associated with localized diseases; however 30 % of RCC will relapse after surgical procedure (Janzen et al., 2003). Moreover, the possibility of reappearance of RCC will be based on different histopathological and clinical characteristics, which were included in up to twenty various scoring systems characterized by heterogeneity (Meskawi et al., 2012). Renal cell carcinoma is classified as chromophobe (5 %), papillary (10–15 %), clear cell (75–80 %) and renal cell carcinoma. Despite recent advances in very early diagnosis and advanced treatment, the individuals associated with RCC are still suffered from increased mortality. In this aspect, continuous effort on early diagnosis of RCC, progression, pathogenesis, important diagnostic markers and novel pharmacological targets are required (Vasudev et al., 2020).
Several genes associated with the modifications of histone have been reported by various sequencing projects in ccRCC and a lower frequency of mutations was reported. These include histone demethylases (JARID1C and UTX) and histone methylases (SETD2 and MLL2) (van Haaften et al., 2009). Epigenetic alterations play a potent role in the progression and development of human cancers. DNA methylation analysis is state of art method utilized widely in cancer disease due to the reversible property of the biological processes underlying DNA methylation process (Pan et al., 2018). DNA methylation is used to demonstrate the importance of clinically relevant adverse pathological parameters. Recent studies revealed the specific associations between unfavourable histopathological characteristics and DNA methylation (Peters et al., 2018), shorter recurrence-free or cancer-specific survival (Atschekzei et al., 2012), metastatic disease (Tezval et al., 2016), and the predicted response to anti-angiogenic therapy (Peters et al., 2014). The main objective of the present study was to analyze the genetic markers to diagnose RCC in earlier stages.
2 Materials and methods
2.1 Specimens
Thirteen RCC patients admitted to renal cancer treatment were subjected to this analysis after written consent by patients. All experimental procedures were approved by the Institutional Ethical Committee (2983/2021–21). The pathological properties of selected patients were analyzed using clinical history. The patient’s age, sex, tumour type, stage and pathological conditions were registered.
2.2 Renal cell culture
Metastatic renal tissues and primary renal patient cancer tissues were derived from the abdomen region between July 2020 and November 2021. A spring scissor was used to cut a small piece of cancer tissue and subjected to collagenase treatment. After 30 min incubation at 32 ± 2 °C, the reaction mixture was centrifuged 500 g for 20 min. The final pellet was mixed with Dulbecco’s Modified Eagle Medium (DMEM) culture medium incorporated with fetal bovine serum (FBS) and antibiotics (streptomycin and penicillin). The prepared renal cancer cells were cultured in DMEM with FBS (10 %) and incubated at 37 °C in a CO2 incubator.
2.3 Quantitative methylation-specific PCR and analysis
The expression of genes with CpG islands in their promoters was quantified in this study using quantitative RT-PCR arrays. The main aim of the study was to analyze the methylation-regulated genes linked with metastasis. Quantitative methylation-specific PCR was carried out as suggested previously (Moller et al., 2017).
2.4 Cathepsin B expression
2.4.1 In vitro culture of human renal cancer cell lines
The cell lines, A498 and 769-P were obtained from American Type Culture Collection and were grown under aseptic condition using the culture medium (RPMI 1640). The tissue culture medium was supplemented with fetal bovine serum (10 %) and incubated with 1 % antibiotics (streptomycin and penicillin). It was incubated at 37 °C in a humidified chamber containing 5 % CO2. The cell lines were analyzed and ensured free of any contamination (Keppler et al., 2000).
2.4.2 Isolation of RNA and Real-Time polymerase chain reaction analysis (RT-qPCR)
RNA was extracted from the renal cancer individuals from both healthy and tumour tissues using a commercial RNA isolation kit (Qiagen, Germany), according to the manufacturer’s instruction. Then complementary DNA was derived from mRNA using a commercial kit (Qiagen, Germany). A nanodrop spectrophotometer was used to determine the quality of the synthesized DNA. Further, the expression of genes was quantitatively analyzed using RT-qPCR by cDNAs as suitable templates. The PCR reaction consists of forward (F-5ˊ-TTCTTGCGACTCTTGGGACTTC-3ˊ) and reverse (R-5ˊTGACGAGGATGACAGGGA ACTA-3ˊ) primers for the determination of Cathepsin B expression. PCR reaction was performed for 35 cycles and the quantification value was determined based on relative expression change of CTSB compared with the internal standard (Patel et al., 2019).
2.5 Transfection analyses
2.5.1 CTSB overexpression in renal cancer cells (A498 and 769-P cells)
A498 and 769-P cells were grown up to 70 % confluence by standard method using RPMI 1640 medium. After 70 % confluence, it was washed with Dulbecco’s phosphate-buffered saline and was digested with trypsin. It was centrifuged and the cells were resuspended in fresh culture medium containing 10 % FBS. The cells were further transfected with pcDNA-3.1-CTSB and an empty plasmid was considered as the control. Then human CTSB cDNA was subcloned into expression vector according to the manufacturer’s instructions (Qiagen, Germany). After transfection, the cells were harvested after 2nd, 3rd and 4th day and transfection potential was tested after four days. The experiment was performed in triplicates and an average value was used for analysis.
2.5.2 Knockdown of CTSB gene in A498 and 769-P cells
Renal cancer cells (A498 and 769-P) were grown in tissue culture medium and grown up to 70 % confluence. It was transfected with short hairpin RNAs (shRNAs) and used for the silencing of CTSB. shRNA sequences used for silencing were, F-5ˊ-TGAATTCCCAA CACGTCACCGGAGAGATAAGATCT3ˊ and R-5ˊ-ATAGTCGACCCAACACGTCAC CGGAGAGATTAGATCTTAT-3ˊ. These sequences were further cloned into the pCI-neo using EcoR1 vector. The selected plasmids were used to transfect A498 and 769-P cell lines for 96 h according to the manufactures instruction. The efficiency of knockdown method was tested using Western blotting analysis. The reduction was considerably high after three days and the experiment was repeated three times and an average value was considered for analysis (Patel et al., 2019).
2.5.3 Knockdown analyses
The cells (A498 and 769-P) after transfections were washed with phosphate buffered saline and harvested. It was treated with lysis buffer with protease inhibitor cocktail. The cell lysates were subjected for the determination of proteins and the proteins were resolved using 12 % sodium dodecyl sulphate polyacrylamide gel electrophoresis. The expression of CtsB was evaluated using primary and secondary antibodies.
2.6 Analysis of A498 and 769-P cells proliferation
The renal cancer cell lines were maintained in 96-well plates and incubated for 12 h. These cells were transfected with specific constructs. After 48 h incubation, the cells were cultured in RPMI-1640 medium supplemented with 10 % FBS and maintained for three days. Then (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) (MTT) was added to each wells and incubated for 6 h. After 6 h incubation, MTT solution was decanted and MTT solvent was applied and vortexed for 15 min. The cell density was measured using a microplate reader at 490 nm (Patel et al., 2019).
2.7 Statistical analysis
Statistical analysis was performed by three different experiments (n = 3) and the result was expressed as mean ± SD. The difference between the experimental and control groups was determined using was student’s t test. The p value < 0.01 was considered as significant.
3 Results
3.1 Clinical history of RCC patients
The age, gender, histopathology and stage of tumour specimens were described in Table 1. Patient’s age ranged between 49 and 82 years. Among the 13 specimens, only 3 were obtained from the female patients and the remaining 10 were sourced from the male patients. Renal-cell carcinomas (RCC) develop from the epithelium of the renal cells and this account for approximately 85 % of renal cancers and have different subtypes. In this study, only 15.4 % samples were papillary RCC type, and the remaining 84.6 % were clear cell RCC type. *Stages were broadly classified into three stages.
Reference No
Age
Sex
Histopathology
Stage*
F0010
49
F
Clear cell RCC
I
M0062
54
M
Papillary RCC
I
M0063
68
M
Clear cell RCC
II
F0079
73
F
Clear cell RCC
I
M0083
82
M
Clear cell RCC
III
M0085
65
M
Papillary RCC
I
F0087
51
F
Clear cell RCC
I
M0091
75
M
Clear cell RCC
I
M0094
69
M
Clear cell RCC
I
M0096
73
M
Clear cell RCC
II
M0099
65
M
Clear cell RCC
I
M0106
81
M
Clear cell RCC
II
M0109
77
M
Clear cell RCC
II
3.2 Analysis of CpG methylation in renal cancer tissues
CpG methylation analysis was performed using metastatic and primary renal cancer cells. The amount of CpG methylation was high in patient’s diagnosed metastatic renal cancer. Tumour size varied between primary and metastasis were statistically significant (p < 0.001). The present findings revealed that increased CpG methylation was associated with renal cancer. In CpG expression, low expression was observed in metastasis, whereas this percentage was increased in primary renal cancer tissues (p < 0.001). High CpG expression was observed in metastasis and decreased expression level was observed in primary renal cancer cells (p < 0.001).
3.3 Down regulation of RUNX3 in metastatic cancer tissues
Gene array was performed to identify the patterns of gene expression regulated by methylation between primary renal cancer and metastatic renal cancer. Immunohistochemical staining revealed the expressions of RUNX3 in metastatic renal cancer and showed positive reactivity (Fig. 1). Down regulation was observed in RUNX3 gene. Western blot analysis revealed the decreased expression of RUNX3 in metastatic cancer compared with primary renal cancer (Fig. 2).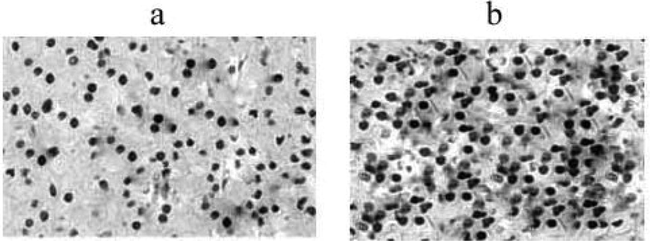
Immunohistochemical staining of the expressions of RUNX3 in primary (a) and metastatic renal cancer (b) (25X magnification).
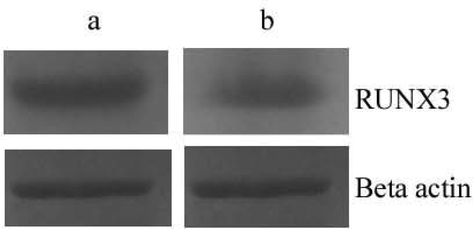
Western blot analysis of RUNX3 in the primary renal cancer (a) and metastatic renal cancer cells (b).
3.4 CtsB mRNA expression in RCC tissues
The amount of CtsB mRNA expression increased in RCC tissues and was associated with other pathophysiological conditions. A total of 13 samples from RCC patients were used in this study and CTSB transcript analyses were performed on kidney tumour cells and surrounding normal kidney tissues. RT-qPCR analysis revealed increased expression of CtsB gene than normal cells in kidneys. In normal cells, expression of CtsB gene was found to be high in older patients (greater than70 ages).
3.5 Cathepsin B expression and correlation with RCC tissues
The expression of CtsB gene was positively correlated with RCC and normal kidney cells. The total expression of CtsB was high in primary renal tissues, protein expressions were decreased in the tumour samples. In certain samples, protein expression was not changed compared with non-cancer tissues (Fig. 3).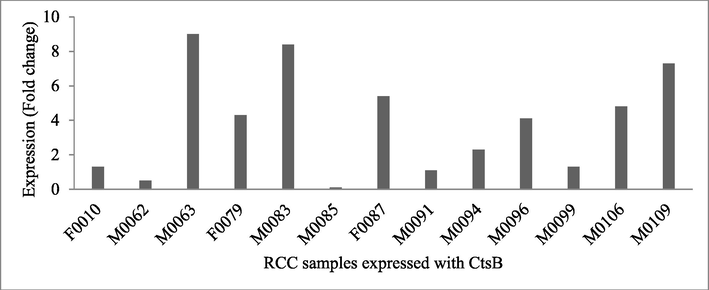
CtsB protein expression in healthy tissues and renal cell carcinoma cells. The expression of CtsB was analyzed in the control and infected tissues by western blot analysis and protein fold changes were analyzed by Log10T/NT. The figure shows the ratio between normal tissue (NT) and cancer tissue of CtsB in all tested 13 samples.
3.6 Influence on CtsB protein expression and cell proliferation
The expression of CtsB gene was confirmed in vitro using A498 and 769-P cancer cells. The present finding revealed overexpression of CtsB and the result was described in Fig. 4. To confirm the expression of CtsB, knocked-down experiment was performed to suppress the expression of CtsB in cancer cell lines, A498 and 769-P using short hairpin RNA vectors (shSTFA and shCTSB). Western blotting analysis revealed that the expression of CtsB protein and reduced after shCTSB treatment (Fig. 5). Down regulation of CtsB protein was observed in our study. CtsB overexpression and knocked-out effects were analyzed on A498 and 769-P cell proliferation and the result was compared with control cells transfected with control vectors. In this study, we analyzed an improved growth in both A498 and 769-P cell lines after overexpression of CtsB (p < 0.01) (Fig. 6).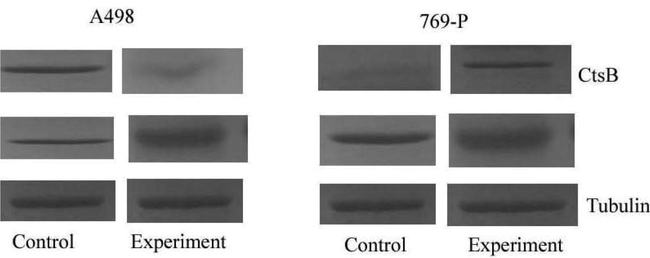
Expression of CtsB in cancer cell lines induced StfA expression in vitro. CtsB expression was analyzed by Western blotting in A498 and 769-P after transfection with plCTSB.
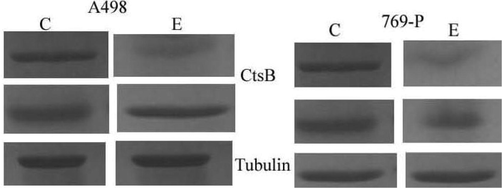
Silencing of cathepsin B in A498 and 769-P cells. CtsB expression was analyzed by Western blotting analysis in A498 and 769-P after transfection with shCTSB. Tubulin was used as the housekeeping protein (C-control; E-experiment).
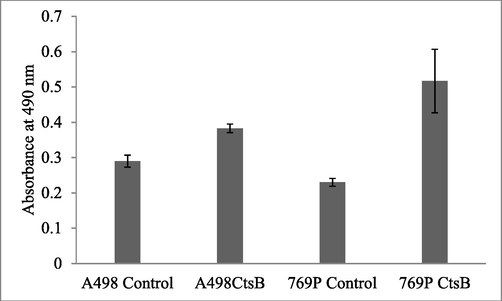
Influence of CtsB overexpression and proliferation of renal cancer cell lines. CtsB overexpression was determined by MTT assay. The cancer cell lines were incubated in 96-well plates and treated with MTT and the cells viability was determined. The result was analyzed and the significant level was determined.
4 Discussion
In the present study, we analyzed the differentially expressed genes in primary and metastatic renal cancer tissues. RUNX3 gene and most of the expressed genes were down-regulated in metastatic renal cancer with hypermethylation in CpG islands. In the present study, RUNX3 expression was significantly reduced in metastatic renal cancer tissues than in primary renal cancer tissues. RUNX3 expression was high in normal renal tissues than in adjacent renal cell carcinoma tissues. RUNX3 effectively suppressed the invasion and migration of renal carcinoma cells and targeted miR-6780a-5p/E-cadherin/EMT signals (Chen et al., 2017). RUNX3 considerably decreased invasion and carcinoma cell migration ability. It has been reported that the expression level of RUNX3 was considerably decreased in clear cell renal carcinoma cases and RUNX3 affected the metastatic abilities and proliferative capacities of RCC by regulating TIMP1 and cyclins (He et al., 2012). RUNX3 regulated the development of RCC and the possible role of RUNX3 was analyzed in this study. RUNX3 gene has been stated to be inactivated by aberrant methylation in colorectal, gastric, pancreatic, bile duct and renal cancers (Wada et al., 2004). The present finding is highly correlated with the previous finding, as RUN3X downregulation observed was due to methylation in CpG islands in metastatic renal cancer tissues.
Renal cell carcinoma is characterized by specific clinical manifestations, unique biology, and prognostic outcomes. The molecular mechanism of RCC involved various molecular pathways and the discovery of new biomarkers is useful for the treatment of cancers (Kato et al., 2013). The low pH-mediated cancer cell development is involved in the degradation of the basement membrane, as stated previously in melanoma, renal cell carcinoma, and ovarian and breast cancer (Rothberg et al., 2013). Cathepsin expression is not regulated in various tumour types and is effectively involved in cancer progression, metastasis, angiogenesis and antibiotic resistance. The activity of cathepsin was regulated by various natural inhibitors and these natural inhibitors interactive with their active site irreversibly or reversibly (Phan et al., 2022). The present study was consistent with previous findings that revealed increased expression level of CtsB was associated with cancer (Rempel et al., 1994).
The expression of CtsB was regulated by natural inhibitors in cells. The present findings show that an increased level of CtsB can effectively involved in tumour development. It was also revealed that CtsB expression was correlated with STFA in normal tissue and tumour cells. The increased expression of CTSB was mainly associated with cancer cases. The decreased inhibitor activity in cancer accompanied by increase of CtsB in cancer can involve the spreading of cancer cells (Xia et al., 2022). The improved level of CtsB has been observed in normal kidney tissues of elder patients, and this could be associated with the age-related role of CtsB that was earlier reported in human serum, liver and rat’s brain (Wyczałkowska-Tomasik and Pączek, 2012; Rudzińska et al., 2020). CtsB has been detected in nucleus, mitochondria and cytosol and regulates cellular division and cell death (Talukdar et al., 2016). In a study, a GFP-tagged CtsB revealed that the optimum regulation and presence of CtsB is critical for various cellular functioning, and any variation in the expression of CtsB can affect the cell homeostasis and the development of malignant tumour (Baici et al., 2006). The increased expression of CtsB in A498 and 769-P cells increased in the development of RCC, and continuous hyperexpression was observed.
5 Conclusions
In recent years various tumour suppressor genes have been identified and Runt-related transcription factor 3 is one of the tumour suppressor genes. The present study revealed that methylation plays an important role in the regulation of metastasis of RCC and is related to Runt-related transcription factor 3 pathway inhibitions. In RCC, CtsB gene is one of the biomarkersand the expression of CtsB was determined in vitro. Runt-related transcription factors 3 and CtsB are important biomarkers in RCC cells.
Acknowledgment
The authors thanks Bule Hora University, Ethiopia for the support. The author MA Rathi, thank Karpagam Academy of Higher Education for the support and encouragement. “The authors would like to thank the Deanship of Scientific Research at Umm Al-Qura University for supporting this work by Grant Code: (22UQU4320141DSR62)”.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- SFRP1 CpG island methylation locus is associated with renal cell cancer susceptibility and disease recurrence. Epigenetics. 2012;7(5):447-457.
- [Google Scholar]
- Regulation of human cathepsin B by alternative mRNA splicing: homeostasis, fatal errors and cell death. Biol. Chem.. 2006;387:1017-1021.
- [Google Scholar]
- RUNX3 regulates renal cell carcinoma metastasis via targeting miR-6780a-5p/E-cadherin/EMT signaling axis. Oncotarget. 2017;8(60)
- [Google Scholar]
- Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer.. 2019;144(8):1941-1953.
- [Google Scholar]
- RUNX3 mediates suppression of tumor growth and metastasis of human CCRCC by regulating cyclin related proteins and TIMP-1. PLoS One. 2012;7(3)
- [Google Scholar]
- Surveillance after radical or partial nephrectomy for localized renal cell carcinoma and management of recurrent disease. Urol. Clin. N. Am.. 2003;30(4):843-852.
- [Google Scholar]
- Acidic extracellular microenvironment and cancer. Cancer Cell Int.. 2013;13(1):1-8.
- [Google Scholar]
- Increased expression of mature cathepsin B in aging rat liver. Cell Tissue Res.. 2000;302(2):181-188.
- [Google Scholar]
- A review of integrated staging systems for renal cell carcinoma. Eur. Urol.. 2012;62(2):303-314.
- [Google Scholar]
- Heterogeneous patterns of DNA methylation-based field effects in histologically normal prostate tissue from cancer patients. Sci. Rep.. 2017;7(1):1-14.
- [Google Scholar]
- DNA methylation profiles in cancer diagnosis and therapeutics. Clin. Exp. Med.. 2018;18(1):1-14.
- [Google Scholar]
- Clinical stage migration and survival for renal cell carcinoma in the United States. Eur. Urol. Oncol.. 2019;2(4):343-348.
- [Google Scholar]
- DNA methylation biomarkers predict progression-free and overall survival of metastatic renal cell cancer (mRCC) treated with antiangiogenic therapies. PloS one. 2014;9(3)
- [Google Scholar]
- DNA methylation of neural EGFL like 1 (NELL1) is associated with advanced disease and the metastatic state of renal cell cancer patients. Oncol. Rep.. 2018;40(6):3861-3868.
- [Google Scholar]
- Discovery of pH-Selective Marine and Plant Natural Product Inhibitors of Cathepsin B Revealed by Screening at Acidic and Neutral pH Conditions. ACS omega. 2022;7(29):25346-25352.
- [Google Scholar]
- Cathepsin B expression and localization in glioma progression and invasion. Cancer Res.. 1994;54(23):6027-6031.
- [Google Scholar]
- Acid-mediated tumor proteolysis: contribution of cysteine cathepsins. Neoplasia. 2013;15(10):1125-IN9.
- [Google Scholar]
- Cysteine cathepsins inhibition affects their expression and human renal cancer cell phenotype. Cancers. 2020;12(5):1310.
- [Google Scholar]
- Release of cathepsin B in cytosol causes cell death in acute pancreatitis. Gastroenterology. 2016;151(4):747-758.e5.
- [Google Scholar]
- Tumor specific epigenetic silencing of corticotropin releasing hormone-binding protein in renal cell carcinoma: association of hypermethylation and metastasis. PloS one. 2016;11(10)
- [Google Scholar]
- Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat. Gen.. 2009;41(5):521-523.
- [Google Scholar]
- Challenges of early renal cancer detection: symptom patterns and incidental diagnosis rate in a multicentre prospective UK cohort of patients presenting with suspected renal cancer. BMJ open. 2020;10(5)
- [Google Scholar]
- Frequent loss of RUNX3 gene expression in human bile duct and pancreatic cancer cell lines. Oncogene. 2004;23(13):2401-2407.
- [Google Scholar]
- Cathepsin B and L activity in the serum during the human aging process: cathepsin B and L in aging. Arch. Gerontol. Geriatr.. 2012;55(3):735-738.
- [Google Scholar]
- CTSV (cathepsin V) promotes bladder cancer progression by increasing NF-κB activity. Bioengineered. 2022;13(4):10180-10190.
- [Google Scholar]







