Translate this page into:
Comparison of the SARS-CoV-2 (2019-nCoV) M protein with its counterparts of SARS-CoV and MERS-CoV species
⁎Corresponding author. afrefaei@ksu.edu.sa (Abdulwahed Fahad Alrefaei)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Coronaviruses M proteins are well-represented in the major protein component of the viral envelope. During the viral assembly, they play an important role by association with all other viral structural proteins. Despite their crucial functions, very little information regarding the structures and functions of M proteins is available. Here we utilize bioinformatic tools from available sequences and 3D structures of SARS-CoV, SARS-CoV2, and MERS-CoV M proteins in order to predict potential B-cell epitopes and assessing antibody binding affinity. Such study aims to aid finding more effective vaccines and recognize neutralizing antibodies. we found some rather exciting differences between SARS-COV-2, SARS-Cov and MERS-CoV M proteins. Two SARS-CoV-2 peptides with significant antigen presentation scores for human cell surface proteins have been identified. The results reveal that N-terminal domains of M proteins of SARS-CoV and SARS-CoV2 are translocated (outside) whereas it is inside (cytoplasmic side) in MERS-CoV.
Keywords
M protein
SARS-CoV-2
SARS-CoV
MERS-CoV
B-cell epitopes
Structural proteins
1 Introduction
Coronaviruses (CoVs) family are mostly responsible for enzootic infections. In the last two decades, CoVs have noticeably arisen in human populations, each species within this family has its unique characteristic features but also shared some similarities. However, after the emergence of severe acute respiratory syndorme coronavirus (SARS-CoV) in 2002, this family has been widely known. They are a group of viruses that cause diseases in mammals and birds (Perlman and Netland, 2009). Unlike other species within this family such as SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV-2 (or 2019-nCoV) has highly spread in infected population (Huang et al., 2020). With the numbers infected rising well above a 56 million and confirmed deaths above 1.3 million as of 19th November 2020, it has noticeably become the paramount healthcare for the global community at present. The high mortality rate of some CoVs, along with their ease of transmission accelerates the demand for more investigation into CoV molecular biology which will help in the development of effective anti-coronaviral drugs. Improvement of effective therapeutic and prevent strategies are clearly limited by the lack of detailed structural information on viral proteins. Thought, such proteins are considered as a good model for this class of proteins (Armstrong et al., 1984).
The shape of the viral envelope is mainly determined by its membrane (M) protein, which is the most abundant structural protein in the CoVs family (Neuman et al., 2011). Analysis of several types of CoVs showed that the viral size presumably depends on the interaction of M protein with spike (S), nucleocapsid (N) proteins and viral genomic RNA (Neuman et al., 2011). It is also considered as the central organiser of CoVs assembly, due to its interaction with all other structural proteins (Masters, 2006). For example, interaction of M protein with S protein is required for retention of S protein in the ER-Golgi intermediate compartment and its integration into new virions (Opstelten et al., 1995). In addition, M protein plays important role in structure-stabilizing of N protein as it is located in the internal core of virions (Mortola and Roy, 2004; Glowacka et al., 2011; Narayanan et al., 2000). It has been demonstrated that M proteins of some CoVs have much higher immunogenicity for T-cell responses than the nonstructural viral proteins (Li et al., 2008). In addition, it plays a critical role in virus-specific B-cell response due to its ability to produce efficient neutralizing antibodies in SARS patients (Pang et al., 2004).
Vaccine advancement is considered as one of the most significant issues to prevent most infectious diseases mainly when treatment is not available yet. The infection rate of CoVs can be limited by developing a potential vaccine. Bioinformatics tools for prediction B-cell epitope candidates are currently being utilized in several applications including vaccine design, development of diagnostics and monitoring of unwanted immune responses against protein therapeutics (Larsen et al., 2010; Lund et al., 2011; Robson, 2020). Antibodies that are produced by B-cells are significant in predicting effective vaccines (Olsson et al., 2007). Even though the ability of the human immune system to mount its antibodies against pathogens, only neutralizing antibodies can completely block the entry of pathogens into the human body (Suarez and Schultz-Cherry, 2000). The body's high ability to produce neutralizing antibodies mainly depends on finding unique epitopic sites on viral surface proteins that those antibodies can bind to.
In this study, we performed bioinformatic, and homology structural modeling analyses of three spices of betacoronaviruses: SARS-Cov, SARS-CoV2, and MERS-CoV. We analysed the homology of M protein sequences of those three species and identified all of the amino acid changes in their M protein sequences. We also used IEDB to predict likely epitopes on the M proteins of those species that are likely to be recognized in humans.
2 Materials and methods
2.1 M protein sequences
In this study, M protein sequences of the three species of CoVs were retrieved from the National Center for Biotechnology Information (NCBI): namely SARS-CoV2 (protein ID: YP_009724393), SARS-CoV (protein ID: NP_828855), and MERS-CoV (protein ID: YP_009047210). These protein sequences were then subjected for comparison using different bioinformatics prediction tools.
2.2 Phylogenetic tree
We used M protein sequences of the three species of CoV as a query to search the NCBI Protein Database to identifying M proteins across diverse Coronaviruses species. Consequently, the full length amino acid sequences of those three species were selected for multiple alignment by using CLUSTALX 2.1 program (Thompson et al., 1997). A bootstrap re-sampling technique was used to ensure the robustness of the generated topological tree. Neighbor Joining (NJ) phylogenetic analysis was conducted in Geneious Prime software (Kearse et al., 2012).
2.3 Structure modeling
The secondary structure of M proteins of the three CoVs were generated using computer-based structure PSIPRED server (Mcguffin et al., 2000). Consequently, a three-dimensional (3D) structures of those proteins were predicted after submitting to Phyre2 server (Kelley et al., 2015).
2.4 B-cell epitope prediction
Immune-Epitope Data-base and Analysis Resource (IEDB) (Vita et al., 2015) have been utilized to list available data that are highly related to coronaviruses. BepiPred method in IEDB was used in order to predict linear B-cell epitopes (Jespersen et al., 2017) from the conserved regions with a default threshold value 0.55(81% Specificity and 29% Sensitivity). The method combines the predictions of a hidden Markov model and the tendency scale approach (Larsen et al., 2006). The complete M protein sequences of the three species were analyzed with BepiPred method to predict the potential B-cell epitopes.
3 Results
3.1 Hydrophobicity and hydrophlicity of M protein
The hydropathy profile shows that M protein obviously consists of three domains-the amino (N)-terminal domain, the transmembrane domain (TMD), and the carboxy (C)-terminal domain. The hydrophobicity analysis of M proteins of SARS-CoV-2, SARS-CoV and MERS-CoV reveals that M protein has short hydrophilic region at N-terminal domain consisting of 10 amino acids, followed by a large hydrophobic region of approximately 90 amino acids at TMD, and ends with long hydrophilic carboxyl terminus consisting of 100 amino acids (Fig. 1a). In addition, we predicted the TMD in M protein using Hidden Markov Model (HMM) (Sonnhammer et al., 1998) the model provides the most probable location and orientation of TMD in the M protein of MERS-CoV, SARS-CoV and SARS-Cov-2 (Fig. 1b-d). The results reveal that M protein has three TMDs. Those regions constitute about 50% of CoV M protein. Which may emphasize the interactions of CoV M protein with other major structural proteins of CoVs family via those regions of the Cov M protein. Interestingly, the results also reveal that N-terminal domains of M proteins of SARS-Cov and SARS-Cov2 are translocated (outside) whereas it is inside (cytoplasmic side) in MERS-CoV.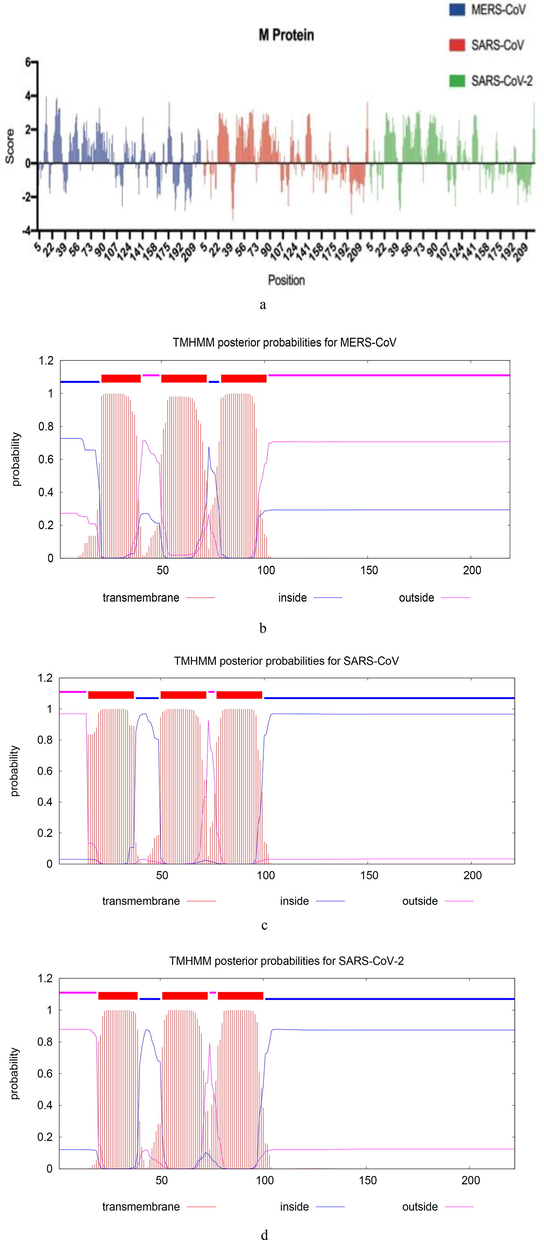
Hydropathy profile and prediction of TMD of M protein (a) Hydropathy profile for M proteins of SARS-CoV-2, SARS-CoV and MERS-CoV. The positive values state hydrophobic and negative values state hydrophilic regions in the protein. (b) The plot shows the posterior probability of inside/outside/ TMD for MERS-CoV M protein. (c) For SARS-CoV M protein. (d) For SARS-CoV-2 M protein.
3.2 Comparison of amino acid identity of the M protein
Comparison of SARS-CoV2 M protein sequence to sequences for SARS-CoV and MERS-CoV revealed a high degree of similarity between SARS-CoV-2 and SARS-CoV, but a more limited similarity with MERS-CoV (Fig. 2). However, the multiple sequence alignment of SARS-CoV-2, SARS-CoV, and MERS-CoV M proteins reveals that this protein contains completely five conserved proline residues (green boxes). Unlike SARS-CoV and MERS-CoV M proteins, the inserted S4 residue can be observed in SARS-CoV-2 M protein, which may reflect a unique feature of SARS-CoV-2 M protein. Another different feature that distinguishes MERS-CoV M protein from SARS-CoV and SARS-CoV-2 M proteins is that Methionine (M4) residue in MERS-CoV M protein corresponding to Aspargine (N4 and N5) residues in both SARS-CoV and SARS-CoV M proteins, respectively. However, the blocking mutation study in this residue of SARS-CoV M protein has not shown any major impacts on virus assembly (Voß et al., 2009). The three M proteins are identical at position 158 with Cysteine (C) residue (blue box) (Tseng et al., 2013), it has been reported that replacing this residue with S residue in SARS-CoV M protein led to a significant reduction in M protein secretion, but not for other C residues within this protein. The multiple sequence alignment analysis also shows the highly conserved dileucine (LL) motif at the C-terminal domains of all the three proteins (orange box). In addition, the highly conserved Phenylalanine (F95) and S110 residues play important role in virus assembly (Tseng et al., 2010).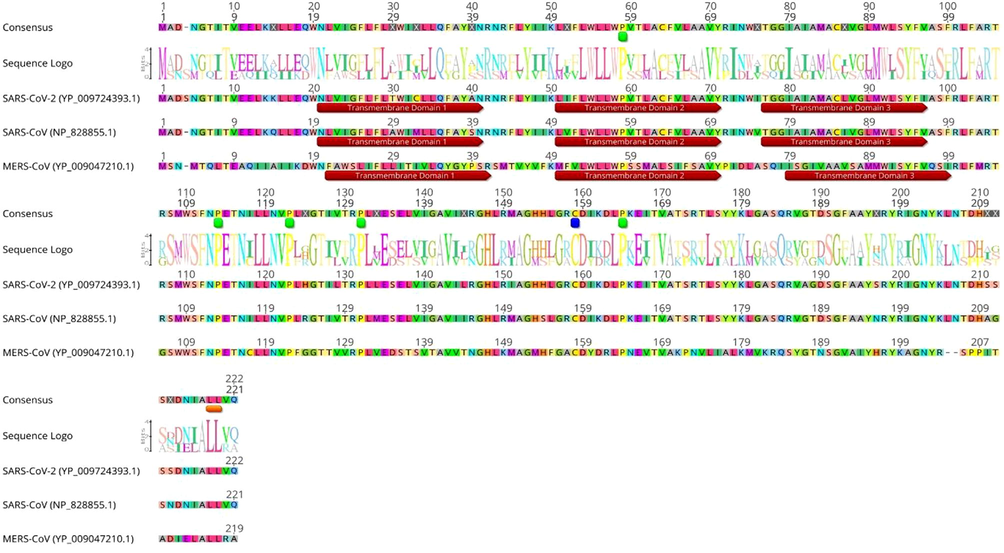
The multiple sequence alignment of the M protein produced by using CLUSTALX 2.1 program. The conserved proline is spotted (green), cysteine (blue) and dileucine (LL) motif (orange). The prediction of transmembrane helices is indicated in brown box.
3.3 Sequence-based phylogenetic analysis of M protein
Taxonomically, SARS-CoV, SARS-CoV2 and MERS-CoV are all demonstrated to be Betacoronaviruses closely related to each others (Fig. 3). The analysis confirmed that all SARS-CoV2 M protein sequences clustered very closely with SARS-CoV M protein sequences, with the closest matching sequence corresponding to a bat coronavirus (bat-CoV) species called Bat-CoV-RaTG13. It is also estimated that the rate of molecular evolution of SARS-CoV and SARS-CoV2 M proteins are 1.9 × 10−3 and 4.5 × 10−4 substitutions per site, respectively. Furthermore, the result demonstrated that MERS-CoV M protein forms its own clade, with the closest matching sequence corresponding to a Human betacoronavirus (human-CoV) species called 2c Jordan-N3 and the rate of molecular evolution of 5.11 × 10−4 substitutions per site. Phylogenetically, M protein sequences from HKU4 and BtCoV form a sub-clade that groups with MERS-CoV M protein sequence.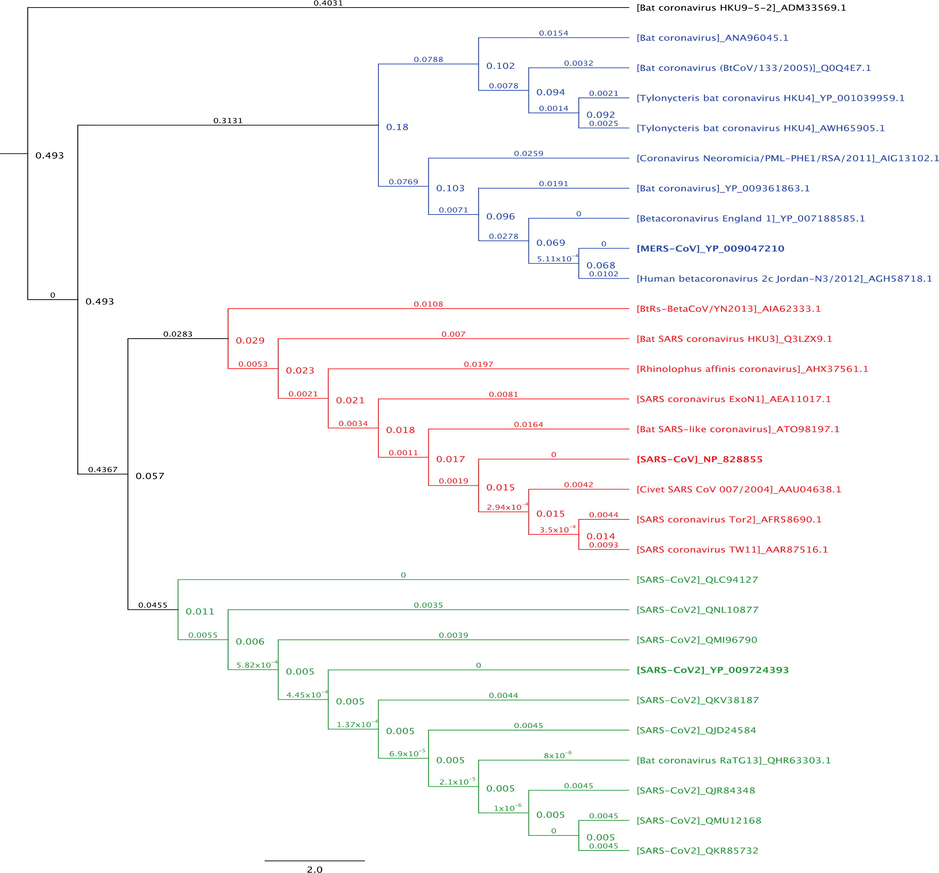
Phylogenetic analysis of M protein sequences from 29 orthologues. The M protein sequences were aligned using MUSCLE alignment (Edgar, 2004). The Neighbor-joining tree was generated based on the alignment. The tree was rooted using Bat-CoV HKU9-5 M protein sequence as the outgroup. The highly related betacoronaviruses MERS-CoV are highlighted (blue color), SARS-CoV (red color), and SARS-CoV2 (green color). Number at nodes indicates bootstrap support (1000 replicates), and the scale bar 2 represents the estimated number of substitutions per site. Accession numbers of sequences used in the analyses are shown next each species.
3.4 Secondary structure of M protein
For assessing the annotation of biological function of M protein, computer-based structure PSIPRED (Mcguffin et al., 2000) server has been used to predict the secondary structures of M proteins of SARS-CoV, SARS-CoV2 and MERS-CoV. The M proteins of SARS-CoV and SARS-CoV2 have been predicted to contain three closely spaced hydrophobic transmembrane helix signatures, followed by a comparatively large C-terminal endodomain (Fig. 4a-b). In contrast, the M protein of MERS-CoV assumes a topology in which part of the C-terminal endodomain forms two more transmembrane segments (Fig. 4c), thereby locating the C-terminal endodomain on the exterior of the virion (Risco et al., 1995). Several β-strands alternated by short coil loops are predicted at 120–200 amino acid positions. Furthermore, M proteins of SARS-Cov and SARS-Cov2 appear to have similar spatial topology compared to the one in MERS-Cov. For example, the results demonstrate the existence of three predicted α-helix domains in the M protein of SARS-CoV and SARS-CoV2, and indicate that the orientation of the TM region has its N-terminus in the extracellular side of the virion and its C-terminus in the cytoplasmic side (Fig. 4d-e). In contrast, the result reveals that M protein of MARS-CoV has four predicted α-helix domains. Interestingly, both N-terminal and C-terminal of MERS-CoV M protein are oriented in the extracellular side of the virion (Fig. 4f). Nevertheless, the determination of such orientation is more difficult but important to the subsequent function analysis as the interactions of M protein are mainly monitored by its transmembrane and endodomain segments (De Haan et al., 2000).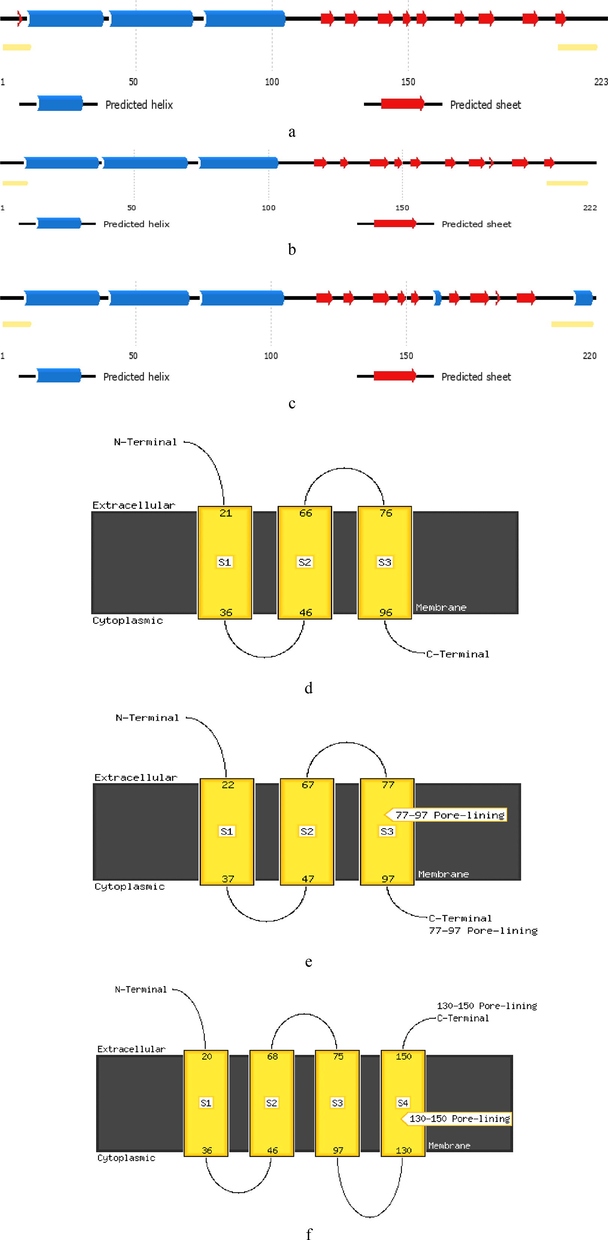
Secondary structures and possible topological transmembrane models of M protein (a) Secondary structure of SARS-CoV2 M protein. (b) Secondary structure of SARS-CoV M protein. (c) Secondary structure of MERS-CoV M protein. The yellow bars at the terminal ends of each sequences indicate possible disordered regions. (d) Topological transmembrane model of SARS-CoV2 M protein. (e) Topological transmembrane model of SARS-CoV Mprotein. (f) Topological transmembrane model of MERS-CoV M protein.
3.5 Production of disorder probabilities of the M protein
Firstly, we looked for longer regions of consecutively predicted disordered amino acids sequence because such regions are more likely to be truly intrinsic disordered regions in M protein. Meta-predictor has predicted two intrinsic disordered regions in SARS-CoV2 M protein: from position 1 to 7 and from position 205 to 222 with the average strength score of 0.82 and 0.65, respectively (Fig. 5a). Whereas, it revealed three intrinsic disordered regions in SARS-CoV M protein: from position 1 to 6 and from position 207 to 210 and position 216 to 221 with the average strength score of 0.82, 0.53 and 0.61, respectively (Fig. 5b). In addition, the predictor revealed that MERS-CoV M protein has two intrinsic disordered regions: from position 1 to 6 and position 216 to 219 with the average strength score of 0.85 and 0.61, respectively (Fig. 5c). The results show that the M-protein of SARS-CoV is characterized by the highest level of intrinsic disordered regions. We could clearly observe that each virus under study is characterized by a unique intrinsic disordered profile. This could be the reason of their differences in transmission modes. It has been shown that M-proteins of human coronaviruses manage to be more disordered than those of the animal coronaviruses (Goh et al., 2012). These differences can be due to the need of such viruses to adapt to the modifications in the environmental changes associated with the process of transmission between the viral hosts.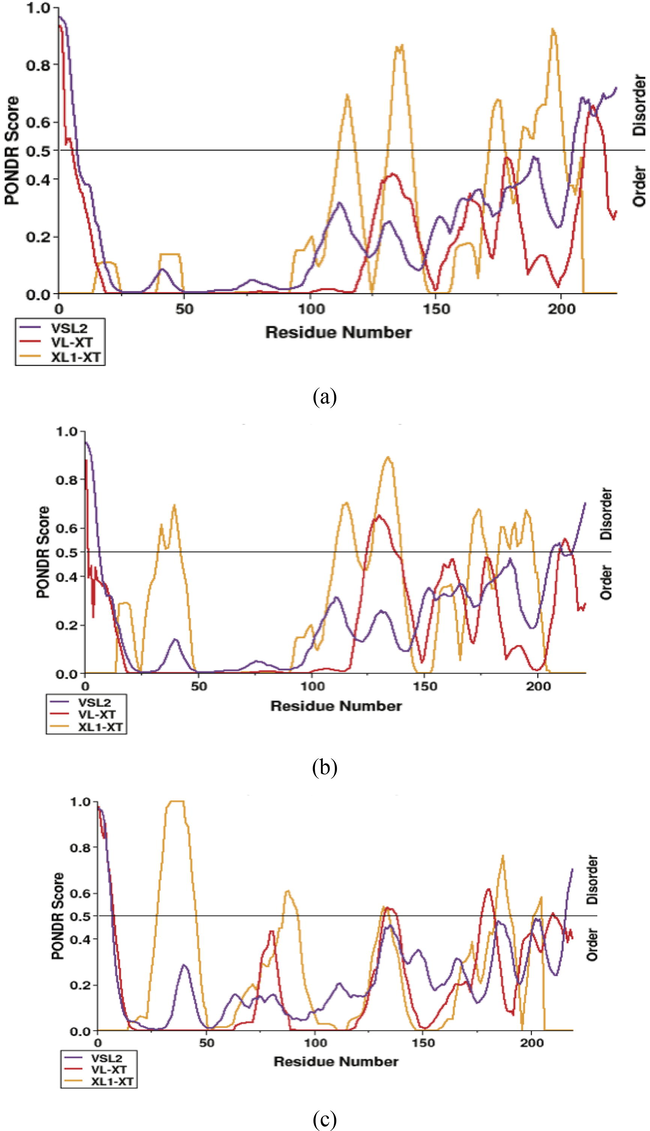
Intrinsic disorder prediction of M proteins, (a) SARS-CoV2 M protein. (b) SARS-CoV M protein. (c) MERS-CoV M protein. The disorder profiles generated using three predictors: PONDR® VSL2 (purple line), PONDR® VL-XT (red line), PONDR® XL1-XT (orange line).
3.6 Potential B-cell epitopes of M protein
M proteins of SARS-CoV, SARS-CoV2 and MERS-CoV were subjected to BepiPred linear epitope prediction (Fig. 6a-c). The prediction analysis performed with BepiPred based on the available M protein PDB structures of SARS-CoV, SARS-CoV2 and MERS-CoV shows two of the likely epitope regions (Fig. 6d-f). The average affinity score of M protein to B cell was >0.55 (corresponding to a specificity cutoff of 81%); all values equal or greater than this threshold was predicted to be potential B-cell eiptopes. The predicted epitope length with start and end positions were mentioned in Table 1. The results revealed that some regions on SARS-CoV M protein are dominant for B cell responses and that those regions also are highly conserved in terms of sequence with SARS-CoV2. Two regions were recognized in SARS-CoV2 (183–189, 200–217), SARS-CoV (183–188, 199–215) and MERS-CoV (180–188, 200–211) M proteins. Furthermore, the results proved that more mutations on these regions might lead to more chemical and physical features in M protein which in turn will induce possible changes of immunogenicity.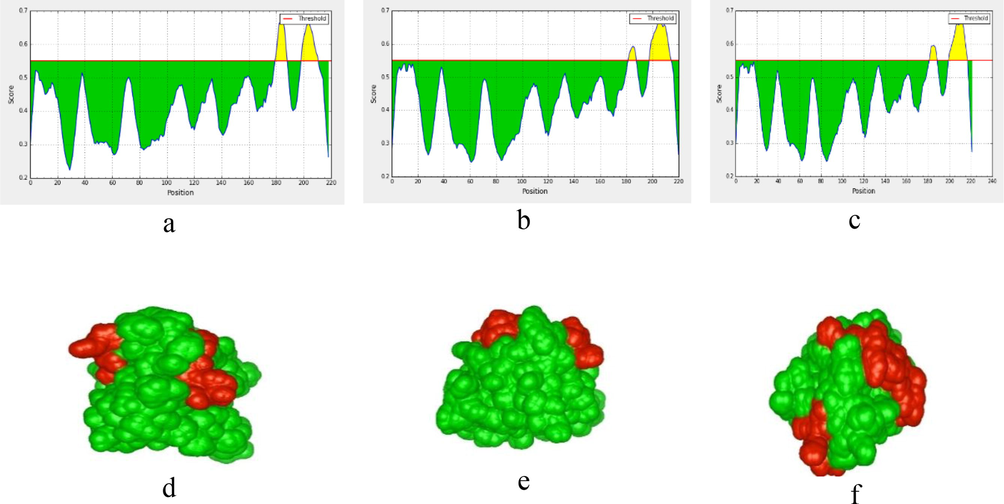
BepiPred linear epitope prediction of M proteins, (a) for MERS-CoV M protein. (b) for SARS-CoV M protein. (c) SARS-CoV2 M protein. The desired epitope residue showed in yellow color. The red horizontal line indicates surface accessibility threshold (0.55). The x-axis and y-axis represent the position and score, respectively. The highest peak region indicates the most effective B-cell epitope. (d) 3D presents linear epitopes on the MERS-CoV M protein surface. (e) for SARS-CoV M protein. (f) for SARS-CoV2 M protein.
Strain
Start
End
Peptide
Length
SARS-CoV2
183
189
ASQRVAG
7
200
217
RIGNYKLNTDHSSSSDNI
18
SARS-CoV
183
188
SQRVGT
6
199
215
RIGNYKLNTDHAGSNDN
17
MERS-CoV
180
188
MVKRQSYGT
9
200
211
AGNYRSPPITAD
12
4 Discussion
The matter of the hydropathy of a particular sequence of amino acids supposes added significance when structural proteins are considered. Structural proteins are characterized by hydrophobicity and hydrophilicity scores using their amino acid sequences, the grand average hydrophicity values (GRAVY) (Kyte and Doolittle, 1982). Which usually differ in range of ±4 where positive values state hydrophobic and negative values state hydrophilic regions in the protein. In order to know further for vaccine development, the hydrophilicity and hydrophobicity of M protein profile was investigated among its amino acid sequence. The hydrophobic sequence of M protein is mainly located at the predicted transmembrane domain, while the hydrophilic sequences at N- and C-terminal domains. Overall, CoVs M proteins differ in their amino acid content, but most share the same basic structural characteristics.
To achieve an initial assessment of shared and specific features of M protein, multiple sequence alignment was performed to compare the M protein sequence of SARS-CoV-2 with that of the SARC-CoV and MERS-CoV. The alignment model was based on the profile HMM. The M protein is conserved across the three coronaviruses. The multiple sequence alignment analysis also shows the highly conserved dileucine (LL) motif at the C-terminal domains of all the three proteins. The mutation in this motif leads to weaken the interaction and packaging between M and N proteins (Tseng et al., 2013, 2010; Saikatendu et al., 2007). In addition, the highly conserved Phenylalanine (F95) and S110 residues play important role in virus assembly (Tseng et al., 2010). We constructed phylogenetic tree by using first structure sequences of M protein of those three species as query to retrieve 29 orthologues sequences derived from various CoV species (Fig. 3), In order to provide important insights into their evolutionary and functional relationships at protein levels.
B-cell epitopes are those sites on the protein that can be recognized by antibodies of the immune system. Determining such regions can be utilised in the design of suitable vaccines and diagnostics tests. The traditional experimental epitopes scanning method obviously not practicable on a genomic scale. Prediction approaches are less time-consuming and more cost effective and dependable methods. This study aimed to apply IEDB software in order to predict the appropriate CoV eptitope vaccine against the well-known world population alleles via M protein and its modification sequence after the pandemic spread of SARS-CoV2 in late 2019. The results of this study revealed that the M proteins and their modified sequences of SARS-CoV, SARS-CoV-2 and MERS-CoV can be regarded as a defensive immunogenic with a strong conservation due to their highly capacities to determine neutralizing antibodies. We predicted likely human antibody binding sites (B-cell epitopes) on SARS-CoV, SARS-CoV2 and MERS-CoV M protein with BepiPred.
Because of the diversity of the existing of intrinsic disorder prediction methods, we decided to combine them into more accurate meta-prediction method (Xue et al., 2010a). In order to infer the potentially intrinsic disorder regions of M protein. In our study on the prediction of intrinsic disorder, three predictors were utilised to predict disordered regions. VSL2 (Peng et al., 2006) (Various Short Long, version 2), XL1-XT (Romero et al., 1997) and VL-XT (Li et al., 1999). All the three predictors employed the same attributes based on amino acid compositions. The amount and the peculiarity of distribution of such regions play important roles in behavior and transmission modes of Coronavirus (Goh et al., 2012, 2008a, 2008b; Xue et al., 2010b). In general, the amounts of disorder regions in the M proteins of coronaviruses are predicted to be less comparing to other structural viral proteins such as N proteins (Li et al., 1999). As the main function of M proteins is to protect the virion, it is strongly appealing to suppose that these diversities in the overall disorder regions of M proteins may associate with its mechanism to protect the viruses from different environment conditions which in return can reflect differences in the viral transmission mode. Further development on how coronavirus will behave in terms of transmission would be extraordinarily effective not just for medical but also fundamental research. Such a model will also provide a tool to aid the implementation of public health policies for dealing with old and even newly emerging pathogenic viruses.
Acknowledgments
We extend our appreciation to the Research Support Project (number RSP-2020/218), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Sequence and topology of a model intracellular membrane protein, E1 glycoprotein, from a coronavirus. Nature. 1984;308:751-752.
- [Google Scholar]
- Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol.. 2000;74:4967-4978.
- [Google Scholar]
- MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res.. 2004;32:1792-1797.
- [Google Scholar]
- Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol.. 2011;85:4122-4134.
- [Google Scholar]
- A comparative analysis of viral matrix proteins using disorder predictors. Virol. J.. 2008;5:126.
- [Google Scholar]
- Protein intrinsic disorder toolbox for comparative analysis of viral proteins. BMC Genomics. 2008;9:S4.
- [Google Scholar]
- Understanding viral transmission behavior via protein intrinsic disorder prediction: coronaviruses. J. Pathogens. 2012;2012
- [Google Scholar]
- Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506.
- [Google Scholar]
- BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res.. 2017;45:W24-W29.
- [Google Scholar]
- Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647-1649.
- [Google Scholar]
- The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc.. 2015;10:845-858.
- [Google Scholar]
- A simple method for displaying the hydropathic character of a protein. J. Mol. Biol.. 1982;157:105-132.
- [Google Scholar]
- Identification of CD8+ T cell epitopes in the West Nile virus polyprotein by reverse-immunology using NetCTL. PLoS One. 2010;5:e12697
- [Google Scholar]
- Predicting protein disorder for N-, C-and internal regions. Genome Inf.. 1999;10:30-40.
- [Google Scholar]
- T cell responses to whole SARS coronavirus in humans. J. Immunol.. 2008;181:5490-5500.
- [Google Scholar]
- Lund, O., Nascimento, E. J., Maciel Jr, M., Nielsen, M., Larsen, M. V., Lundegaard, C., Harndahl, M., Lamberth, K., Buus, S. & Salmon, J., 2011. Human leukocyte antigen (HLA) class I restricted epitope discovery in yellow fewer and dengue viruses: importance of HLA binding strength. PLoS One, 6, e26494.
- Efficient assembly and release of SARS coronavirus-like particles by a heterologous expression system. FEBS Lett.. 2004;576:174-178.
- [Google Scholar]
- Characterization of the coronavirus M protein and nucleocapsid interaction in infected cells. J. Virol.. 2000;74:8127-8134.
- [Google Scholar]
- A structural analysis of M protein in coronavirus assembly and morphology. J. Struct. Biol.. 2011;174:11-22.
- [Google Scholar]
- Induction of immune memory following administration of a prophylactic quadrivalent human papillomavirus (HPV) types 6/11/16/18 L1 virus-like particle (VLP) vaccine. Vaccine. 2007;25:4931-4939.
- [Google Scholar]
- Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol.. 1995;131:339-349.
- [Google Scholar]
- Protective humoral responses to severe acute respiratory syndrome-associated coronavirus: implications for the design of an effective protein-based vaccine. J. Gen. Virol.. 2004;85:3109-3113.
- [Google Scholar]
- Length-dependent prediction of protein intrinsic disorder. BMC Bioinf.. 2006;7:208.
- [Google Scholar]
- Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol.. 2009;7:439-450.
- [Google Scholar]
- Risco, C., Antón, I. M., Suñé, C., Pedregosa, A. M., Martin-Alonso, J. M., Parra, F., Carrascosa, J. L. & Enjuanes, L., 1995. Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion. Journal of virology, 69, 5269-5277.
- Computers and viral diseases. Preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput. Biol. Med.. 2020;119:103670.
- [Google Scholar]
- Sequence data analysis for long disordered regions prediction in the calcineurin family. Genome Inf.. 1997;8:110-124.
- [Google Scholar]
- Ribonucleocapsid formation of severe acute respiratory syndrome coronavirus through molecular action of the N-terminal domain of N protein. JVI. 2007;81:3913-3921.
- [Google Scholar]
- A hidden Markov model for predicting transmembrane helices in protein sequences. Ismb 1998:175-182.
- [Google Scholar]
- Immunology of avian influenza virus: a review. Dev. Comp. Immunol.. 2000;24:269-283.
- [Google Scholar]
- The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res.. 1997;25:4876-4882.
- [Google Scholar]
- Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J. Biol. Chem.. 2010;285:12862-12872.
- [Google Scholar]
- Identifying SARS-CoV membrane protein amino acid residues linked to virus-like particle assembly. PLoS One. 2013;8:e64013
- [Google Scholar]
- Studies on membrane topology, N-glycosylation and functionality of SARS-CoV membrane protein. Virol. J.. 2009;6:1-13.
- [Google Scholar]
- Xue, B., Dunbrack, R. L., Williams, R. W., Dunker, A. K. & Uversky, V. N., 2010. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochimica et Biophysica Acta (BBA)-Proteins and Proteomics, 1804, 996-1010.
- Xue, B., W Williams, R., J Oldfield, C., Kian-Meng Goh, G., Keith Dunker, A. & N Uversky, V., 2010. Viral disorder or disordered viruses: do viral proteins possess unique features? Protein and peptide letters, 17, 932-951.







