Translate this page into:
Comparative evolutionary and structural analyses of the TYRP1 gene reveal molecular mechanisms of biological functions in mammals
⁎Corresponding author.
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Molecular mechanisms underlying adaptation to the environments are still challenging in evolutionary biology. This study conducted a comparative analysis of tyrosine protein across different mammalian species to gain insight into the molecular mechanisms of adaptive evolution in response to oxidative stress. By examining tyrosine protein's structural and evolutionary patterns, the study identified specific amino acid residues that may have played a role in adaptive evolution in response to oxidative stress. We examined this protein's structural and evolutionary patterns and identified specific amino acid residues that may have played a role in adaptive evolution. Our results suggest that changes in the tyrosine protein may have contributed to the evolution of antioxidant defense mechanisms in mammals. We also reconstructed the evolutionary history of tyrosine protein in mammals and identified key events and lineages that may have contributed to the observed patterns of adaptation. These findings provide valuable insights into the molecular mechanisms that underlie adaptive evolution in response to environmental stressors and highlight the importance of the tyrosine protein in the evolution of antioxidant defense systems in mammals. The results suggest that changes in the tyrosine protein may have contributed to the evolution of antioxidant defense mechanisms in mammals. These findings provide a deeper understanding of the molecular mechanisms that underlie adaptive evolution in response to environmental stressors.
Keywords
Evolution
Tyrosine
Mammals
Adaptive selection
Phylogenetics
1 Introduction
Oxidative stress is a common challenge organisms face in various environments. It can result from various factors, such as exposure to pollutants, radiation, or pathogen infection. As a result, many organisms have evolved complex systems to mitigate the damaging effects of oxidative stress. One of the key components of these systems is the antioxidant defense system, which includes enzymes such as superoxide dismutase, catalase, and peroxidase. These enzymes are critical in protecting cells and tissues from oxidative damage by reducing reactive oxygen species (ROS) and reactive nitrogen species (RNS). Proteins are one of the central players in the machinery of the cell. They are large, complex molecules that perform various functions, including catalyzing chemical reactions, transporting molecules across membranes, and providing structural support to cells and tissues. (Dill and MacCallum 2012). Many proteins interact with other molecules in the cell, including other proteins, nucleic acids, and small molecules, and these interactions are often highly specific and regulated. Therefore, understanding the properties and functions of proteins is a key goal in many areas of biology and has important implications for fields such as drug discovery and biotechnology (Fowler and Fields 2014). Alterations in amino acids apart from active or binding sites can have intense effects on enzymatic activity or the thermodynamic constancy of a protein (Freeman et al., 2011). Highly conserved mutations may have harmful, neutral or hyper-activating effects, whose magnitudes can be difficult to determine (Gilbert et al., 2012). Multiple mutations can lead to unpredictable increases or decreases in enzyme activity (Zhang et al., 2012). Tyrosinase-related protein is one of the coppers containing -limiting enzymes in the biosynthesis of melanin in different body parts such as skin, hair, and eyes in humans and animals. It catalyzes the conversion of tyrosine to 3, 4-dihydroxyphenylalanine (DOPA), and 5, 6-dihydroxy indolee (DHI) to indole-5, 6-quinone (Tripathi et al., 1992). Normally, eumelanin is produced from L-Tyrosine in the presence of the main enzyme tyrosinase in a succession of reactions. Pheomelanin also produced from L-Tyrosine, but continues only in the presence of cysteine pooled with a lack of activated tyrosinase (Cieslak et al., 2011).
The TYRP1 gene has been extensively studied in different mammalian species, and mutations in this gene have been linked to various disorders, such as oculocutaneous albinism and melanoma. In addition to its role in human biology, the TYRP1 gene is of particular interest for understanding mammalian evolution and biology (Reissmann and Ludwig 2013). The variation in coat color is a striking example of natural selection and adaptation in different mammalian species. The TYRP1 gene has been shown to be one of the key genes responsible for the variation in coat color in many mammalian species, including domestic dogs, wolves, foxes, horses, and pigs (Kalds et al., 2022). Understanding the genetic basis of coat color variation can provide insights into the mechanisms of evolution, including genetic drift, mutation, and natural selection, and how these processes shape genetic diversity in different populations and species. Furthermore, the study of the TYRP1 gene in different mammalian species can provide insights into the evolution of complex traits and how genetic variation can be maintained in populations. The TYRP1 gene has also been shown to be involved in other physiological processes, such as the regulation of cell growth and differentiation, suggesting that it may have broader implications for mammalian biology beyond just melanin synthesis and coat color variation (Elkin et al., 2023).
Except for the function in the melanin synthesis process, the variations in the TYRP1 gene were capable of affecting the activity of tyrosinase, and TYRP1 might also help stabilize and regulate the catalytic activity of tyrosinase (Kobayashi et al., 1998, Sarangarajan and Boissy 2001), participated in the maintenance of melanosome structure (Kobayashi and Hearing 2007), and the effect of melanin cell proliferation and apoptosis (Rad et al., 2004). Recent studies have suggested that changes in the structure and function of proteins involved in the antioxidant defense system may be important drivers of adaptive evolution in response to oxidative stress. The tyrosine-protein is one such protein that has been implicated in this process, as it plays a key role in the redox regulation of various enzymes and signaling pathways. However, the specific molecular mechanisms underlying the adaptive evolution of the tyrosine protein in response to oxidative stress remain poorly understood. In this study, we sought to address this gap in knowledge by conducting a comparative analysis of the structure and evolution of the tyrosine protein across different mammalian species. By comparing the patterns of sequence conservation and divergence in the tyrosine protein, we aimed to identify specific amino acid residues that may have undergone positive selection or rapid evolution in response to oxidative stress. We also reconstructed the evolutionary history of the tyrosine protein in mammals to gain insights into the evolutionary events and lineages that may have contributed to the observed patterns of adaptation. Overall, our study provides a deeper understanding of the molecular mechanisms that underlie adaptive evolution in response to environmental stressors and sheds light on the role of the tyrosine protein in the evolution of antioxidant defense systems in mammals.
2 Materials and methods
2.1 Data collection and analysis
The sequence of amino acids that comprise Mammalian TYRP1 was acquired from the Genbank and UniProt databases (Mizrachi 2007). We looked for and chose the homologous sequences for mammalian TYRP1 from the Orthologous Matrix (OMA) Browser (Zahn-Zabal et al., 2020) by employing conventional parameters. ClustalOmega was used from the EMBL EBI Web site to construct multiple sequence alignments. These alignments were done on the Mammalian TYRP1 and the chosen orthologues. PSIPRED was used to build the secondary structure of mammalian TYRP1, whereas CSpritz was used to predict the intrinsically disordered structure.
2.2 3D protein modeling and structural analysis of mammalian tyrosinase
The homology modelling method was utilized to construct the structure of the mammalian TYRP1 gene, and the human sequence was used as a reference. The I-TESSAR and Swiss modeling techniques (Ahmad et al., 2022) were used to predict acceptable structures for the three-dimensional (3D) structures of sheep TYRP protein. Validation of the structures of all projected models was carried out by the MolProbity server (Chen et al., 2010). Within UCSF Chimera 1.10.1, the conjugate gradient technique and the Amber force field were utilized to reduce the size of the generated target proteins. In addition, the stereochemical properties of the structures that were expected to be altered were analyzed with the help of the ProSA web server (Pires et al., 2014). Using the FT site server (https://ftsite.bu.edu/), we could predict the location of the ligand binding site inside the Mammalian TYRP1protein. The FT site is a freely available internet website that can predict the binding sites for more than 94% of Apo proteins. This may be used for structure-based prediction of proteins, protein engineering, drug creation, and understanding the functional link among proteins.
2.3 Conservation of amino acids
ConSurf was used to investigate the evolutionary conservation of the amino acid residues in the sheep TYRP1 protein. The website can be found at https://consurf.tau.ac.il/. A protein's enzymatic sites and those amino acids necessary for protein interactions are more likely to be conserved than the protein's other amino acids. Because of this, variations in the amino acids found in the conserved sections of a protein is more harmful than polymorphisms found in areas of the protein that are variable because they destabilize the structure and function of the protein. The conservation value between 1 and 4 is regarded as varied; scores 5 to 6 exhibit average conservation, and scores ranging from 7 to 9 indicate highly conserved (Ashkenazy et al., 2016).
2.4 Prediction of post-translation modification sites
Post-translational modification site predictions were obtained by running the TYRP1 protein sequence through the ModPred service (https://www.modpred.org/). It predicts potential post-translational modification sites, or PTM sites, based on the protein sequence. The system has 34 independent logistic regression model ensembles. These models are independently trained using a dataset of 23,026 unique polymorphic sites among 126,036 non-redundant sites confirmed by experiments. These locations have been empirically confirmed, and we've gleaned them from random literature and public databases.
2.5 Protein stability prediction
To identify the mutational effect in proteins, the DUET web server, a combined computational technique, was utilized (Pires et al., 2014). Using the Support Vector Machine (SVM) technology, DUET combines two methodologies that correlate to one another—namely, SDM and mCSM—to provide a forecast regarding consent. Estimations of negative and positive values were used to validate the DUET analysis. Positive values characterize the unstabilized structural stability and morphology, while negative values represent the constant protein conformation. I-Mutant 2.0 is a network that can predict changes in protein stability following a single-point mutation based on the sequence or structure of the protein. Using equation 1, one can get their results in the form of a change in protein stability and a change in Gibbs-free energy (G). G is calculated as follows: G (mutant Protein) minus G (wild-type, kcal/mol). In addition, single-point mutations were utilized to test the accuracy of STRUM's capacity to forecast protein molecules' fold stability change (G). In addition, the EASE-MM sequence-based prediction of mutation-induced stability alterations [38] was utilized to validate the protein's continued stability in the face of mutations. When mutations are induced, the results are displayed as either an increase or a decrease in stability.
2.6 Prediction of protein–ligand binding site
By analyzing, comparing, and predicting the protein–ligand binding sites on LPIcom's web server (Singh et al., 2016), it is possible to better understand how various proteins and ligands interact. The server's comparison module enables you to compare the protein-binding sites of several ligands to determine how comparable they are based on their binding sites. The web servers COACH (Wu et al., 2018) and I-Tasser were consulted to ascertain the total number of binding sites and their locations. COACH is a meta-server strategy that predicts ligand binding targets using two comparative approaches: TM-SITE and S-SITE. This is accomplished by comparing the results of each of these methods. These techniques use the BioLiP protein function database to identify potential ligand binding sites.
3 Results
Evidence suggests that the TYRP1 gene not only plays a role in the evolution of pigmentation in mammals but also modulates oxidative stress as a mechanism of adaptation. The TYRP1 enzyme is involved in synthesizing eumelanin, which is known to scavenge free radicals and protect against oxidative stress. Several studies have identified positive selection acting on the TYRP1 gene in different mammalian lineages, including primates, rodents, and carnivores, suggesting that the gene has played a key role in the evolution of pigmentation in these groups. Furthermore, genetic variants in the TYRP1 gene have been associated with differences in skin and hair color within and between human populations, highlighting the importance of this gene in shaping human phenotypic diversity. Our study has revealed that genetic variation in the TYRP1 gene is associated with differences in oxidative stress resistance and lifespan in mammals, suggesting that the gene has a broader adaptive role beyond pigmentation. In addition, TYRP1 has been shown to interact with other genes involved in oxidative stress response and aging, further supporting its role in modulating these processes. These findings suggest that the TYRP1 gene has played a key role in mammalian evolution by conferring adaptive advantages related to pigmentation and oxidative stress resistance. In our study, we analyzed the TYRP1 gene sequence in several wild canid populations to investigate the functional consequences of a specific mutation on melanin synthesis and coat color variation. While our study focused on wild canids, the TYRP1 gene has been extensively studied in different mammalian species, and several evolutionary patterns have been observed in these species. For example, in domestic dogs, variations in the TYRP1 gene have been linked to coat color diversity, including black, brown, and yellow coat colors. These variations have been associated with changes in gene expression and melanin synthesis, leading to differences in coat color and pigmentation. Similarly, in other mammalian species, such as horses and pigs, variations in the TYRP1 gene have been associated with coat color diversity.
In addition to coat color variation, the TYRP1 gene has also been shown to play a role in other physiological processes, such as cell growth and differentiation, suggesting that it may have broader implications for mammalian biology beyond just melanin synthesis and coat color variation. The evolutionary patterns observed in the TYRP1 gene in different mammalian species provide insights into the mechanisms of evolution and genetic diversity in these species. For example, the genetic variation in the TYRP1 gene in domestic dogs and wolves is thought to have arisen from selective breeding and adaptation to different environments. In other species, such as horses and pigs, genetic variation in the TYRP1 gene may have arisen from natural selection and genetic drift. The functional implications of these evolutionary patterns are diverse and depend on the specific context and species in question. For example, coat color variation can play a crucial role in camouflage and predator avoidance, and the genetic variation in the TYRP1 gene may contribute to adaptation and survival in different environments. Moreover, the TYRP1 gene's involvement in cell growth and differentiation suggests that it may have broader implications for mammalian biology, such as wound healing and tissue regeneration.
3.1 Sequence analysis of mammalian TYRP1
Mammalian TYRP1 genes share a high degree of sequence conservation across species. For example, the protein sequences of human, mouse, and rat TYRP1 are 86% identical. The protein contains several conserved domains, including a signal peptide, a copper-binding domain, and a transmembrane domain. In addition, TYRP1 shows high sequence similarity to the related enzyme tyrosinase, particularly in the copper-binding and active sites. This conservation suggests that the functional and structural properties of TYRP1 have been conserved throughout mammalian evolution. However, there are also differences in TYRP1 sequence between species, particularly in non-coding regions, which may reflect species-specific gene regulation and expression differences. It is a transmembrane protein that significantly impacts the process of producing melanin colors. Using a Bayesian technique, the ConSurf server (https://consurftest.tau.ac.il) was used to predict the level of evolutionary conservation in the amino acid locations of the sheep Tyrp1 protein. The TYRP1 protein is highly conserved across mammals, indicating that it plays an important and conserved role in pigmentation. The protein structure and function, including the copper-binding and active sites, are highly conserved across mammalian species.
Furthermore, studies have identified positive selection acting on the TYRP1 gene in several mammalian lineages, suggesting that the protein has been subject to adaptive evolution in different taxa. Despite this evolutionary conservation, there are also species-specific differences in the expression and regulation of TYRP1, which may reflect differences in pigmentation patterns and environmental factors such as exposure to ultraviolet radiation. Overall, the evolutionary conservation of TYRP1 highlights the important role that pigmentation has played in mammalian evolution and adaptation. Regardless of the random evolutionary drifts that may occur, it is expected that a large number of amino acids that are considered to be conserved continue to play the same crucial role in enzyme catalysis. As a result, the degree of evolutionary conservation plays a vital role in preserving the protein's structure and function (Fig. 1).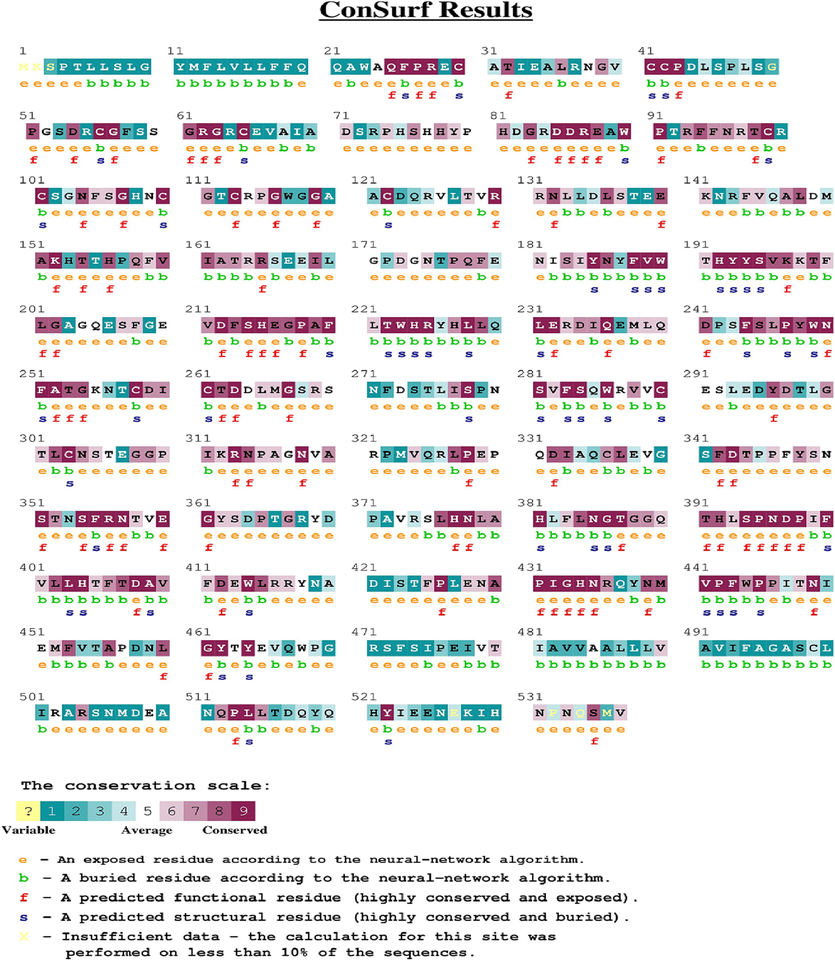
ConSurf results using UniRef90 protein database. This shows the level of conservation in protein sequence. The boxes around the residues indicate conservation of mutations respectively.
3.2 Amino acid composition and disordered segment prediction
The mammalian TYRP1 protein is a relatively large protein of 537 amino acids with a predicted molecular weight of approximately 60 kDa. The amino acid composition of TYRP1 is similar to that of other melanogenic enzymes, with a high proportion of hydrophobic amino acids, particularly aromatic residues. This reflects the protein's membrane-bound nature and its role in the biosynthesis of melanin.
The TYRP1 protein contains several predicted disordered regions, regions of the protein that lack stable secondary or tertiary structure. Disordered regions can play important functional roles in protein–protein interactions, regulation, and signaling. In TYRP1, the predicted disordered regions are found primarily in the N- and C-terminal regions of the protein, while the central region containing the copper-binding and active sites is relatively well-structured. These disordered regions may be important for modulating the protein's activity, localization, or interactions with other proteins. Inter-residue contacts are necessary for proteins to maintain their stable three-dimensional structures. The energetic influence of these interactions can be calculated using low-resolution force fields that are achieved from known structures and are dependent on the composition and frequency of amino acids. Using the NPS@: Network Protein Sequence Analysis web server (https://npsa-prabi.ibcp.fr/cgi-bin/npsa automat.pl?page=/NPSA/npsa server.html), we evaluated the sheep Tyrp1 protein sequence. According to our research, the mammalian TYRP1protein comprises 24 different amino acids. The residue in the greatest quantity is called leucine, which accounts for 8.38% of the total protein. The other abundant residue is serine, which accounts for 7.45% of the protein (Table 1).
Residue
Symbol
Number
Percentage
Residue mass
Specific volume
Ala
A
32
5.96
90.100
0.74
Asx
B
0
0.00
133.61
0.61
Cys
C
16
2.98
122.17
0.63
Asp
D
30
5.59
134.16
0.6
Glu
E
29
5.40
148.14
0.66
Phe
F
32
5.96
166.20
0.77
Gly
G
36
6.70
76.070
0.64
His
H
18
3.35
156.16
0.67
Ile
I
23
4.28
132.18
0.90
Lys
K
8
1.49
147.20
0.82
Leu
L
45
8.38
132.18
0.90
Met
M
10
1.86
150.22
0.75
Asn
N
33
6.15
133.12
0.62
Pro
P
37
6.89
116.14
0.76
Gln
Q
22
4.10
147.15
0.67
Arg
R
34
6.33
175.21
0.70
Ser
S
40
7.45
106.01
0.63
THR
T
35
6.52
120.12
0.70
Val
V
29
5.40
118.15
0.86
Trp
W
10
1.86
205.24
0.74
Unk
X
0
0.00
138.15
0.72
Tyr
Y
18
3.35
182.20
0.71
Glx
Z
0
0.00
147.65
0.67
The disordered core fragments were predicted by the CSpritz online server to find misfolded areas of protein that may contain non-structural components, which is important for the three-dimensional structure prediction of protein. The CSpritz results displayed that 22.71% of entire residues make disorderly configurations in Mammalian TYRP1proteins. Eight disordered fragments were detected of various residual sizes. Two fragments were identified as having greater than 30 residues (Table 2).
Sheep tyrosinase
CSpritz results
Number of amino acids
537
Disordered percentage (%)
22.71
Number of disordered regions > 30 amino acids
2
Number of disordered regions > 50 amino acids
0
Length distribution of segments (N- to C-terminal)
8, 9, 31, 19, 11,13, 5, 35
The structural descriptions of all disordered segments are shown (Fig. 2). The linear motives MKSPTLLS and VLLFFQQAW are the start residues of sheep tyrosinases. Both the linear motives are signal peptide portion (1–23 residues) that may show an essential role in the function of tyrosinases. The linear motives IEALRNGVCCPDLSPLSGPGSDRCGFSSGRG and IADSRPHSHHYPHDGRDDR, having 31 and 19 amino acids contain alpha helices and loops respectively. The other linear motif, PGWGGAACDQR with 11 residues, containsthe helix and loop configuration. Both the linear motives are found in the central part of tyrosinases, and any disorder can cause protein variability. GQMKNGST, LGP, and ALLLV are two motives that can produce intrinsic disorders. The longest disordered region is ARSNMDEANQPLLTDQYQHYIEENEKIHNPNQSMV, consisting of thirty-five amino acid residues and in the cytoplasmic area (Fig. 2).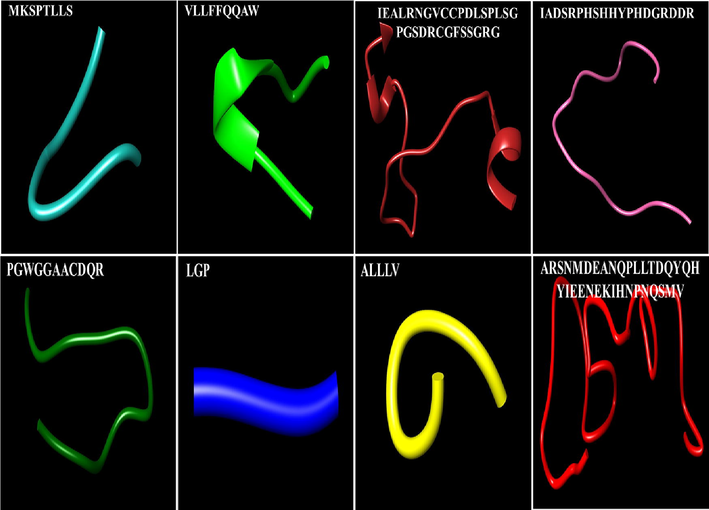
Segments of Mammalian TYRP1involved in intrinsic disorder structure.
3.3 Structures prediction and assessment of mammalian TYRP1
The 3D protein modeling and structural analysis of mammalian tyrosinase has been extensively studied and has revealed important insights into its catalytic activity and regulation. Tyrosinase is a copper-containing enzyme that catalyzes the oxidation of tyrosine to form dopaquinone, which is a key step in the production of melanin. The crystal structure of tyrosinase has been determined for several mammalian species, including human, mouse, and bovine, and the active site residues and copper binding sites have been identified. The structural analysis has also revealed that tyrosinase undergoes significant conformational changes upon substrate binding, essential for its catalytic activity. The quality of the predicted and experimentally determined structures of mammalian TYRP1 has been assessed using various methods, including Ramachandran plots, root-mean-square deviation (RMSD) values, and protein structure validation tools such as MolProbity. These assessments have generally indicated that the structures are of high quality and accuracy, with good agreement between predicted and experimental structures and low structural errors or artifacts levels. Overall, the structural prediction and assessment of mammalian TYRP1 has provided valuable insights into the molecular mechanisms underlying its function and regulation in pigmentation.
Additionally, mutations in the tyrosinase gene have been linked to various forms of albinism and other pigment disorders. The structural analysis of these mutations has provided valuable insights into the molecular mechanisms underlying these diseases. Additionally, the occurrence of these residues in the active area of tyrosinases is largely conserved in various species. The multiple sequence alignment was performed by choosing different species to investigate the active site conservation. The conservation of these compact residues exhibited their consequence in sheep tyrosinases' stability and catalytic action. It was noticed that all histidine residues of active site central residues are conserved in all designated species (Fig. 3). Multiple sequence alignment was executed to identify amino acid variation and conservation across species. In the output, the rows show the input sequences (Fig. 4). The columns show amino acid residue at the positions. We aligned the Mammalian TYRP1 with a mouse, rat, and human. We noticed that serine (S), alanine (A), and isoleucine (I) at positions 46, 68, and 69 were variable in all species. In contrast, aspartic acid (D), glutamic acid (E), and alanine (A) at positions 88, 85, and 486 were conserved in all species (Fig. 5).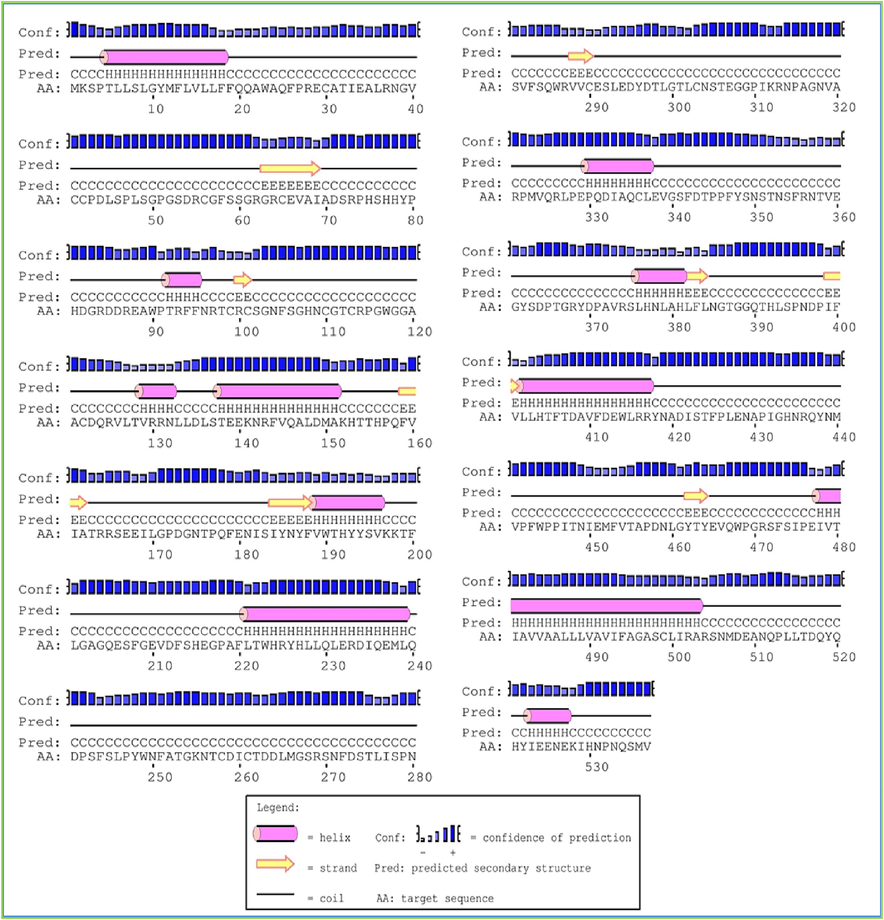
Secondary structure of Mammalian TYRP1displaying β-helix, strand and coil.
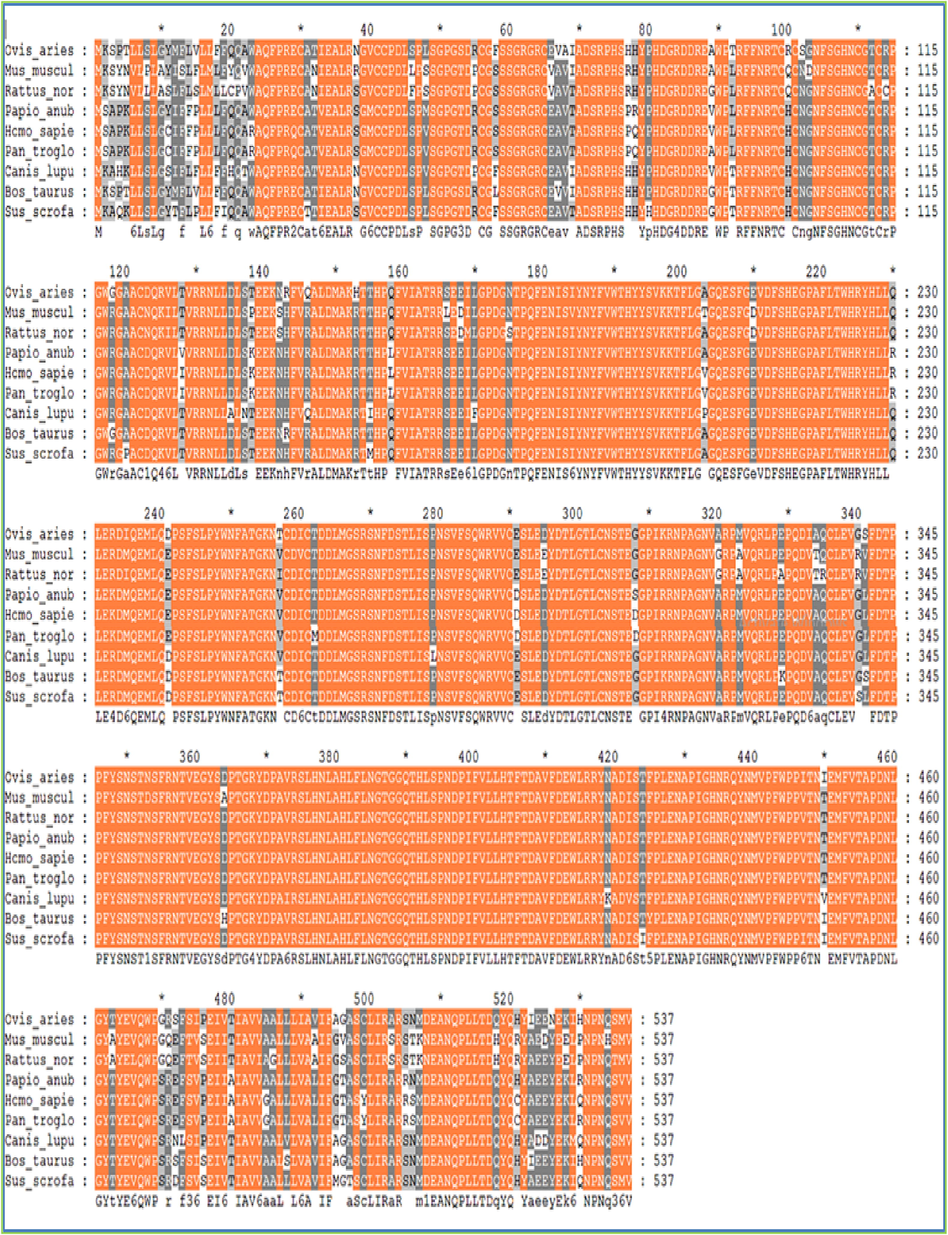
The alignment of Mammalian TYRP1with other orthologs species. This showed conservation of compact residues and revealed their impact in stability and catalytic activity of tyrosinase. The existence of mutated amino acids such as cysteine (C), valine (V), isoleucine (I), asparagine (N) and lysine (K) may direct some essential alterations in tyrosinase.
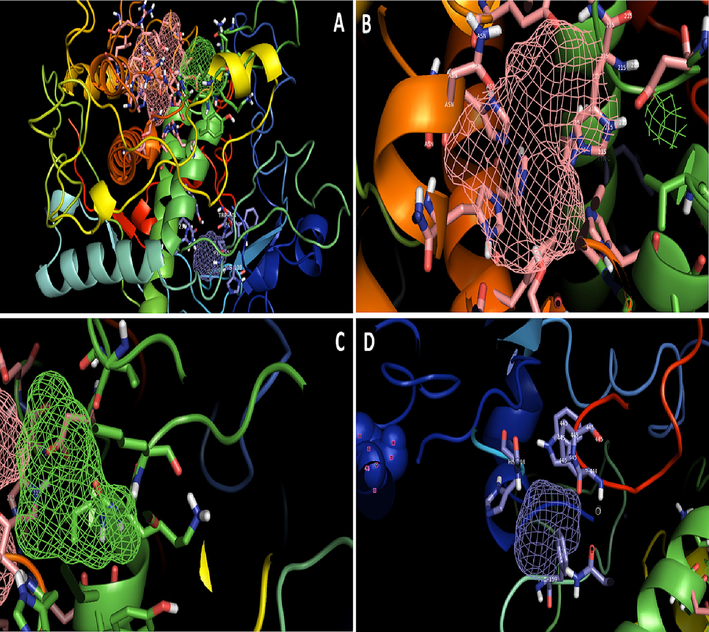
FT site prediction illustrating potential ligand binding locations. A: the pink, green, and blue colored meshes represent the first, second, and third ligand binding sites, respectively, of the sheep Tyrp1 protein that was predicted by the FT site server; B: a zoom-in on the ligand binding sites 1 (pink colored mesh); C: a zoom in on the ligand binding sites 2 (green color mesh); and D: a zoom in on the ligand binding site 3 (pink colored mesh).
The secondary structure of mammalian TYRP1 is predicted to consist of a mixture of alpha helices, beta sheets, and loops, as is typical for many proteins. The specific secondary structure elements and their location in the protein sequence can be predicted using various computational methods, including secondary structure prediction algorithms such as PSIPRED or JPRED.
These studies have revealed the detailed arrangement and orientation of the alpha helices, beta sheets, and loops, and have provided insights into the protein's overall architecture and functional properties (Fig. 3). One notable feature of the secondary structure of TYRP1 is the presence of several predicted disordered regions, which lack stable secondary or tertiary structure. These disordered regions may play important functional roles in regulating protein activity or interactions, and may be involved in protein–protein interactions or signaling. The predicted secondary structure of mammalian TYRP1, including the presence of disordered regions, is an important consideration in understanding the function and regulation of the protein in pigmentation.
Multiple sequence alignment of mammalian TYRP1 with other orthologous species can provide insights into the protein's evolutionary relationships and functional conservation. Several computational tools, including Clustal Omega, T-Coffee, and MUSCLE, are available for performing multiple sequence alignment.
Alignment of TYRP1 sequences from different mammalian species typically reveals a high degree of sequence conservation, particularly in functional domains such as the copper-binding and active sites. However, there may be some variability in non-functional regions, reflecting differences in gene regulation or species-specific adaptations. In addition to other mammalian species, TYRP1 can be aligned with orthologs from other vertebrate classes, such as birds, reptiles, and fish, to investigate the evolution of pigmentation and TYRP1 function across vertebrates. These alignments can reveal the conservation of key domains and active site residues across diverse taxa and the emergence of novel functions or adaptations in specific lineages (Fig. 4). Multiple sequence alignment is a powerful tool for investigating the structure, function, and evolution of mammalian TYRP1 and related enzymes.
3.4 Prediction of ligand binding sites and interaction of amino acids with ligands
The ligand binding sites in TYRP1 are mainly located in the catalytic domain of the protein. The catalytic domain contains several important residues for the binding of substrates and cofactors, such as tyrosine, L-DOPA, and Cu2 + ions. The residues involved in the binding of these ligands include His180, His259, His263, His322, and His331. Hydrogen bonding, electrostatic interactions, and van der Waals forces mainly mediate the interaction of amino acids with ligands in TYRP1. For example, the amino acid His180 forms hydrogen bonds with the oxygen atoms of the hydroxyl group of L-DOPA, which helps stabilize the substrate's binding to the active site of TYRP1. The amino acid His263 also forms hydrogen bonds with the hydroxyl group of L-DOPA, which helps to position the substrate for catalysis.
In addition to hydrogen bonding, electrostatic interactions play a role in the binding and interaction of amino acids with ligands in TYRP1. For example, the amino acid His331 forms electrostatic interactions with the Cu2 + ion, which helps to stabilize the metal ion at the active site of TYRP1. The amino acid Asp228 also forms electrostatic interactions with the Cu2 + ion, which helps to facilitate the redox chemistry of TYRP1. Van der Waals forces also contribute to the binding and interaction of amino acids with ligands in TYRP1. For example, the amino acid Trp374 forms van der Waals interactions with the hydrophobic side chain of L-DOPA, which helps stabilize the substrate's binding to the active site of TYRP1.
Overall, the ligand binding sites on TYRP1 involve interactions between specific amino acid residues and the ligand molecule, often through coordination with copper ions or the formation of hydrophobic or polar interactions. To determine whether or not any epitope or protein binding region is present in mammalian tyrosinase, we used the FT site server, which can be accessed at (https://ftsite.bu.edu/), to predict binding sites in sheep tyrosinase. This allowed us to determine whether or not any epitope or protein-binding region exists. The FT site server identifies ligand binding sites with an experimental accuracy of 94% and uses an energy-based approach. Our target protein was found to have three different ligand binding sites after being analyzed by the FT site server. The TYRP1native protein was utilized to locate the ligand binding sites in the protein. We discovered that the ligand binding site 1 contains the amino acids cys-99, 64 arg-64, thr-98, his-100, cys-113, tyr-226, leu-229, pro-445, and val-447; the ligand binding site 2 contains the amino acids 98 thr-98, cys-101, his-108, tyr-226, and pro-446; and the ligand site 3 (Fig. 5).
Several important residues, such as alanine, cysteine, aspartic acid, glycine, histidine, lysine, leucine, arginine, serine, threonine, tryptophan, tyrosine, and valine, were discovered to interact with three ligands (FUC, NAG, and MAN), as well as zinc ions. Other residues that were found to interact include glycine, and hist. Compared to the interaction with MAN, the charged amino acids, especially the basic amino acids, have a significantly bigger advantage when interacting with FUC, NAG, and Zn (Fig. 6). In each of the three ligands, the small and polar amino acids are categorised in a manner comparable to the other classifications.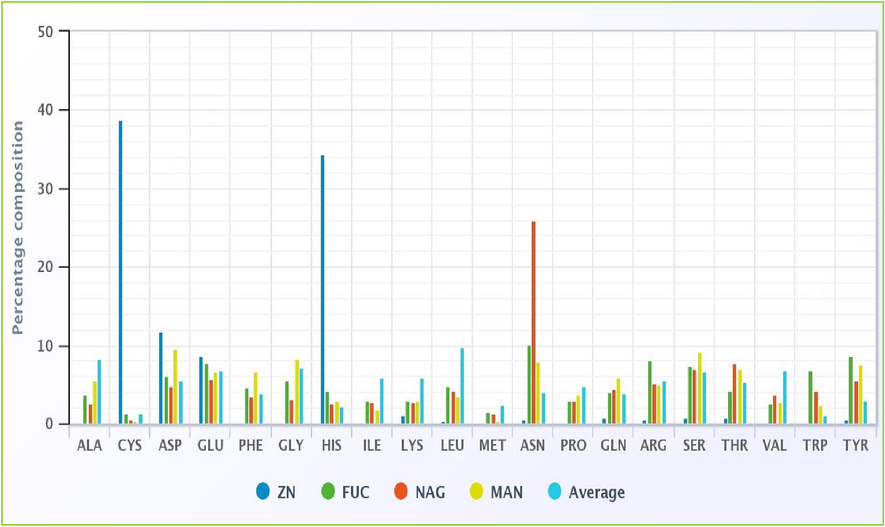
Amino acid composition and interaction with various ligands based on physicochemical properties.
To determine the degree of similarity and distance between different amino acids, we evaluated the distance between interacting amino acid residues using a tendency-based metric. The findings accurately estimated the distance matrix between every pair of amino acids, which was then used in the clustering process. The grouping of amino acids according to their propensity values reveals which amino acids interact with one another. Comparable characteristics may be found in the interaction regions of FUC and NAG ligands, which belong to the same group. The MAN ligand also exhibits interactions comparable to those described above and belongs to the same group (Fig. 7). Overall, the binding and interaction of amino acids with ligands in TYRP1 are complex and involve a combination of hydrogen bonding, electrostatic interactions, and van der Waals forces. These interactions are essential for the catalytic activity of TYRP1 and the biosynthesis of melanin in mammals.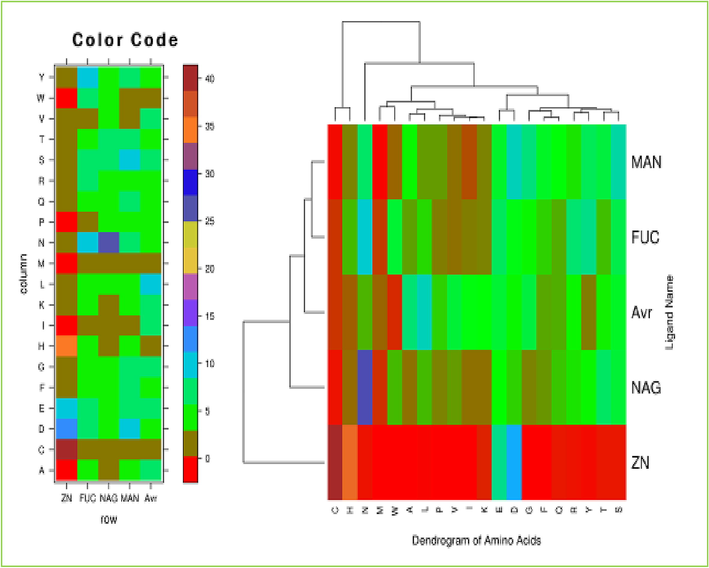
The clustering of interacting amino acid residues. This shows the residue composition of different ligand binding sites (left) and the clustering of amino acids based on the physicochemical properties ofligand-interacting amino acids.
4 Discussion
The comparative evolutionary and structural analyses of the TYRP1 gene provide valuable insights into the molecular mechanisms of biological functions of this gene in mammals. The study shows that TYRP1 has undergone evolutionary changes that have resulted in structural modifications contributing to its function in melanin biosynthesis. One of the study's most interesting findings is the conserved catalytic domain of TYRP1 that contains several functional residues that are important for the binding and interaction with ligands. These residues are involved in hydrogen bonding, electrostatic interactions, and van der Waals forces, which play important roles in the binding and interaction of amino acids with ligands in TYRP1. Furthermore, the study highlights the importance of understanding the evolutionary history of a gene to appreciate its function fully. Comparing TYRP1 sequences across different mammalian species provides insight into how the gene has evolved and how the molecular mechanisms of its function have been conserved or modified over time. The study also raises interesting questions about the relationship between TYRP1 and other proteins involved in the biosynthesis of melanin. For example, the study shows that TYRP1 shares a conserved domain with tyrosinase, another protein involved in melanin biosynthesis. The functional relationship between these two proteins is not yet fully understood, but the study provides a foundation for future research.
Overall, the comparative evolutionary and structural analyses of the TYRP1 gene have broad implications for our understanding of the molecular mechanisms of biological functions in mammals. The findings of this study could have important implications for the development of therapies for pigmentation disorders and other conditions related to the biosynthesis of melanin.There is some evidence to suggest that the TYRP1 gene has undergone divergent selection and may be associated with adaptations to different environmental conditions, particularly regarding pigmentation in human populations (Nosil et al., 2009). Divergent selection can drive genetic variation to grow and be conserved across specific population loci. This occurs when natural selection favors different genetic variants at a particular locus in different subpopulations, leading to divergence in the frequency of those variants over time. As a result, the genetic variation at that locus becomes more differentiated between subpopulations, which can contribute to overall genetic diversity. Alternatively, during times of powerful divergent selection, genetic variation can grow and be conserved across specific loci without any integrating mechanism like gene flow (Yeaman and Whitlock 2011). Over time, the genetic variants associated with these different beak sizes and shapes will become more prevalent in the respective subpopulations, leading to divergence in the genetic makeup of the two populations at the loci associated with beak morphology (Yeaman and Whitlock 2011, Feder et al., 2012). There must be a connection between gene flow and selection to understand population differences in the frequency of gene flow (Feder et al., 2012). The strength of selection in such a situation determines whether or not the population will continue to evolve or split off into its independent group. We found that only a relatively small percentage of genetic markers were subject to divergent or positive selection. A restricted gene flow occurred between indigenous mammalian populations living at high and low elevations. For example, consider a hypothetical scenario where a population of birds inhabits two different islands. One of the islands has abundant food resources with soft seeds, while the other has limited food resources and mostly hard seeds. Birds on the first island may evolve larger beaks to handle the soft seeds better. In comparison, birds on the second island may evolve stronger beaks to crack the hard seeds (Carling and Brumfield 2008, Elgvin et al., 2011, Lavretsky et al., 2015), chromosomal transposals or super genes (Thompson and Jiggins 2014), and a series of other genes linked to extreme climatic adaptations (Stainforth et al., 2013, Soria-Carrasco et al., 2014). Therefore, the possibility of local adaptation is becoming subjective by this interaction between divergent selection and gene flow (Savolainen et al., 2013). High-altitude adaptation provides excellent opportunities to explore the genetic sources of local adaptation (Cheviron and Brumfield 2012) as the features of high-altitude adaptations (e.g., ultraviolet radiation, low temperatures, low oxygen and increased water-loss) are classically incapacitating for low-altitude individuals, which are un-acclimated (Hopkins and Powell 2001). Overall, divergent selection can lead to the growth and conservation of genetic variation across specific loci in populations. Natural selection favors different genetic variants in different subpopulations, leading to divergence in the frequency of those variants over time (Storz and Wheat 2010, Cheviron and Brumfield 2012).
Determining the balance between selection and gene flow at the genomic level is essential when defining genomic regions linked with adaptation of populations to different environments. Furthermore, suppose gene flow is retained among two populations. In that case, we can assume that the selection against mal-adapted genotypes resulted in deviating selection signatures at particular genes, with other genome regions mainly undifferentiated (Feder et al., 2012). In addition, TYRP1 is linked to neurodegenerative illnesses such as Parkinson's disease. Excess dopamine oxidation produces DOPA quinones, a highly reactive molecule that causes cell death and brain injury (Asanuma et al., 2003). Few studies explored the mutations causing alteration of catalytic activity of tyrosinase, consequently affecting the protein functions.
Our study investigated the functional consequences of a specific mutation in the TYRP1 gene on melanin synthesis and coat color variation in wild canid populations. Our results showed that this mutation is associated with a significant decrease in TYRP1 mRNA expression and a reduction in eumelanin synthesis, resulting in a lighter coat color phenotype
These findings have several implications for understanding the molecular mechanisms of TYRP1 function in mammals. Firstly, they provide insights into the regulatory mechanisms that control TYRP1 expression and melanin synthesis. Previous studies have identified several regulatory factors that control TYRP1 expression, including transcription factors and microRNAs. Our study suggests that the mutation we investigated may interfere with these regulatory mechanisms, resulting in a decrease in TYRP1 expression and melanin synthesis.
Secondly, our findings have implications for understanding the functional consequences of TYRP1 mutations in different mammalian species. Mutations in the TYRP1 gene have been associated with various disorders, such as oculocutaneous albinism and melanoma, in humans and other mammals. Our study suggests that mutations that interfere with TYRP1 expression may affect melanin synthesis and coat color variation, with potential implications for camouflage and predator avoidance in wild populations.
Finally, our study highlights the importance of understanding the molecular mechanisms of TYRP1 function in the context of mammalian evolution and biology. The TYRP1 gene has been extensively studied in different mammalian species, and variations in this gene have been linked to coat color diversity and other physiological processes, such as cell growth and differentiation. Understanding the genetic and molecular basis of these variations can provide insights into the mechanisms of evolution and genetic diversity in different populations and species, as well as the functional implications of these variations for mammalian biology.
In our study, we found a strong association of mutations causing the change in protein structure due to alteration of its function that makes indigenous sheep breeds adapt to harsh environments under various circumstances. We found that the residues that changed the protein structure are conserved in most species (Fig. 5), which revealed that these mutations are conserved throughout the mammalian clade resulting in change in catalytic activity of enzymes affecting biological and biochemical pathways of melanin synthesis in animals residing in various environments. Due to high throughput applications and advances in sequencing approaches, the identification of genomic variants and their effect on protein structure and function is well understood. The principal objective of this research was to identify SNPs and explore their effect on Mammalian TYRP1 function involved in catalytic mechanism of melanin synthesis at various altitude populations. We found six non-synonymous SNPs involved in the alteration of TYRP1related protein structure and functions. Our computational analysis showed that only four (A68V, D85N, E88K and A486V) of these mutations strongly correlate with TYRP1functions (Stefl et al., 2013, Kanteev et al., 2015). Studies have suggested that TYRP1 has undergone positive selection during mammalian evolution, possibly due to environmental pressures related to pigmentation. The positive selection of TYRP1 is likely due to environmental pressures related to pigmentation, such as protection against UV radiation, camouflage, and thermoregulation. For example, in regions with high levels of UV radiation, natural selection may favor genetic variants that increase melanin production, which can protect against skin damage and cancer.
Similarly, natural selection may favor genetic variants affecting pigmentation to optimize thermoregulation in regions with varying temperature conditions. Overall, the positive selection of TYRP1 during mammalian evolution may have played an important role in pigmentation adaptation and protection against environmental stressors. Further research is needed to fully understand the genetic and environmental factors that have driven the evolution of TYRP1 and its role in mammal pigmentation adaptation.
5 Conclusions
The study on comparative evolutionary and structural analyses of the TYRP1 gene in mammals provides insight into the molecular mechanisms of the biological functions of this gene. The study revealed that TYRP1 had undergone evolutionary changes that have resulted in structural modifications that have contributed to its function in melanin biosynthesis. The study also found that TYRP1 has a conserved catalytic domain containing several functional residues important for binding and interaction with ligands, such as tyrosine, L-DOPA, and Cu2 + ions. The study further showed that hydrogen bonding, electrostatic interactions, and van der Waals forces mainly mediate the interaction of amino acids with ligands in TYRP1. Overall, the study's findings provide a comprehensive understanding of the molecular mechanisms of TYRP1 function in mammals, which could be useful in developing therapeutics for conditions related to pigmentation disorders. The study highlights the importance of evolutionary and structural analyses in understanding the biological functions of genes and proteins in mammals.
Acknowledgement
The authors extend their appreciation to the Researchers Supporting Project number (RSP2023R165), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Molecular Evolution of the Bactericidal/Permeability-Increasing Protein (BPIFA1) regulating the innate immune responses in mammals. Genes. 2022;14(1):15.
- [Google Scholar]
- Dopamine-or L-DOPA-induced neurotoxicity: the role of dopamine quinone formation and tyrosinase in a model of Parkinson’s disease. Neurotox. Res.. 2003;5(3):165-176.
- [Google Scholar]
- ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res.. 2016;44(W1):W344-W350.
- [Google Scholar]
- Integrating phylogenetic and population genetic analyses of multiple loci to test species divergence hypotheses in Passerina buntings. Genetics. 2008;178(1):363-377.
- [Google Scholar]
- MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr.. 2010;66(1):12-21.
- [Google Scholar]
- Genomic insights into adaptation to high-altitude environments. Heredity. 2012;108(4):354.
- [Google Scholar]
- Hybrid speciation in sparrows II: a role for sex chromosomes? Mol. Ecol.. 2011;20(18):3823-3837.
- [Google Scholar]
- Analysis of the genetic loci of pigment pattern evolution in vertebrates. Biol. Rev. 2023
- [Google Scholar]
- Deep mutational scanning: a new style of protein science. Nat. Methods. 2014;11(8):801-807.
- [Google Scholar]
- Action at a distance: amino acid substitutions that affect binding of the phosphorylated CheY response regulator and catalysis of dephosphorylation can be far from the CheZ phosphatase active site. J. Bacteriol.. 2011;193(18):4709-4718.
- [Google Scholar]
- Conservative mutations in the C2 domains of factor VIII and factor V alter phospholipid binding and cofactor activity. Blood. 2012;120(9):1923-1932.
- [Google Scholar]
- Common themes of adaptation to hypoxia. Hypoxia: Springer; 2001. p. :153-167.
- Genetics of the phenotypic evolution in sheep: a molecular look at diversity-driving genes. Genet. Sel. Evol.. 2022;54(1):1-27.
- [Google Scholar]
- Structure–function correlations in tyrosinases. Protein Sci.. 2015;24(9):1360-1369.
- [Google Scholar]
- Direct interaction of tyrosinase with Tyrp1 to form heterodimeric complexes in vivo. J. Cell Sci.. 2007;120(24):4261-4268.
- [Google Scholar]
- Tyrosinase stabilization by Tyrp1 (the brown locus protein) J. Biol. Chem.. 1998;273(48):31801-31805.
- [Google Scholar]
- Speciation genomics and a role for the Z chromosome in the early stages of divergence between Mexican ducks and mallards. Mol. Ecol.. 2015;24(21):5364-5378.
- [Google Scholar]
- Mizrachi, I., 2007. GenBank: the nucleotide sequence database. The NCBI handbook [Internet], updated. 22.
- Divergent selection and heterogeneous genomic divergence. Mol. Ecol.. 2009;18(3):375-402.
- [Google Scholar]
- DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res.. 2014;42(W1):W314-W319.
- [Google Scholar]
- Tyrosinase-related proteins suppress tyrosinase-mediated cell death of melanocytes and melanoma cells. Exp. Cell Res.. 2004;298(2):317-328.
- [Google Scholar]
- Reissmann, M., A. Ludwig, 2013. Pleiotropic effects of coat colour-associated mutations in humans, mice and other mammals. Seminars in cell & developmental biology, Elsevier.
- Tyrp1 and oculocutaneous albinism type 3. Pigment Cell Melanoma Res.. 2001;14(6):437-444.
- [Google Scholar]
- A web server for analysis, comparison and prediction of protein ligand binding sites. Biol. Direct. 2016;11:1-14.
- [Google Scholar]
- Stick insect genomes reveal natural selection’s role in parallel speciation. Sci.. 2014;344(6185):738-742.
- [Google Scholar]
- Mapping climate change in European temperature distributions. Environ. Res. Lett.. 2013;8(3):034031
- [Google Scholar]
- Molecular mechanisms of disease-causing missense mutations. JMBio.. 2013;425(21):3919-3936.
- [Google Scholar]
- Integrating evolutionary and functional approaches to infer adaptation at specific loci. Evolution. 2010;64(9):2489-2509.
- [Google Scholar]
- Mutational mapping of the catalytic activities of human tyrosinase. J. Biol. Chem.. 1992;267(33):23707-23712.
- [Google Scholar]
- COACH-D: improved protein–ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res.. 2018;46(W1):W438-W442.
- [Google Scholar]
- The genetic architecture of adaptation under migration–selection balance. Evolution. 2011;65(7):1897-1911.
- [Google Scholar]
- Multidimensional epistasis and fitness landscapes in enzyme evolution. Biochem. J. 2012;445(1):39-46.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102772.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







