Translate this page into:
Comparative evaluation of the performance of the PCR assays commonly used for the determination of sex in avian species
⁎Corresponding author. ikonomop@aua.gr (John Ikonomopoulos)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Due to the significance of accurate genetic determination of sex of birds, the aim of this study was to assess comparatively the performance of the methods of Griffiths et al. (1998) and Fridolfsson and Ellegren (1999), and to clarify the specifications that could improve their reliable application in practice. The reference material consisted of samples (n = 354) of whole blood, oral swabs and feathers, collected from 118 individuals of known sex and 15 avian species, including several that have not been tested before. The investigation was conducted with respect to ISO17025, including assessment of DNA quality and investigation for PCR inhibitors, use of controls for all the stages of the analysis, and the assessment of method accordance.
The determination of sex using the PCR assay developed by Griffiths et al. produced the expected results for 100 of the individuals and all the samples (n = 300) that were collected from them. The respective values recorded for the assay developed by Fridolfsson and Ellegren were 98 and 294. Method accordance was 100% for both assays under study. Overall relative specificity of the method of Griffiths et al. was 97.1 ± 3.25%, corresponding to 94.2 ± 6.34% and 100% respectively, for male and female individuals. Relative specificity of the method of Fridolfsson and Ellegren was 83.1 ± 6.78%, corresponding to 100% and 65.5 ± 12.23%, respectively, for male and female individuals. Despite the fact that the birds studied were of known sex, in terms of phenotype and reproductive record, inconsistent results were recorded with both methods in some species examined. The difference between the relative specificities was statistically significance in connection with the biological samples in general (p < 0.01) and with the female individuals (p < 0.01). In conclusion, the method of Griffiths et al. proved in most cases, more reliable for the determination of sex of birds. However when testing for the first time a species of birds, a preliminary study that would include method calibration and application on control samples is highly recommended before selecting the method of choice.
Keywords
Avian sex
Bird sexing
PCR
Comparative evaluation
Avian species
CHD gene
1 Introduction
Definite determination of sex in avian species can be a challenging task, especially in the absence of sexual dimorphism or when morphometric and behavioral traits are ambiguous. In these cases laparoscopy can be used, but it requires general anesthesia and is usually conducted only in large birds. Expectedly the polymerase chain reaction (PCR) was put to use from early on, in order to solve this problem. Within this context, sex can be determined based on the outcome of PCR amplification targeting the CHD gene, which encodes the chromodomain helicase DNA binding protein (Ellegren, 1996; Griffiths et al., 1996) and is located in both chromosomes of almost all species of birds, except ratites (Ellegren, 1996; Fridolfsson and Ellegren, 1999; Griffiths et al., 1998).
The PCR assays more commonly used for the determination of sex in birds are those reported by Fridolfsson and Ellegren (1999), Griffiths et al. (1998) and Kahn et al. (1998). However the latter, which spams the same intron with that of Griffiths et al., has been reported to be significantly less specific and was therefore not included in this investigation (Jensen et al., 2003).
The oligonucleotide primers incorporated in the assays of Griffiths et al., and Fridolfsson and Ellegren are designed to amplify specific fragments of the CHD gene introns belonging to the Z and W sex chromosomes. Given that males are homogametic (ZZ), this approach results to a single amplification product corresponding to the Z chromosome of individuals of the specific sex, and two amplification products in heterogametic (ZW) females. In practice however, sex determination using the specific PCR assays proved in several cases inaccurate, which in some cases was associated with polymorphisms in the CHD-Z gene leading to a heterozygote genotype (ZZ’) of males (Dawson et al., 2001; Lee et al., 2002; Robertson and Gemmell, 2006). Furthermore, the method proposed by Fridolfsson and Ellegren has been reported to fail to produce any amplification products for specific species of birds (Çakmak et al., 2017) or to produce a single DNA product in both sexes (Dubiec and Zagalska-Neubauer, 2006; Ong and Vellayan, 2008). Similarly, false results were recorded with the method of Griffiths et al. for certain avian species, due to unspecified reasons (Çakmak et al., 2017; McDonald and Griffith, 2011; Vucicevic et al., 2013) or the incomplete separation of the amplification products (Cheng et al., 2006; Dawson et al., 2001; Han et al., 2009).
The comparative assessment of the performance of the two PCR assays commonly used for the determination of sex of birds was the subject of seven more studies (Table 1). In four of these studies, the assessment relied on only one type of sample (Dawson et al., 2001; Leppert et al., 2006; Ong and Vellayan, 2008; Vucicevic et al., 2013) whereas the rest (Çakmak et al., 2017; Khaerunnisa et al., 2013; Sulandari and Zein, 2012), referred to a comparative analysis of samples of blood and feathers. Oral swabs have never been included in this type of investigation. Three of the relevant studies (Dawson et al., 2001; Khaerunnisa et al., 2013; Leppert et al., 2006) were performed on a small number of avian species (n ≤ 7) whereas the relevant number analysed within the context of the rest, varied from 32 (Ong and Vellayan, 2008) to 77 (Çakmak et al., 2017).
Reference
Number of species
Number of individuals
Type(s) of samples
Conclusion
Dawson et al. (2001)
4
768
Blood
G and F should be combined after pre-assessment, depending on the species of birds
Leppert et al. (2006)
4
701
Blood
G and F should be combined after pre-assessment, depending on the species of birds
Ong and Vellayan (2008)
32
72
Feathers
F is recommended, except for species of the Anatidae that should be tested with G
Sulandari and Zein (2012)
62
339
Blood/Feathers
F
Khaerunnisa et al. (2013)
7
21
Blood/Feathers
G and F should be combined depending on the species of birds
Vucicevic et al. (2013)
58
284
Feathers
F after chemical/thermal calibration, depending on the species of birds
Çakmak et al. (2017)
77
230
Blood/Feathers
G and F should be combined after pre-assessment, depending on the species of birds
Considering the significance of accurate genetic determination of sex of birds and the fact that the outcome of the main PCR assays used today for this purpose can be influenced by several factors, the aim of this study was to assess comparatively the performance of the methods of Griffiths et al. and Fridolfsson and Ellegren, and to clarify the specifications that could improve their reliable application in practice. Within this context, the specific issue was addressed for the first time with respect to ISO17025 requirements and with regards to many avian species, including several that have not been tested before, and all the types of samples (blood, feathers, oral swabs) commonly used for sex determination.
2 Material and methods
For reasons of reliability, this investigation was designed and conducted with respect to the provisions foreseen within the context of the quality standard ISO17025. These provisions were applied in connection with the structure of the laboratory with reference to levels of safety, the accreditation of equipment, sample flow starting from the assessment of sample quality upon its arrival to the laboratory until the conclusion of the analysis, the assessment of DNA quality including the investigation for PCR inhibitors, the use of control samples for all the stages of the analysis i.e. DNA isolation, PCR, submerged gel electrophoresis, and the assessment of method accordance.
2.1 Biological samples
The investigation presented here was conducted on samples (n = 354) of whole blood (n = 118) collected from the right jugular or the ulnar vein, oral swabs (n = 118), and feathers (n = 118) plucked from the abdominal region (Table 2). These samples were collected during routine testing from 118 adult individuals, the sex of which was determined based on mating behavior and ovulation. The sample population consisted of captive live birds admitted to veterinary clinics or hosted in collaborating pet shops in the general geographic area of Athens, Greece, and the Attica Zoological Park.
Species
Number of individuals
Male
Female
Total
Agapornis roseicollis
12
6
6
36
Alopochen aegyptiacus
10
3
7
30
Anas bahamensis
2
2
0
6
Aratinga solstitialis
7
3
4
21
Calocitta colliei
7
3
4
21
Chalcopsitta duivenbodei
7
3
4
21
Dacelo leachii
8
4
4
24
Entomyzon cyanotis
2
2
0
6
Gallus gallus domesticus
12
6
6
36
Myiopsitta monachus
7
4
3
21
Psittacula eupatria
6
4
2
18
Sarcops calvus
9
4
5
27
Serinus canaria
10
5
5
30
Sturnia pagodarum
11
5
6
33
Taeniopygia guttata
8
6
2
24
Total
118
60
58
354
2.2 DNA isolation
DNA isolation was performed from all types of samples within 12 h after their collection, using the Nucleospin Tissue Kit (Macherey-Nagel GmbH & Co. KG, Germany). Based on the instructions provided by the manufacturer, samples of high keratin content and/or viscosity such as avian whole blood and feathers were incubated with proteinase K overnight. Feathers were previously homogenized and grinded with liquid nitrogen using mortar and pestle. Isolated DNA was stored at −20 °C until use.
The quality of the DNA isolated was assessed with regards to purity and integrity by submerged gel electrophoresis followed by image analysis using a Bio-Rad ChemiDoc XRS + Molecular Imager (Bio-Rad Laboratories Inc., U.S.A.), and by spectrophotometry at 260/280 nm, using a NanoDrop 8000 Spectrophotometer (Thermo Fisher Scientific Inc., U.S.A.). The presence of inhibitors in the samples was assessed by a PCR assay targeting a housekeeping gene (cytochrome), (Abdulmawjood et al., 2003).
DNA isolation and PCR analysis were conducted in compliance with ISO17025 accreditation requirements.
2.3 PCR analysis
The PCR assays were prepared using an Invitrogen Taq DNA Polymerase kit protocol (Thermo Fisher Scientific Inc., U.S.A.), and consisted of 2.5 μl of PCR buffer (1X), 0.15 μl of Taq polymerase (0.75 U), 0.5 μl of dNTPs (0.2 mM), 1 μl of MgCl2 (1.5 mM) and 3 μl of template DNA. Reaction mixtures were completed with 0.5 μl (0.5 μΜ) of each of the primers previously reported, and PCR grade water to a final volume of 25 μl; the thermal profiles of both PCR assays were those originally suggested by Griffiths et al. (1998) and Fridolfsson and Ellegren (1999) with minor modifications (Table 3).
Reference
Primers
Nucleotide Sequence1
Thermal profile
Griffiths et al. (1998)
P2
5′-TCTGCATCGCTAAATCCTTT-3′
Initial step of 94 °C for 2 min. 30 cycles of 94 °C for 30 s, 50 °C2 for 45 s, 72 °C for 45 s. Final step of 72 °C for 5 min.
P8
5′-CTCCCAAGGATGAGRAAYTG-3′
Fridolfsson and Ellegren (1999)
2550F
5′-GTTACTGATTCGTCTACGAGA-3′
Initial step of 94 °C for 2 min. 10 cycles of 94 °C for 30 s, 60 °C-51 °C (−1 °C/cycle) for 30 s, 72 °C for 30 s. 35 cycles of 94 °C for 30 s, 50 °C for 30 s, 72 °C for 30 s. Final step of 72 °C for 5 min.
2718R
5′-ATTGAAATGATCCAGTGCTTG-3′
PCR was performed in an Applied Biosystems Verity 96-well Thermal Cycler (Thermo Fisher Scientific Inc., U.S.A.). PCR products were analyzed by submerged electrophoresis using 3.5% agarose gels stained with ethidium bromide (0.5 μg/mL), and visualized using a Bio-Rad ChemiDoc XRS + Molecular Imager (Bio-Rad Laboratories Inc., U.S.A.). For confirmation of the specificity of the amplification process, approximately 20% of the PCR products were submitted to sequence analysis, which was conducted on both strands using the Applied Biosystems BigDye Terminator Cycle Sequencing Kit and PRISM 377 DNA Sequencer (Thermo Fisher Scientific Inc., U.S.A.). Results were analyzed and compared to deposited sequences in the GenBank database using the Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI).
For the assessment of method accordance, both PCR assays under study were used for the analysis of six (n = 6) samples of DNA isolated from blood that was collected from 3 male, and from an equal number of female individuals of Gallus gallus domesticus. The specific set of samples was divided in five (n = 5) aliquots that were marked with coded numbers and were tested five (n = 5) times (once a day); the analysis was conducted by the same operator who was not aware of the true identity of the samples, using the same equipment of the same laboratory. Accordance was recorded for both PCR assays as the percentage of correct results over the total number of samples, using the following formula (Langton et al., 2002; Scotter et al., 2001): where CM the total number of correct results recorded with regards to male individuals (max number = 15 i.e. 3 samples of male individuals tested 5 times); CF, the relevant number corresponding to females; N, the total number of samples (N = 30).
2.4 Statistical analysis
Relative specificities are presented as 95% confidence intervals of proportions. Differences in sex determination between the method of Griffiths et al. and the method of Fridolfsson and Ellegren were assessed comparing the corresponding relative specificities with the non-parametric McNemar test, which is used for comparing two proportions when the data are paired. Statistical analysis was performed using the statistical package SPSS (version 16.0); the significance level was set at 5%.
3 Results
The determination of sex using the PCR assay developed by Griffiths et al. produced the expected results for 100 of the individuals and all the samples (n = 300) that were collected from them. The respective values recorded for the PCR assay developed by Fridolfsson and Ellegren were 98 and 294, with regards to individuals and samples (Table 4). The outcome of the PCR assay targeting cytochrome did not provide indications of the presence of PCR inhibitors for any of the samples tested, and the result of the sequence analysis of the PCR amplification products was confirmatory of their specificity. Method accordance was 100% for both assays under study.
Species
Number of Individuals
PCR assay
Method(s) recommended
G
F
Male
Female
Male
Female
Male
Female
G
F
Agapornis roseicollis
6
6
6
6
6
6
+
+
Alopochen aegyptiacus
3
7
3
7
3
0*
+
Anas bahamensis
2
0
2
0
2
0
+
+
Aratinga solstitialis
3
4
3
4
3
4
+
+
Calocitta colliei
3
4
3
4
3
4
+
+
Chalcopsitta duivenbodei
3
4
3
4
3
4
+
+
Dacelo leachii
4
4
4
4
4
4
+
+
Entomyzon cyanotis
2
0
−
−
2
0
+
Gallus gallus domesticus
6
6
6
6
6
6
+
+
Myiopsitta monachus
4
3
4
3
4
3
+
+
Psittacula eupatria
4
2
4
2
4
0*
+
Sarcops calvus
4
5
4
5
4
0*
+
Serinus canaria
5
5
2
−
5
5
+
Sturnia pagodarum
5
6
5
6
5
0*
+
Taeniopygia guttata
6
2
−
−
6
2
+
Overall relative specificity of the method of Griffiths et al., with a 95% confidence interval was 84.7 ± 6.49% (100 of 118 individuals correctly identified), corresponding to 81.7 ± 9.79% and 87.9 ± 8.38% respectively, for male (49 of the 60 correctly identified) and female (51 of the 58 correctly identified) individuals. Fifteen of the 118 individuals did not react with the method of Griffiths et al., eight of which corresponded to male, and seven to female birds (Fig. 1). After excluding these samples, the overall relative specificity of the method of Griffiths et al., with a 95% confidence interval was 97.1 ± 3.25% (100 of 103, for which sex was correctly determined), corresponding to 94.2 ± 6.34% and 100% respectively, for male (49 of the 52, for which sex was correctly determined) and female (51 of the 51, for which sex was correctly determined) individuals. Relative specificity of the method of Fridolfsson and Ellegren with a 95% confidence interval was 83.1 ± 6.78% (98 of 118 individuals correctly identified), corresponding to 100% and 65.5 ± 12.23%, respectively, for male (60 of the 60, for which sex was correctly determined) and female (38 of the 58, for which sex was correctly determined correctly identified) individuals (Fig. 2), (Table 4).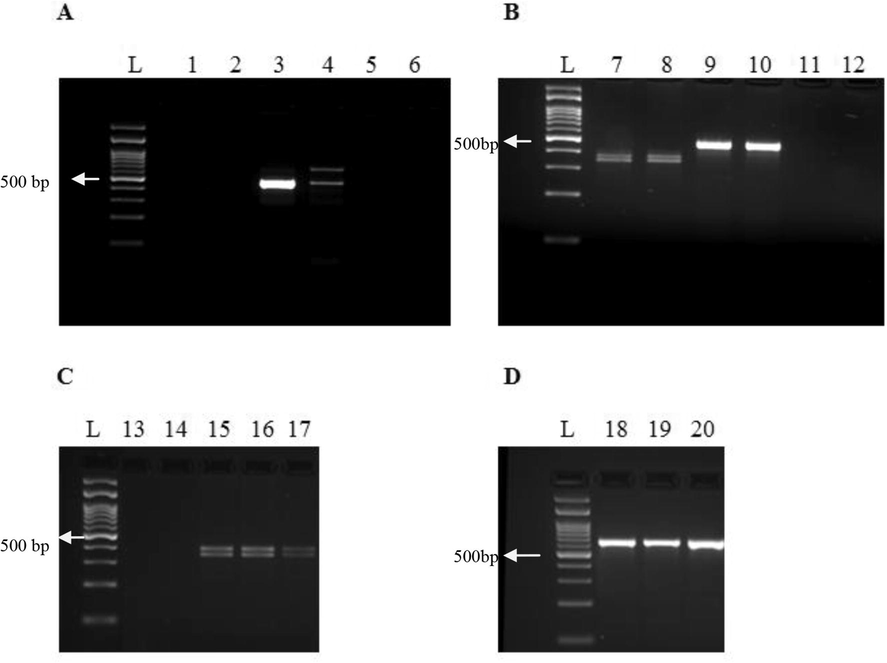
Representative submerged gel electrophoresis of the PCR amplification products of the methods of Griffiths et al. and Fridolfsson and Ellegren corresponding to samples of blood indicating inconsistency: amplification failure (A) and false reactions (B–D). Lanes L: 100 bp DNA ladder (Nippon Genetics Europe GmbH, Germany). Lanes 5, 6, 11, 12, 13, 14: Negative controls for DNA isolation (lanes 5, 11, 13) and PCR (lanes 6, 12, 14). Lanes 1–4: PCR amplification products of the method of Griffiths et al. (lanes 1, 2) and Fridolfsson and Ellegren (lanes 3, 4), from a male (lanes 1, 3) and a female (lanes 2, 4) individual of Taeniopyrgia guttata, indicating amplification failure for both individuals with regards to the method of Griffiths et al. (lanes 1, 2). Lanes 7–10: PCR amplification products of the method of Griffiths et al. (lanes 7, 8) and Fridolfsson and Ellegren (lanes 9, 10), from two female individuals of Alopochen aegyptiaca indicating in both cases false reaction, i.e. females reacting as male with regards to the method of Fridolfsson and Ellegren (lanes 9, 10). Lanes 15–20: PCR amplification products of the method of Griffiths et al. (lanes 15–17) and Fridolfsson and Ellegren (lanes 18–20), from three female individuals of Sarcops calvus indicating false reaction i.e. females reacting as male with regards to the method of Fridolfsson and Ellegren (lanes 18–20).
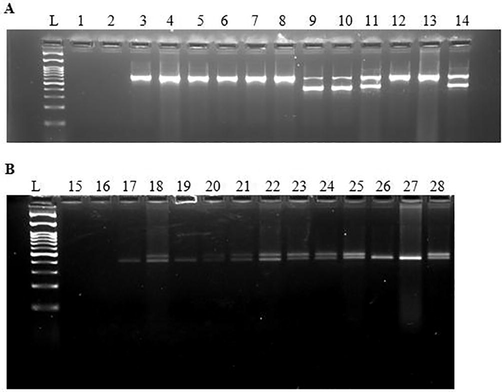
Representative submerged gel electrophoresis of the PCR amplification products of the methods of Fridolfsson and Ellegren (A) and Griffiths et al. (B), corresponding to samples of blood collected from avian species not tested before; specific results indicate false reactions corresponding to female individuals (lanes 18, 20–22) of Sturnia pagodarum reacting as male (lanes 4, 6–8). Lanes L: 100 bp DNA ladder (Nippon Genetics Europe GmbH, Germany). Lanes 1, 2, 15, 16: Negative controls for DNA isolation (lanes 1, 15), and PCR (lanes 2, 16). Lanes 3, 5, 17, 19: Two male individuals of Sturnia pagodarum. Lanes 4, 6, 7, 8, 18, 20, 21, 22: Four female individuals of Sturnia pagodarum. Lanes 9, 10, 23, 24: Two female individuals of Dacelo leachii. Lanes 11, 14, 25, 28: Two female individuals (n = 2) of Calocitta colliei. Lanes 12, 13, 26, 27: Two male individuals (n = 2) of Calocitta colliei.
Based on the non-parametric McNemar test, the difference between the relative specificity recorded with regards to each of the PCR protocols under evaluation is statistically significant (p < 0.01). The method of Griffiths et al., failed to determine the sex in a proportion that corresponds to 2.9 ± 3.25% of the samples; the relevant proportion recorded with regards to the method of Fridolfsson and Ellegren was 17.0 ± 6.78%. Comparing the relative specificities recorded with regards to female individuals indicated that the difference between the two methods is also statistically significant (p < 0.01), and the relevant failure rates were 0% and 34.5 ± 12.23% for the methods of Griffiths et al., and Fridolfsson and Ellegren, respectively. The outcome of the specific investigation was not statically significant with regards to male individuals. The corresponding failure rates for males were estimated to 5.8 ± 6.34% and 0% for the methods of Griffiths et al., and Fridolfsson and Ellegren, respectively (p > 0.05).
4 Discussion
With regards to the avian species included in this study Anas bahamensis, Calocitta colliei, Dacelo leachii, Entomyzon cyanotis, Sarcops calvus and Sturnia pagodarum have not been tested before. Agapornis roseicollis, Serinus canaria and Taeniopygia guttata were tested very recently for the first time by Çakmak et al. (2017), who used both methods under study here on a limited number of individuals [Agapornis roseicollis (1 male and 1 female), Serinus cararia (1 male) and Taeniopygia guttata (1 male and 1 female)], whereas Chalcopsitta duivenbodei, Myiopsitta monachus and Psittacula eupatria were previously tested using only one of the methods under study (Bosnjak et al., 2013; Gábor et al., 2014; Sulandari and Zein, 2012).
In terms of measures of quality assurance, the assessment of DNA quality and the confirmation of the specificity of the amplification products were included only in the investigations that were conducted respectively by Khaerunnisa et al. (2013), Leppert et al. (2006) and Çakmak et al. (2017). Notably, the presence of PCR inhibitors has not been investigated in any of the relevant studies conducted in the past, in spite indications that inhibition may have a negative impact on only one of the amplicons produced by each of the PCR assays, which can be misleading given that PCR is not completely inhibited (Vucicevic et al., 2013). The structure of the laboratory that can influence the level of false positive reactions associated with the carry-over effect (contamination of amplicons from previous reaction), the use of control samples incorporated in all the stages of the analysis and the accreditation of the equipment used to conduct it, are also parameters that have not been considered in the past. Overall it can be stated that the evidences provided here can address effectively some of the gaps of the relevant research and improve the reliable application of PCR used for the determination of sex of birds, in practice.
The methods under investigation produced consistent results on all the types of samples that were tested i.e. there was no test-result variation within each method, associated with the type of sample that was used to conduct the analysis. The efficiency of the amplification assessed on the basis of the intensity of the signal generated in gel electrophoresis by the PCR products was higher in decreasing order, for whole blood, oral swabs and feathers, which is in agreement with the findings of other (Harvey et al., 2006; Sacchi et al., 2004). In this respect it is noteworthy to draw attention to the comparative advantage of oral swabs over blood and feathers for the determination of sex in birds, since their collection is very simple, non-invasive and of low cost. At the same time oral swabs are processed more easily compared to feathers for DNA isolation and they produce more DNA, which is less likely to contain PCR inhibitors. Band size variations were more easily assessed for PCR products of blood samples, especially when using the method of Fridolfsson and Ellegren that produces DNA amplicons the size of which results to more distinct bands in submerged agarose gel electrophoresis compared to those of Griffiths et al. (150–250 base pairs versus 10–80 base pairs). Considering the size of the amplification products, the density of the agarose gel acquires critical significance in terms of being able to differentiate with certainty the two bands produced with both PCR assays for female individuals, from the single band corresponding to the males. Indeed as suggested by Griffiths et al., using agarose gels of 3% w:v proved adequate in this study for the differentiation of the PCR products, with the exception of Agapornis roseicollis, Alopochen aegyptiacus, Aratinga solstitialis and Psittacula eupatria. In the specific cases, distinguishing the amplicons produced by the method of Griffiths et al. was possible using an agarose gel of no less than 3.5% w:v. Therefore the density of the agarose gels used to conduct this investigation was in all cases 3.5% w:v, which was considered preferable in terms of the reliability of the assessment, especially when a specific species of birds is being analysed with the two PCR assays under study, for the first time. Alternatively, capillary or pulsed field gel electrophoresis can be used to improve separation of PCR amplicons of similar size, though their application in practice is not easy for reasons of complexity and cost (Çakmak et al., 2017).
The methods under investigation did not produce consistent results with regards to sex determination of the following species: Alopochen aegyptiacus, Psittacula eupatria, Sarcops calvus and Sturnia pagodarum. In all cases, the inconsistency was recorded in connection with female individuals that reacted as male using the method of Fridolfsson and Ellegren, which implies that its cause could be preferential amplification (Table 4). It is noteworthy that for the specific species of birds, the issue of false determination of sex was not resolved by increasing the density of the agarose gel to 3.5%, which as mentioned above proved efficient in connection to the method of Griffiths et al. With regards specifically to sex determination of females, false results were also reported previously for one individual of Alopochen aegyptiacus (Vucicevic et al., 2013) and Taeniopygia guttata (Çakmak et al., 2017) reacting in both cases as male with the method of Griffiths et al., though in the relevant studies the density of the agarose gel that was used for the electrophoretic separation of the amplification products was 2% and 3% respectively.
Failure to produce any amplification products was recorded consistently using the method of Griffiths et al. on Agapornis roseicollis, Entomyzon cyanotis, Serinus canaria and Taeniopygia guttata, in spite the negative outcome of the PCR assay conducted for the detection of PCR inhibitors (Table 4). The failure of PCR amplification was resolved only with regards to Agapornis roseicollis, by increasing the annealing temperature from 50 °C to 51 °C, as suggested by Griffiths et al., 1998, which however did not prove effective for the rest of the avian species reported above (Entomyzon cyanotis and Taeniopygia guttata). Amplification failure was also reported by Çakmak et al. (2017) with regards to the method of Fridolfsson and Ellegren, in connection with Alopochen aegyptiacus (2 male individuals) and Serinus canaria (1 male individual), which however was not confirmed by our findings. It has to be noted that the biological samples included in this investigation was collected from individuals with typical phenotypic and behavioral sex-characteristics. Considering that post-mortem examination of the individuals included in this study was not possible or ethically justified, we provide evidences from wild bird species that were examined by our team having been found dead in their natural habitat, indicating that phenotypic and genotypic sex in avian species might differ significantly. In fact cases of anatomical genetic abnormalities, such as hermaphrodism, which might generate inconsistent genetic determination results for reasons not associated with the reliability of the methods under investigation, were recorded (Fig. 3). Therefore in assessing the inconsistent results referred to above, it can be reported that for all those cases that PCR optimization does not resolve the problem, its cause could be associated, as reported by others, with genetic variation at the species level (Çakmak et al., 2017; Dawson et al., 2001; Dubiec and Zagalska-Neubauer, 2006; Lee et al., 2002; Ong and Vellayan, 2008; Robertson and Gemmell, 2006), or as indicated above, with anatomical deviations of certain individuals. In any case, the selection of the PCR method that will be used to determine sex of birds must be done with consideration to the specific avian species (Table 4).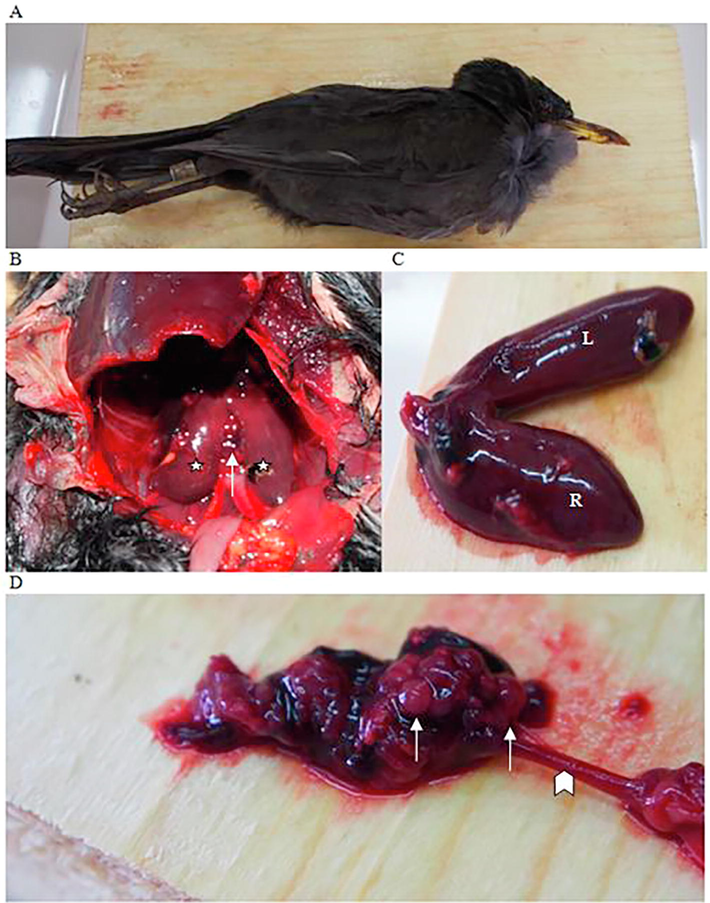
Case of anatomical genetic abnormality in adult common blackbird (Turdus merula). Α. Common blackbird is a species with evident sexual dimorphism; the phenotype of this bird was characteristic of a male. B. Following dissection, removal of the liver and the gastrointestinal tract, both testes (white stars) and an ovary (white arrow) were observed; indicative of haermaphroditism. C. The testes, differed from normal in shape; they were rather elongated and not bean shaped, and the left testis (L) was longer than the right (R), in consistency with the normal asymmetry present in many avian species. Furthermore, both testes were attached to each other at their dorsal apex were not connected with seminiferous tubules. D. The ovary consisted of many oocytes (white arrows), resided in the left part of the body cavity (as in most female avian species), and was attached to a rudimentary oviduct (white arrowhead) leading to the cloaca. There is no evidence of anatomically defined parts such as the magnum or the uterus of the oviduct; yet the most caudal parts appear as intertwined tubes dorsally to the cloaca. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In summarizing with regards to the avian species not tested before, molecular determination of sex produced consistent results for Anas bahamensis, Calocitta colliei and Dacelo leachii using both methods; for Entomyzon cyanotis the method of Griffiths et al. failed to produce any amplification product, whereas for Sarcops calvus and Sturnia pagodarum the method of Fridolfsson and Ellegren produced false results (females reacting as male). With regards to the avian species tested before by only one of the methods under study i.e. Psittacula eupatria, Myiopsitta monachus and Chalcopsitta duivenbodei, sex was determined correctly using both methods, which is in agreement with the reports of Gábor et al. (2014) and Sulandari and Zein (2012), who however tested only the method of Griffiths et al., in connection respectively with Myiopsitta monachus and Chalcopsitta duivenbodei. With regards to Psittacula eupatria, sex was determined correctly using the method of Griffiths et al., but not that of Fridolfsson and Ellegren, which produced false results (all females reacting as male). The only relevant report available in the literature indicates the exact opposite, i.e. sex determined correctly with the method of Fridolfsson and Ellegren, which however referred to only one female individual (Bosnjak et al., 2013).
5 Conclusion
Determination of sex of birds using the methods of Griffiths et al. (1998) and Fridolfsson and Ellegren (1999) is simple, but can be influenced by several factors. The method of Griffiths et al., proved more reliable in connection with most avian species included in this study, except Entomyzon cyanotis, Taeniopygia guttata and Serinus canaria. When testing for the first time a species of birds, a preliminary study that would include method calibration and application on control samples is highly recommended before selecting the method of choice.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Conflicts of interest
No conflict of interest exists.
References
- Toward an international standard for PCR-based detection of Escherichia coli O157. J. Microbiol. Methods. 2003;55:775-786.
- [Google Scholar]
- Feasibility of non-invasive molecular method for sexing of parrots. Pakistan J. Zool.. 2013;45(3):715-720.
- [Google Scholar]
- Comparison of three different primer sets for sexing birds. J. Vet. Diagnostic Invest.. 2017;29:59-63.
- [Google Scholar]
- A critique of avian CHD-based molecular sexing protocols illustrated by a Z-chromosome polymorphism detected in auklets. Mol. Ecol. Notes. 2001;1:201-204.
- [Google Scholar]
- First gene on the avian W chromosome (CHD) provides a tag for universal sexing of non-ratite birds. Proc. R. Soc. B Biol. Sci.. 1996;263:1635-1641.
- [Google Scholar]
- A simple and universal method for molecular sexing of non-ratite birds. J. Avian Biol.. 1999;30:116-121.
- [Google Scholar]
- Sex determination of superorder neognathae (class Aves) by molecular genetics methods. J. Anim. Sci. Biotechnol.. 2014;47:69-72.
- [Google Scholar]
- Sex identification in birds using two CHD genes. Proc. R. Soc. B Biol. Sci.. 1996;263:1251-1256.
- [Google Scholar]
- Sex determination by PCR-RFLP in the oriental white stork Ciconia boyciana. Zool. Stud.. 2009;48:619-624.
- [Google Scholar]
- A comparison of plucked feathers versus blood samples as DNA sources for molecular sexing. J. Field Ornithol.. 2006;77(2):136-140.
- [Google Scholar]
- Conditions for rapid sex determination in 47 avian species by PCR of genomic DNA from blood, shell-membrane blood vessels, and feathers. Zoo Biol.. 2003;22:561-571.
- [Google Scholar]
- Chromosome-Specific intron size differences in the avian CHD gene provide an efficient method for sex identification in birds. Auk. 1998;115:1074-1078.
- [Google Scholar]
- Avian sex determination based on Chromo Helicase DNA-binding (CHD) genes using Polymerase Chain Reaction (PCR) Media Peternak.. 2013;36:85-90.
- [Google Scholar]
- Analysing collaborative trials for qualitative microbiological methods: Accordance and concordance. Int. J. Food Microbiol.. 2002;79:175-181.
- [Google Scholar]
- Sex and death: CHD1Z associated with high mortality in moorhens. Evolution. 2002;56:2548-2553.
- [Google Scholar]
- Sex identification in four owl species from Idaho: DNA and morphometrics. J. Raptor Res.. 2006;40:291-294.
- [Google Scholar]
- To pluck or not to pluck: the hidden ethical and scientific costs of relying on feathers as a primary source of DNA. J. Avian Biol.. 2011;42:197-203.
- [Google Scholar]
- An evaluation of CHD-Specific primer sets for sex typing of birds from feathers. Zoo Biol.. 2008;27:62-69.
- [Google Scholar]
- PCR-based sexing in conservation biology: Wrong answers from an accurate methodology? Conserv. Genet.. 2006;7:267-271.
- [Google Scholar]
- A non-invasive test for sex identification in Short-toed Eagle (Circaetus gallicus) Mol. Cell. Probes. 2004;18:193-196.
- [Google Scholar]
- Validation of ISO method 11290 Part 1—Detection of Listeria monocytogenes in foods. Int. J. Food Microbiol.. 2001;64:295-306.
- [Google Scholar]
- Application of two molecular sexing methods for Indonesian bird species: implication for captive breeding programs in Indonesia. HAYATI J. Biosci.. 2012;19:183-190.
- [Google Scholar]
- Sex determination in 58 bird species and evaluation of CHD gene as a universal molecular marker in bird sexing. Zoo Biol.. 2013;32:269-276.
- [Google Scholar]







