Translate this page into:
Conjugation in the eukaryotic single-celled organism Euplotes aediculatus (Protozoa, Ciliophora): A focus on nuclear divisions, morphogenesis and pheromones
⁎Corresponding author. wangyurui@snnu.edu.cn (Yurui Wang)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Ciliates are unique in having nuclear dimorphism (i.e., possessing both a germline micronucleus and a somatic macronucleus in one cell), and undergo dramatic changes during their sexual process (known as conjugation). Here we investigate the nuclear events and morphogenetic processes during conjugation in the freshwater species Euplotes aediculatus. In addition, three conjugation-related pheromone genes were sequenced and analyzed. The results indicate that: (i) the 65-hour-long conjugation process includes five nuclear divisions, i.e., three prezygotic divisions (mitosis, meiosis I and II) and two postzygotic divisions; (ii) the ciliature reorganizes twice during conjugation with the frontoventral-transverse cirral anlagen regenerating during both morphogenetic events and breaking apart in a 3:3:3:2:2 pattern; and (iii) each of three pheromone genes contains a 444 bp coding region and encodes 147 amino acid polypeptides. These results suggest a broadly shared morphogenetic pattern during conjugation in Euplotes species and provide new information for studies on the evolutionary history of pheromone genes.
Keywords
Ciliate
Conjugation
Euplotes aediculatus
Morphogenesis
Nuclear events
Pheromone genes
1 Introduction
In ciliates, a group of single-celled eukaryotes possessing nuclear dimorphism, conjugation is a sexual process that results in the exchange of genetic information between two individuals (Raikov, 1982; Wang et al., 2021; Zhao et al., 2020). In sessile ciliates such as sessilid peritrichs, conjugation involves the permanent fusion of a motile microconjugant with a sessile macroconjugant. In contrast, conjugation in free-swimming ciliates is a process of reciprocal fertilization between two morphologically similar individuals of compatible mating types. The process involves the two individuals coming into close contact followed by partial membrane fusion, exchange of haploid gametic pronuclei, karyogamy of the stationary and migratory pronuclei to form a diploid synkaryon within each conjugant and, following the separation of the conjugants, the generation of a new somatic macronucleus (MAC) from the newly formed synkaryon (Orias et al., 2011). Each of these steps is regulated by complex mechanisms that are of great interest to researchers investigating cell biology and epigenetics. Unfortunately, the complete time course and detailed depiction of conjugation are known for only a small number of species and the process remains unknown for the vast majority of ciliates (Asghar et al., 2021; Gong et al., 2020).
Euplotes is one of the most attractive model organisms for investigating cell signaling, sexual processes, morphogenesis, mating type determination and inheritance, mainly because of its ease of collection and cultivation (Luporini et al., 2016). Previous studies indicate that the pattern of nuclear change during conjugation in Euplotes is relatively conserved except for two traits: 1) the presence, e.g., in E. octocarinatus (Kuhlmann and Heckmann, 1991), E. woodruffi (Rao, 1964), E. eurystomus (Turner, 1930), E. cristatus (Wichterman, 1967), E. affinis (Geng et al., 1992), E. minuta (Siegel and Heckmann, 1966), and E. charon (Valbonesi et al., 1987), or absence, e.g., in E. raikovi (Gong et al., 2020) and E. vannus (Jiang et al., 2019), of an additional prezygotic division (mitotic division) after meiosis II of the micronucleus; and 2) the fusion between the posterior fragment of the old MAC and the newly formed MAC, e.g., in E. eurystomus (previously misidentified as E. patella) and E. raikovi (Gong et al. 2020; Pierson, 1943; Turner, 1930). Although the nuclear events and morphogenesis in Euplotes during conjugation have been intensively investigated (Asghar et al., 2021; Gao et al., 2020; Gong et al., 2020; Jiang et al., 2019; Kuhlmann and Heckmann, 1991; Rao, 1964; Turner, 1930; Wichterman, 1967), most studies were carried decades ago and did not link nuclear events to morphogenesis.
In the present study, we provide a detailed description of the dynamic changes of the nuclei and ciliature during conjugation in Euplotes aediculatus and report the time course for each step of the nuclear events. Three pheromone genes in E. aediculatus are also sequenced and analyzed in the context of the phylogenetic relationships among Euplotes species.
2 Materials and methods
Euplotes aediculatus was collected from a freshwater lake (water temperature 25 °C) in Zhongshan Park (36°04′ N, 120°21′ E), Qingdao, China, in July 2020. The identification was based on morphological characters observed in vivo and after protargol and wet silver nitrate staining, as previously described by Curds (1975).
Cells were maintained at room temperature (ca. 25 °C) in sterilized distilled water using Escherichia coli as a food source. Eight strains with four mating types, determined by pairwise mixture, were used in this study (for details, see Table 1). Conjugation induction, mating pair sampling and staining followed Gong et al. (2020).
Strains
Strains
Mating type
Pheromone genes
Sources
C2
1B
C1
2B
C2
–
I
Ea-1
mating pair splitting
1B
+
–
II
Ea-2
conjugation progeny
C1
++
++
–
I-III
Ea-1, Ea-3
mating pair splitting
2B
+
++
+
–
I-II
Ea-1, Ea-2
conjugation progeny
DNA extraction, PCR amplification and sequencing were according to Lian et al. (2021) and Zhang et al. (2020). The coding regions of pheromone genes were amplified with primers Ea-17F (5′-CTGCTTACTCCTCGCTATCC-3′) and Ea-549R (5′-CTTTGAAACAAGGTTGAGCC-3′), which were designed based on the isolated pheromone gene contigs from genomic data (unpublished). The 3′ UTR (untranslated region) and partial telomere sequence were obtained by telomere suspension PCR (Curtis and Landweber, 1999). The sequence similarities were calculated using Geneious Prime v. 2021.1.1. Three pheromone amino acid (aa) sequences of E. aediculatus and another 30 Euplotes pheromone aa sequences were aligned using MUSCLE algorithm (https://www.ebi.ac.uk/Tools/msa/muscle/). Accession numbers are shown in Table 2. The phylogenetic tree without distance corrections was constructed using neighbor-joining methods on the multiple sequence alignment website (https://www.ebi.ac.uk/Tools/msa/muscle/) (Madeira et al., 2019). The phylogenetic analysis based on 18S rDNA was carried out according to Gong et al. (2020).
Species
Pheromone
Accession number
Species
Pheromone
Accession number
E. aediculatus
Ea-1
MZ488504
E. nobilii
En-1
ACQ66088
Ea-2
MZ488505
En-2
ACQ66089
Ea-3
MZ488506
En-6
ABK15649
E. crassus
Ec-α
AHX71996
En-A1
ACQ66090
Ec-1
AEC04940
En-A2
ACQ66091
Ec-2
AHX71994
En-A3
ACQ66092
Ec-3
AHX71995
En-A4
ACQ66093
E. octocarinatus
Eo-1
O15823
E. raikovi
Er-1
AXL95182
Eo-2
O15825
Er-2
AXL95183
Eo-3
P28717
Er-4
AXL95187
Eo-4
P28716
Er-5
AXL95186
Eo-5
CAA76771
Er-6
AXL95188
E. petzi
Ep-1
ANY30848
Er-8
AXL95185
Ep-2
ANY30849
Er-10
AXL95189
Ep-3
ANY30850
Er-11
AXL95184
Ep-4
ANY30851
Er-20
P26888
Er-22
AXL95190
3 Results
3.1 Nuclear changes during conjugation in Euplotes aediculatus
3.1.1 Nuclear events in conjugant cells
Conjugation was induced by mixing together cells of compatible mating types. The formation of mass mating pairs, which occurred about 3 h after mixing, was taken as the starting point (time 0) of the mating process (Fig. 1G). Samples were collected every hour after time 0 and 20–30 conjugating pairs were recorded at each time point. This enabled us to provide a detailed time course of the conjugation process.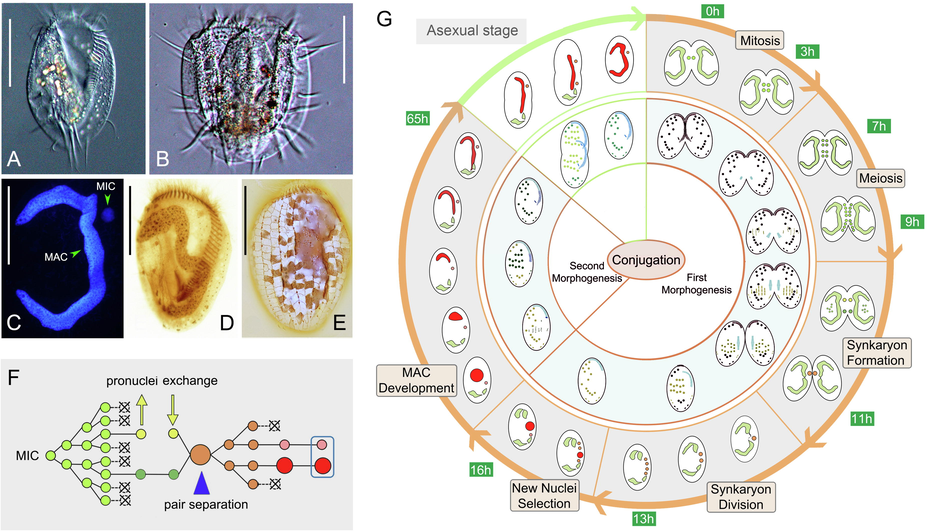
Morphology of vegetative cell and nuclear and morphogenetic events during conjugation in Euplotes aediculatus. (A) Ventral view of a representative vegetative cell. (B) Ventral view of a mating pair. (C) Vegetative cell after Hoechst 33342 staining. (D) Vegetative cell after protargol staining. (E) Silverline system on dorsal side after silver nitrate staining. (F) Diagram showing nuclear events during conjugation. (G) Morphogenesis and nuclear change during conjugation. Light green nuclei = the diploid MIC before gametic nuclei formation; dark green and yellow nuclei = stationary and migratory nuclei, respectively; orange nuclei = synkaryon and synkaryon division products; pink and red nucleus = new MIC and MAC, respectively. Abbreviations: MAC, macronucleus; MIC, micronucleus. Scale bars = 60 μm.
After forming pairs, the micronucleus (MIC) begins to swell and undergoes mitosis (first prezygotic division), which lasts about 3 h (Fig. 2A, B). The two MICs in each conjugant then undergo a classical two-step meiosis (Fig. 2B–I). The first meiotic division (second prezygotic division) takes about 4 h to produce four haploid nuclei (Fig. 2B–G). The second meiotic division (third prezygotic division) takes about 2 h to produce eight haploid nuclei (Fig. 2G–I). It is noticeable that micronuclear division is not always synchronous in a given mating pair (Fig. 2F, H). For example, by the time one of the two conjugating cells has completed the first meiotic division and is equipped with four haploid nuclei, the other cell may have only just finished mitosis (first prezygotic division) (Fig. 2F).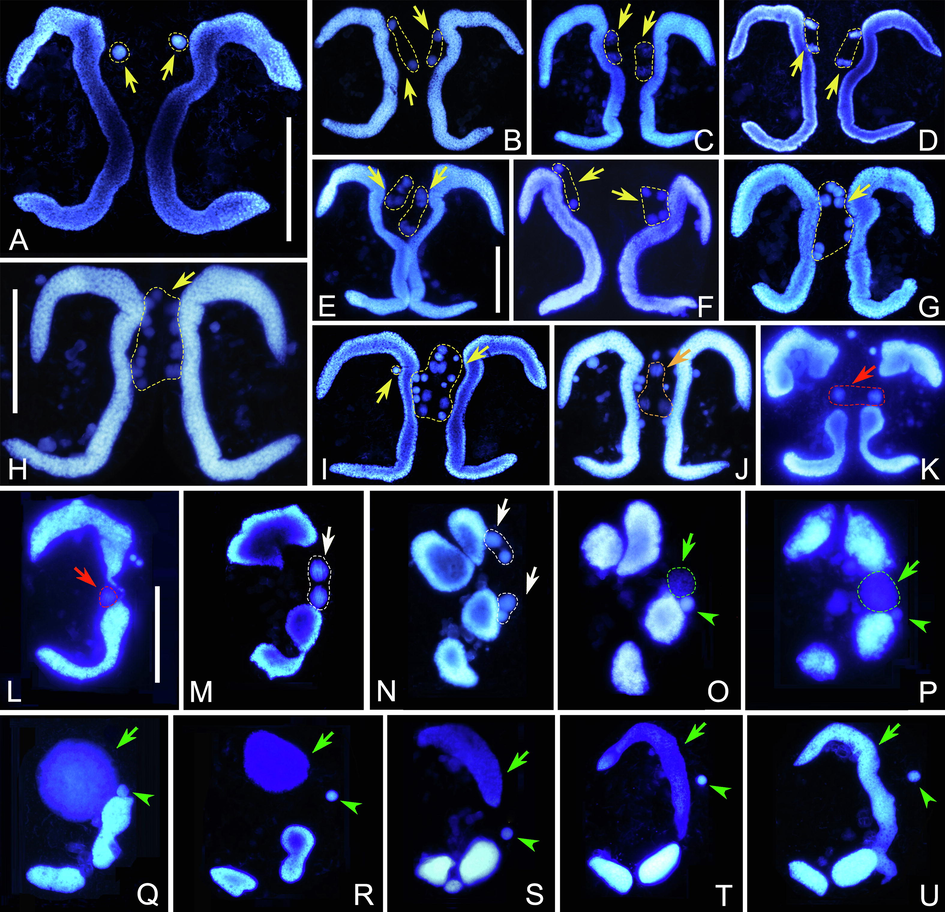
Nuclear events of Euplotes aediculatus after Hoechst 33342 staining. (A) MIC of each conjugant swells after pair formation. (B) The first prezygotic division. (C–G) Various stages of the first meiotic division. In F, cells have different numbers of nuclei due to asynchronous division. (H, I) The third prezygotic division. (J) Each cell has two pronuclei (orange arrow) while the other nuclei degenerate. (K) Synkaryon formation after exchange and fusion of pronuclei (red arrow). (L) After synkaryon formation, the conjugants separate. (M, N) The synkaryon divides twice producing four nuclei (white arrows). (O–Q) One out of four nuclei swells and becomes the new MAC anlage, one become the new MIC. (R–U) The MAC anlage differentiates into the new MAC. Yellow arrows indicate nuclei derived from prezygotic divisions. Green arrows and arrowheads mark the MAC anlage and the new MIC, respectively. Scale bars = 60 μm.
In each conjugant, two of the eight haploid nuclei swell while the other six become pyknotic and degenerate (Fig. 2J). The two swollen nuclei represent one migratory and one stationary pronucleus. Following the exchange of migratory pronuclei, fusion with the local stationary pronucleus occurs forming a synkaryon (Fig. 2K). At the same time, the parental MAC begins to break into two parts (Fig. 2K). During this approximately 2 h process, the pyknotic nuclear products are still visible in some pairs.
3.1.2 Nuclear events in post-conjugant cells
The conjugant cells usually separate from each other after synkaryon formation. The synkaryon then undergoes two rounds of mitosis (first and second postzygotic divisions), forming four nuclear products, two of which localize in the anterior part and the other two in the posterior part of the cell (Fig. 2L–N). Concurrently, the parental MAC continues to degrade into four fragments (Fig. 2N). This process takes about 2 h. Following the second postzygotic division, one posterior nucleus becomes the new MIC and the other swells and differentiates into the macronuclear anlage (Fig. 2O), while the two anterior nuclei degenerate. The new MAC anlage increases in size and gradually moves from the mid-body to the anterior part of the cell (Fig. 2O–Q). Meanwhile, the two anterior fragments of the parental MAC progressively degrade (Fig. 2O–Q). After about 50 h from the formation of the MAC anlage (65 h after the mixing of compatible mating types), the new MAC develops into a hook-shaped structure that eventually becomes C-shaped during first cell division after mating. The posterior fragments of the parental MAC neither degenerate nor fuse with the new MAC (Fig. 2R–U) but instead are retained until the first vegetative division after mating.
3.2 Cortical morphogenesis during conjugation in Euplotes aediculatus
3.2.1 Morphogenesis in conjugant cells
The parental adoral membranelles are gradually resorbed anteriad and only a few apical membranelles are retained (Fig. 3A, B). The new oral primordium (OP) appears as a small patch of kinetosomes in the mid-body region near the left margin marking the beginning of stomatogenesis. The parental paroral membrane is resorbed (Fig. 3C). The above process will be completed approximately 3 h after the conjugants form pairs. With the continuous expansion of the OP, the kinetosomes progressively assemble into new membranelles (Fig. 3D–F). These newly formed membranelles continue to extend and migrate upward and eventually reach half of the length of the old adoral zone of membranelles (AZM) thereby replacing it (Fig. 3H). After the mating pairs separate, the old apical membranelles will be resorbed completely. No paroral membrane was detected during conjugational morphogenesis.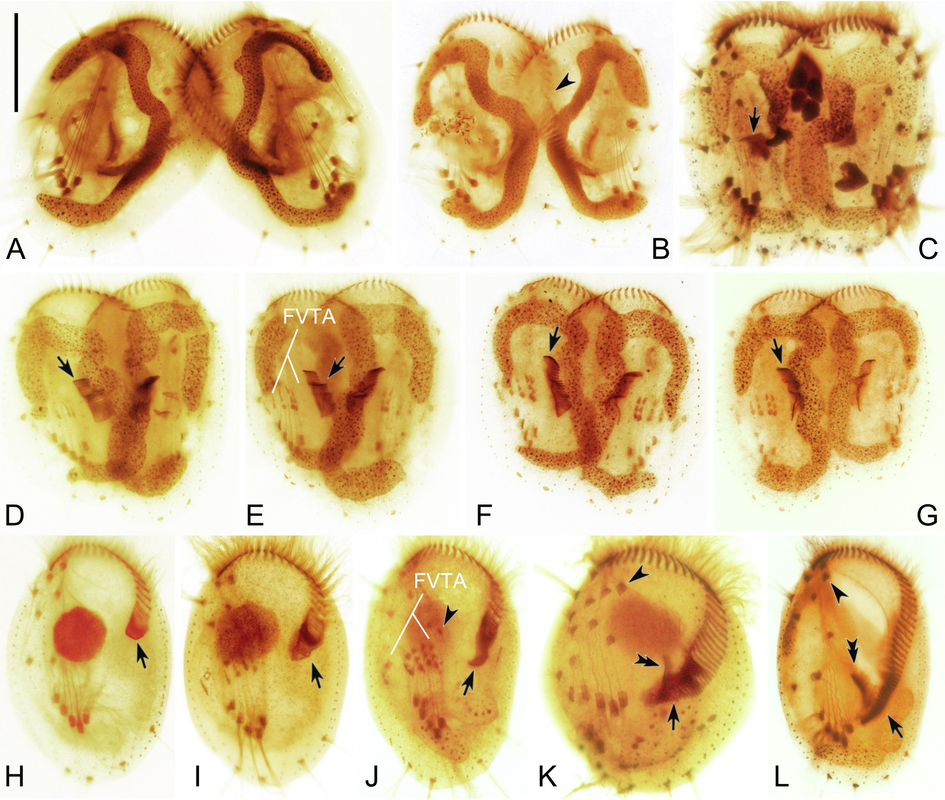
Morphogenesis during conjugation in Euplotes aediculatus. (A) Early stage of conjugation. (B) Slightly later stage showing the degraded old adoral membranelles (arrowheads). (C–G) Series of later stages, to show the new adoral membranelles (arrows) and the new frontoventral-transverse cirral anlagen (FVTA). (H) Exconjugant, arrow indicates the formation of new adoral membranelles during the second round of morphogenesis. (I–L) Exconjugants at later stages, showing the development of adoral membranelles (arrows), cirral anlagen including cirrus I/1 (arrowheads in J, K and L), and paroral membrane (double-arrowheads). Abbreviation: FVTA, frontoventral-transverse cirri anlagen. Scale bar = 60 μm.
At the same time, a large number of basal bodies are generated de novo on the right side of the new OP forming the frontoventral-transverse cirral anlagen (FVTA), which subsequently develop into five streaks (Fig. 3D, E). These streaks extend in both directions, broaden, and then break apart in a 3:3:3:2:2 pattern (Fig. 3F). Finally, they develop into frontoventral-transverse cirri and migrate to their final positions. The first round of morphogenesis lasts about 16 h.
3.2.2 Cortical morphogenesis in post-conjugant cell
When the conjugants separate, the posterior part of the AZM, the paroral membrane, and the leftmost frontal cirrus have not yet regenerated (Fig. 3H). After a period of time, some kinetosomes appear below the incomplete AZM, assemble into new adoral membranelles, and complete the AZM (Fig. 3I–K). Later, the paroral membrane primordium appears near the proximal end of the new AZM. This elongates and eventually forms the paroral membrane (Fig. 3K, L).
Similar to the first round of reorganization, the anlagen of the FVTA originate de novo to the right of the AZM (Fig. 3I). These anlagen broaden, develop into five streaks in a 3:3:3:2:2 pattern, and eventually give rise to the FVT cirri (Fig. 3J, K). At the same time, an anlage appears de novo to the left of the five streaks, and this develops into the leftmost frontal cirrus (Fig. 3J, K). Eventually all the cirri migrate to their final positions and replace the old ones (Fig. 3L). The second round of morphogenesis takes approximately 15 h.
3.3 The pheromone genes of Euplotes aediculatus
Three pheromone genes (Ea-1, Ea-2 and Ea-3) were obtained from four mating types of E. aediculatus, respectively (Table 1). The nucleotide sequence of each was deposited in GenBank with accession numbers, lengths and GC contents as follows: MZ488504, 1907 bp, 29.2% (Ea-1); MZ488505, 1935 bp, 30.7% (Ea-2); MZ488506, 1927 bp, 30.7% (Ea-3). As shown in Table 3, the identities between the full-length nucleotide sequences are 83.1% (Ea-1/Ea-2), 80.0% (Ea-1/Ea-3) and 93.2% (Ea-2/Ea-3), respectively. By comparing with transcriptome data (unpublished) and searching the classical GT-AG intron boundaries, three introns were detected in each gene. Excluding introns, 3′ UTR, and partial telomeres, the coding region of each gene is 444 bp and the nucleotide sequence identities between them are 81.3% (Ea-1/Ea-2), 82.2% (Ea-1/Ea-3) and 94.4% (Ea-2/Ea-3), respectively (Table 3).
Homologous parts of the chromosomes
Length (bp)
GC content (%)
Identity (%)
Ea-1
Ea-2
Ea-3
Ea-1
Ea-2
Ea-3
Ea-1/Ea-2
Ea-1/Ea-3
Ea-2/Ea-3
Macronuclear chromosome
1907
1935
1927
29.2
30.7
30.7
83.1
80.0
93.2
Intron 1
335
335
335
31.6
33.7
32.5
96.4
78.7
80.5
Intron 2
419
420
420
19.6
22.9
23.8
59.6
59.8
94.5
Intron 3
77
104
102
27.3
26.9
26.5
54.8
56.9
97.1
3′ UTR + Telomere
632
632
626
29.6
29.2
29.3
98.7
97.8
97.7
Coding region
444
444
444
34.2
36.9
36.9
81.3
82.2
94.4
Each gene encodes 147 aa peptides (Fig. 4). By comparing with other Euplotes pheromones, the pre (signal-peptide) and pro segments of these three pheromones are excluded, leaving secreted segments with 93 aa peptides. The aa sequence identities among secreted segments are 60.2% (Ea-1/Ea-2), 62.4% (Ea-1/Ea-3) and 87.1% (Ea-2/Ea-3), respectively. The full length of aa sequence identities between these are 72.1% (Ea-1/Ea-2), 74.8% (Ea-1/Ea-3) and 88.4% (Ea-2/Ea-3), respectively.
Alignment of the full length of amino acid sequences of pheromones in six Euplotes species. The three conserved cysteine residues are indicated by arrowheads.
To elucidate the phylogenetic relationships among Euplotes pheromones, we constructed phylogenetic tree based on the full length of Euplotes pheromone aa sequences, and compared it with the tree based on 18S rDNA (Fig. 5A). The pheromone protein tree shows that: 1) pheromones from the same species group together forming a homologous protein family; and 2) E. aediculatus exhibits a closer relationship to E. octocarinatus than to other species. Consistently, E. aediculatus groups together with E. octocarinatus with full support (100% ML, 1.00 BI) in the 18S rDNA tree.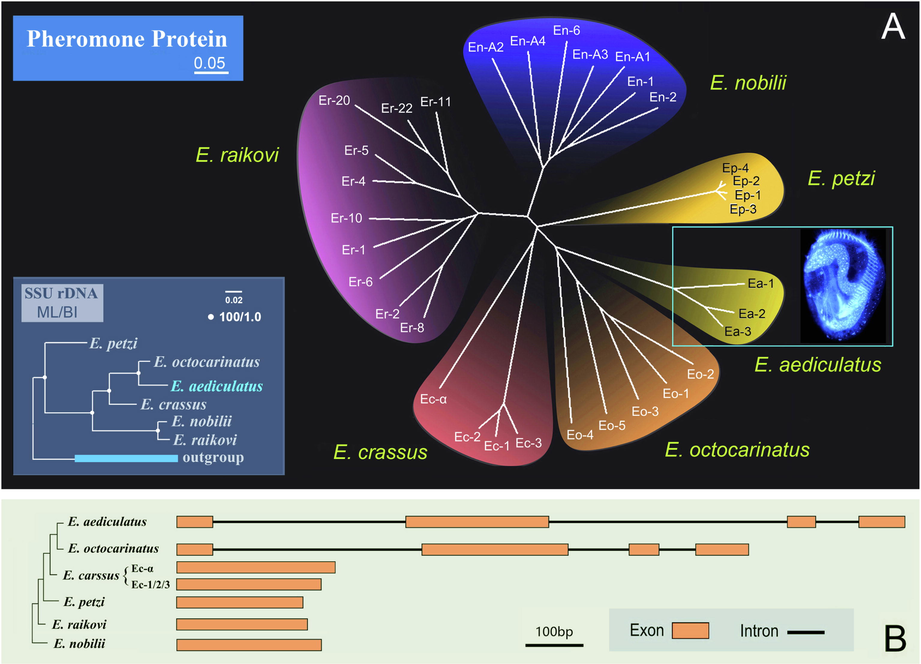
The phylogenetic relationships and coding region composition of Euplotes pheromones. (A) Neighbor-joining (NJ) tree of Euplotes pheromones (from six Euplotes species) based on the full length of amino acid sequences. The scale bar represents 5 variations per 100 aa residues. The bottom left insert shows the maximum likelihood (ML) tree based on 18S rDNA of six Euplotes species. (B) Comparison of the coding region of pheromone genes from Euplotes. The structures for other Euplotes pheromones are modified from Pedrini et al. (2017), focusing on the coding regions only and ignoring introns outside the coding region. Orange boxes depict exon sequences; solid lines depict intron sequences.
4 Discussion
4.1 Comparisons of nuclear change and morphogenesis during conjugation in Euplotes
The duration of conjugation in Euplotes aediculatus (65 h) is shorter than that in E. vannus (75 h) and E. octocarinatus (100 h) but longer than that in E. raikovi (50 h) (Gong et al., 2020; Jiang et al., 2019; Kuhlmann and Heckmann, 1991). During conjugation in E. aediculatus, the nuclear change contains three prezygotic divisions and two postzygotic divisions. The former character is also shared by E. vannus, E. raikovi and certain strains of E. crassus (Gong et al., 2020; Jiang et al., 2019; Lueken, 1973), whereas the MIC in other congeners (e.g., E. crassus, E. woodruffi, E. patella, E. octocarinatus, E. affinis, E. minuta, E. cristatus, and E. charon) undergoes an additional prezygotic division (Geng et al., 1992; Heckmann, 1964; Kuhlmann and Heckmann, 1991; Rao, 1964; Siegel and Heckmann, 1966; Turner, 1930; Valbonesi et al., 1987; Wichterman, 1967). Notably, the prezygotic divisions of E. aediculatus are asynchronous, which is similar to E. raikovi but unknown in other species of Euplotes (Gong et al., 2020). Two postzygotic divisions occur in most Euplotes species, the exceptions being E. cristatus and E. charon (Valbonesi et al., 1987; Wichterman, 1967). Intriguingly, in E. crassus the four prezygotic divisions and two postzygotic divisions are not only found during conjugation, but also during autogamy (Dini et al., 1999).
The retained posterior fragments of the parental MAC in E. aediculatus appear to have the following potential roles: 1) when the new MAC anlage is damaged, the parental MAC fragments have the ability to regenerate a functional vegetative MAC (Kloetzel, 1981); and 2) irradiated parental MAC fragments at the beginning of cortical morphogenesis could result in an immature new MAC and no cortical reorganization, demonstrating the important roles of old MAC fragments in nuclear and cortical development (Fidler et al., 1985). The retention of parental MAC fragments is also observed in E. patella and E. woodruffi (Sato, 1985; Wang et al., 1991). In addition to retention, there are two other potential fates of the parental MAC among Euplotes species: 1) in E. affinis, E. charon, E. cristatus, E. minuta, E. octocarinatus and E. vannus, the parental MAC fragments degenerate completely (Geng et al., 1992; Jiang et al., 2019; Kuhlmann and Heckmann, 1991; Siegel and Heckmann, 1966; Valbonesi et al., 1987; Wichterman, 1967); and 2) in E. raikovi, they fuse with the new MAC (Gong et al., 2020).
Two rounds of ciliature morphogenesis, i.e., conjugational and post-conjugational, occur during conjugation in E. aediculatus. The second round of morphogenesis is similar to the process of opisthe formation during asexual cell division (Zhang et al., 2017), except the dorsal kineties do not reorganize. Morphogenesis during conjugation has been widely investigated in Euplotes, e.g., E. raikovi (Asghar et al., 2021), E. vannus (Gao et al., 2020), E. woodruffi (Wang and Shi, 1989), E. affinis (Geng et al., 1992), E. eurystomus (Katashima, 1959), and E. patella (Hammond and Kofoid, 1937; Turner, 1930). All these species exhibit a highly conserved pattern of morphogenesis with two exceptions: 1) there is only one round of reorganization in E. eurystomus; and 2) the dorsal kineties are not reorganized in E. affinis (Geng et al., 1992; Katashima, 1959). In both cases, however, the apparently missing processes might simply have been overlooked.
4.2 The pheromones of Euplotes aediculatus
Pheromones are secretory chemical factors that are produced by one individual to affect the behavior of other individuals of the same species. In Euplotes, the pheromones comprise three sections, signal peptide, pro segment and secreted form (as shown in Fig. 4, Luporini et al., 2016). The secretory pheromone proteins could: 1) promote the vegetative growth by acting as autocrine (autologous) growth factors; and 2) induce conjugation between compatible mating types by governing non-self recognition (Luporini et al., 2016). In addition, when suspended with a pheromone synthesized by a different mating type, selfing could be detected in E. patella (Kimball, 1942). Euplotes pheromones were first identified in E. patella by Kimball (1942). Subsequently, they were identified in other Euplotes species, such as E. raikovi (Luporini and Miceli, 1986), E. octocarinatus (Möllenbeck and Heckmann, 1999), E. nobilii (Pedrini et al., 2007), E. crassus (Alimenti et al., 2011) and E. petzi (Pedrini et al., 2017). The structure and function of pheromones have been investigated in detail (Luporini et al., 2016). All 33 pheromone aa sequences have conserved cysteine residues (Fig. 4), indicating that the pheromone proteins have a similar spatial structure (Luporini et al., 2016).
There are two kinds of pheromone patterns in Euplotes, i.e., the “E. patella” pattern and the “E. crassus” pattern. In the “E. patella” pattern, the mating type is determined by a single locus (possessing multiple alleles). The homozygote expresses one pheromone while the heterozygote produces two structurally homologous pheromones encoded by two alleles and could mate with both homozygous counter-partners (Luporini and Miceli, 1986). This same pattern is detected in E. patella, E. petzi, E. raikovi, E. octocarinatus and E. nobilii (Pedrini et al., 2017; Vallesi et al., 2014). For several decades, the “E. crassus” pattern was thought to be serially dominant (Heckmann, 1964). The studies by Vallesi et al. (2014), however, indicated that in E. crassus there are two distinct macronuclear pheromone gene families, due to gene duplication at the micronuclear mating-type locus. The alleles of one gene family are codominantly expressed and encode for pheromones (Ec-1, Ec-2 and Ec-3) which are mating-type specific just like the E. patella pheromones. Of the other gene family, only one allele is known, encoding for pheromone Ec-α which is shared among all the analyzed mating types. This pheromone might play a role in supplementing or assisting the mating-type specific pheromones.
The full length of pheromone aa sequences among different Euplotes species varies from 73 to 160 aa. In E. petzi, the length of pheromone sequences is 73 aa. In E. raikovi, except Er-22 for which only a partial sequence is available, the lengths of nine pheromone sequences are relatively conserved (74–76 aa). The length ranges of pheromone amino acids in E. crassus and E. nobilii are 84–91 aa and 83–94 aa, respectively. The three newly sequenced E. aediculatus pheromones are relatively long (all 147 aa), which is within the length range of the E. octocarinatus group (127–160 aa) (Möllenbeck and Heckmann, 1999). Furthermore, in both E. aediculatus and E. octocarinatus, each of the coding regions of their pheromone genes has three introns. Therefore, based on their length and structure, the pheromones of E. aediculatus are much more similar to those of E. octocarinatus than to other Euplotes pheromones (Fig. 5B) (Möllenbeck and Heckmann, 1999).
5 Conclusion
In this study, we report the nuclear changes and morphogenetic processes of conjugation in E. aediculatus and give a detailed time-course for each stage of conjugation. In addition, the nucleotide and amino acid sequence of three pheromone genes in E. aediculatus are reported for the first time. This work indicates the Euplotes species share a conserved pattern of conjugation and provides new information for the investigation of mating type determination in Euplotes.
Author contributions
YW conceived and supervised the study. RG, YC and CS performed experiment and drafted the manuscript. QY, YL, AW and YW revised and improved the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgements
This work was support by Natural Science Foundation of China (No. 32030015 and 32070428), the China Postdoctoral Science Foundation Grant (2021M692010) and the Natural Science Foundation of Shaanxi province (2022JQ-196). The authors would like to thank Prof. Weibo Song, Feng Gao, Ying Yan (OUC, China) and Prof. Pierangelo Luporini (University of Camerino, Italy) for their help during drafting this manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Isolation and structural characterization of two water-borne pheromones from Euplotes crassus, a ciliate commonly known to carry membrane-bound pheromones. J. Eukaryot. Microbiol.. 2011;58:234-241.
- [Google Scholar]
- Morphogenesis of the ciliature during sexual process of conjugation in the ciliated protist Euplotes raikovi. Front. Mar. Sci.. 2021;7:615377
- [Google Scholar]
- A guide to the species of the genus Euplotes (Hypotrichida, Ciliata) Bull. Brit. Mus. Nat. Hist. (Zool.). 1975;28:1-68.
- [Google Scholar]
- Evolution of gene scrambling in ciliate micronuclear genes. Ann. N. Y. Acad. Sci.. 1999;870:349-350.
- [Google Scholar]
- Nuclear phenomena during autogamy in the marine ciliate Euplotes crassus: a tangled cytogenetic process fostering evolutionary conservatism. Acta Protozool.. 1999;38:39-48.
- [Google Scholar]
- Nuclear roles in the post-conjugant development of the ciliate Euplotes aediculatus. III. roles of old macronuclear fragments in nuclear and cortical development. J. Protozool.. 1985;32:429-436.
- [Google Scholar]
- Morphogenetic characters of the model ciliate Euplotes vannus (Ciliophora, Spirotrichea): notes on cortical pattern formation during conjugational and postconjugational reorganization. Eur. J. Protistol.. 2020;73:125675.
- [Google Scholar]
- The morphological changes in conjugation of Euplotes affinis. J. East China Norm. Univ. (Nat. Sci.). 1992;2:81-89.
- [Google Scholar]
- Conjugation in Euplotes raikovi (Protista, Ciliophora): new insights into nuclear events and macronuclear development from micronucleate and amicronucleate cells. Microorganisms. 2020;8:162-174.
- [Google Scholar]
- The continuity of structure and function in the neuromotor system of Euplotes patella during its life cycle. Proc. Am. Philos. Soc.. 1937;77:207-218.
- [Google Scholar]
- Experimentelle untersuchungen an Euplotes crassus. Z. Vererbungsl.. 1964;95:114-124.
- [Google Scholar]
- Time-course analysis of nuclear events during conjugation in the marine ciliate Euplotes vannus and comparison with other ciliates (Protozoa, Ciliophora) Cell Cycle. 2019;18:288-298.
- [Google Scholar]
- A correlation between morphogenesis and old macronucleus during sexual reproduction in Euplotes eurystomus. J. Sci. Hiroshima Univ. (B1). 1959;19:99-107.
- [Google Scholar]
- The nature and inheritance of mating types in Euplotes patella. Genetics. 1942;27:269-285.
- [Google Scholar]
- Nuclear roles in the post-conjugant development of the ciliate Euplotes aediculatus II. Experimentally induced regeneration of old macronuclear fragments. J. Protozool.. 1981;28:108-116.
- [Google Scholar]
- Nuclear processes in Euplotes octocarinatus during conjugation. Eur. J. Protistol.. 1991;26:370-386.
- [Google Scholar]
- Systematic positions and taxonomy of two new ciliates found in China: Euplotes tuffraui sp. nov. and E. shii sp. nov. (Alveolata, Ciliophora, Euplotida) Syst. Biodivers.. 2021;19:359-374.
- [Google Scholar]
- A marine Euplotes (Ciliophora, Hypotrichida) with reduced number of prezygotic micronuclear divisions. J. Protozool.. 1973;20:143-145.
- [Google Scholar]
- Ciliate communication via water-borne pheromones. In: Witzany G., Nowacki M., eds. Biocommunication of Ciliates. Berlin: Springer; 2016. p. :159-174.
- [Google Scholar]
- Mating pheromones. In: Gall J.G., ed. The Molecular Biology of Ciliated Protozoa. Orlando: Academic Press; 1986. p. :263-299.
- [Google Scholar]
- The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res.. 2019;47:W636-W641.
- [Google Scholar]
- Characterization of two genes encoding a fifth so far unknown pheromone of Euplotes octocarinatus. Eur. J. Protistol.. 1999;35:225-230.
- [Google Scholar]
- Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res. Microbiol.. 2011;162:578-586.
- [Google Scholar]
- Cold-adaptation in sea-water-borne signal proteins: sequence and NMR structure of the pheromone En-6 from the Antarctic ciliate Euplotes nobilii. J. Mol. Biol.. 2007;372:277-286.
- [Google Scholar]
- Molecular structures and coding genes of the water-borne protein pheromones of Euplotes petzi, an early diverging polar species of Euplotes. J. Eukaryot. Microbiol.. 2017;64:164-172.
- [Google Scholar]
- A comparative morphological study of several species of Euplotes closely related to Euplotes patella. J. Morphol.. 1943;72:125-165.
- [Google Scholar]
- The Protozoan Nucleus: Morphology and Evolution. Vienna: Springer-Verlag; 1982.
- Nuclear behavior of Euplotes woodruffi during conjugation. J. Eukaryot. Microbiol.. 1964;11:296-304.
- [Google Scholar]
- Nuclear behavior during reconjugation in Euplotes patella (Ciliophora, Hypotrichida) J. Protozool.. 1985;32:485-490.
- [Google Scholar]
- Inheritance of autogamy and the killer trait in Euplotes minuta. J. Protozool.. 1966;13:34-38.
- [Google Scholar]
- Division and conjugation in Euplotes patella Ehrenberg with special reference to the nuclear phenomena. Univ. Calif. Publs Zool.. 1930;33:193-258.
- [Google Scholar]
- Observations on the biology of Euplotes charon (Hypotrichida, Ciliophora) Ital. J. Zool.. 1987;54:111-118.
- [Google Scholar]
- Evidence for gene duplication and allelic codominance (not hierarchical dominance) at the mating-type locus of the ciliate, Euplotes crassus. J. Eukaryot. Microbiol.. 2014;61:620-629.
- [Google Scholar]
- The fate of posterior fragments of old macrunucleus during conjugation in Euplotes woodruffi. Curr. Zool.. 1991;37:402-407.
- [Google Scholar]
- Studies on morphogenesis accompanying binary fission and conjugation in Euplotes woodruffi (Ciliata, Protozoa) Acta Zool. Sin.. 1989;35:353-359.
- [Google Scholar]
- A new hypotrich ciliate, Oxytricha xianica sp. nov., with notes on the morphology and phylogeny of a Chinese population of Oxytricha auripunctata Blatterer & Foissner, 1988 (Ciliophora, Oxytrichidae) Mar. Life Sci. Technol.. 2021;3:303-312.
- [Google Scholar]
- Mating types, breeding system, conjuation and nuclear phenomena in the marine ciliate Euplotes cristatus Kahl from the Gulf of Naples. J. Eukaryot. Microbiol.. 1967;14:49-58.
- [Google Scholar]
- Reconsideration of the taxonomy of the marine ciliate Neobakuella aenigmatica Moon et al., 2019 (Protozoa, Ciliophora, Hypotrichia) Mar. Life Sci. Technol.. 2020;2:97-108.
- [Google Scholar]
- Morphology, ontogeny and molecular phylogeny of Euplotes aediculatus Pierson, 1943 (Ciliophora, Euplotida) Biodivers. Sci.. 2017;25:549-560.
- [Google Scholar]
- Functional analysis of the methyltransferase SMYD in the single-cell model organism Tetrahymena thermophila. Mar. Life Sci. Technol.. 2020;2:109-122.
- [Google Scholar]







