Translate this page into:
Broad-spectrum antibacterial and antibiofilm activity of biogenic silver nanoparticles synthesized from leaf extract of Phyllanthus niruri
⁎Corresponding author. fhusain@ksu.edu.sa (Fohad Mabood Husain)
⁎⁎Co-Corresponding author. mohammeds@agu.edu.bh (Mohammad Shahid),
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Multidrug resistance (MDR) among pathogenic bacteria is a global health concern as it has rendered the current antibiotic therapy ineffective. A recent and rapidly developing research area in the biomedical sector is the use of biogenic nanoparticles as antibacterial agents. In the current investigation, aqueous leaf extract of Phyllanthus niruri (Pn-AgNPs) were employed for the biogenic synthesis of silver nanoparticles, further characterized by SEM, EDX, FTIR, TEM, UV–Vis and XRD techniques. Electron microscopy images of the green-synthesized Pn-AgNPs showed them as evenly dispersed spherical particles having an average size of about ∼ 20 nm. The Pn-AgNPs were further tested for potential antibacterial activity against isolated drug-resistant pathogenic bacteria such as Burkholderia cepacia complex, carbapenem-resistant Enterobacterales (CRE), Citrobacter freundii, Enterococcus faecalis, Escherichia coli, Enterobacter clocae, MRSA, Salmonella typhi, Streptococcus spp, and vancomycin-resistant Enterococci (VRE).The Pn-AgNPs significantly inhibited growth all of the studied bacterial isolates, including antibiotic-resistant isolates such as MRSA, VRE, and CRE. Electron micrographs clearly showed the enhanced penetration of AgNPs leading to disruption of cell membrane causing death of the cell. Further, Pn-AgNPs dramatically reduced the ability of P. aeruginosa, Bacillus cereus, E. coli, and S. aureus to form biofilms. Leakage of protein from bacterial cells could be main reason for the effective bactericidal action of synthesized AgNPs. This antibacterial and antibiofilm potential of Phyllanthus niruri biosynthesized AgNPs against clinically significant drug-resistant bacteria is impressive and can be developed as a novel antimicrobial agent to combat the threat of drug-resistant bacterial infections.
Keywords
Green Synthesis
Phyllanthus niruri
Silver nanoparticles
Antibacterial
Drug-resistant bacteria
Biofilm inhibition
1 Introduction
Emergence and rapid spread of multidrug resistance (MDR) among pathogenic bacteria is a big issue globally as it is a threat to the public health and incurs huge financial losses (Maheshwari et al., 2016). The unabated and uncontrolled use of the antibiotics is the one of the major causes for the increasing resistance among pathogenic bacteria (Qais et al., 2019). Moreover, the development of new antibiotics is a slow, time taking process that requires tremendous labour and huge capital investment (Lee et al., 2019). Bacteria such as Acinetobacter baumanii, ESβL-producing E. coli, MβL-producing Pseudomonas sp., vancomycin resistance Enterococcus (VRE), Methicillin resistant S. aureus (MRSA) are the most common persistent infection causing drug-resistant pathogens (Alekshun and Levy, 2007). Emergence of these MDR bacteria have rendered the current antibiotic therapy ineffective and there is an urgent need to develop or discover antibacterial agents with broad-spectrum activity.
Nanotechnology is a discipline that focuses on the creation and manipulation of nanoparticles that can be used in a variety of contexts, including biomedical applications. The growing field of nano-scale applications has currently transformed the entire world (Prasad et al., 2017). The metallic nanomaterials/nanoparticles possess distinct, genuine biochemical and physical characteristics that differ from their counterpart bulk materials. Nanoparticles of silver, gold, zinc, copper etc has been of much interest in the past few years for various biomedical applications (Rafique et al., 2017). Silver nanoparticles (AgNPs) have attracted most attention and are reported to demonstrate antibacterial, antifungal, antiviral, antioxidant and anti-inflammatory properties (Khandel et al., 2018). Traditionally AgNPs of desired shapes and sizes were synthesized using physical and chemical methods to produce nanoparticles but the use of highly toxic chemicals, high pressure and energy along with the formation of toxic end products has limited the use of such protocols and compelled the researchers to produce biologically compatible nanoparticles (Bala and Rani, 2020). Therefore, the stress has been to produce metallic nanoparticles using biological methods that are affordable, efficient, eco-friendly, and beneficial in the global effort to eliminate hazardous by-products (Burdușel et al., 2018).
The biological synthesis of metal nanoparticles using plant extract has gained lot of focus in the recent years as phytomediated fabrication of NPs is achieved at room temperature, it is cost effective and rapid as compared to the production of NPs using microbial cultures (Bahrulolum et al., 2021). Plant materials help in reduction of silver ion, act as capping and stabilising agents due to the presence of various metabolites as well as complex biomolecules (Urnukhsaikhan et al., 2021).
Phyllanthus niruri Linn (family: Euphorbiaceae) is a common herb of India. Locally called “Bhoomi aavla”, it is a popular medicinal plant whose leaves have been used historically in Ayurveda as a medicine to treat various gastroenteritis issues. It is also reported for anti-inflammatory, Hypoglycaemic, antiviral, antioxidant, hepatoprotective, and anti-calculus formation properties (Lee et al., 2016). The plant is rich in secondary metabolites such as acidic diterpenes, arabinogalactan, alkaloids, flavonoids, lignans, terpenes, and tannins (Shilpa et al., 2018). Its phytoconstituents have the capacity to reduce and stabilize the silver nanoparticles.
Considering the well documented medicinal value of Phyllanthus niruri and limited work on the silver nanoparticle synthesis from this plant, leaves of this plant were used to produce green AgNPs. Further, since finding novel alternatives to currently used antibiotics has become a critical challenge, the synthesized AgNPs were subjected to detailed characterization and assessed for antibacterial potential against clinically important drug resistant strains of both gram-positive and gram-negative bacteria.
2 Materials and methods
2.1 Extract preparation
The Phyllanthus niruri leaves were gathered from the Aligarh Muslim University's Botany Department in Aligarh, India. In the shade, plant leaves were dried. Fine powder from the dried plant leaves was prepared, and 10 gm of powder was stirred into 100 mL of filtered sterilized water. Subsequently, aqueous extract was filtered using 0.22 mm filter paper and Whatman No. 1 filter paper (Maidstone, UK) (Millipore). The heavy biomaterials were extracted in an aqueous state that could be stored at 4 °C for subsequent usage after centrifugation at 10000 rpm for 5 min (Bose and Chatterjee, 2015).
2.2 Synthesis of silver nanoparticles (PA-AgNPs)
In a nutshell, 10 mL of aqueous extract was combined with 100 mL of a 1 mM AgNO3 solution and maintained for 5–6 h at 50 °C until brown precipitate had formed. This coloured precipitate was centrifuged for 15 min at 3000 rpm; the pellet was put on a glass plate and dried in an oven for 24–48 h at 45° Celsius. Once completely dry, the powder was pulverized using a mortar and pestle (Fig. 1).
Diagrammatic representation of the overall process of formation of AgNPs using Phyllanthus niruri herb.
2.3 Uv–vis spectral analysis of Ag-NPs
Synthesized Pn-AgNPs were tentatively characterized by recording the absorbance of the UV–Vis spectrum over the 200–800 nm scan range. A UV–Vis dual beam spectrophotometer (Lasany LI295) were observed to record nanoparticle changes in surface plasmon resonance (SPR). To verify that AgNO3 ions have been reduced, all spectra were obtained in 1 cm optical path quartz cell at room temperature. The scan range of the sample UV–Visible spectrophotometer was 200–800 nm, with a scanning speed of 480 nm per minute. The spectrophotometer was equipped with “UVProv” software for recording and evaluating data. A blank reference was used to perform baseline correction for the spectrophotometer (Anandalakshmi et al., 2016).
2.4 FTIR analysis
In order to locate functional groups, the material was examined using Fourier transform infrared spectroscopy (FTIR) (Zafar et al., 2020). It is based on the absorption precisely corresponding to the bonds within the molecule. The solid powder sample was used for FTIR analysis using a FTIR spectrophotometer (PerkinElmer Spectrum). The petri dish was put in a forced-air oven, and the samples were dried at 60° C for 96 hrs. One milligram of powder sample was mixed with 100 mg KBr powder (FTIR grade) and finely grinded in a pestle and mortar. The pellets were then prepared using a hydraulic press and tested in permeation mode compared to plain KBr pellets (used as blanks) in 400–4000 cm−1.
2.5 Scanning electron microscopy (SEM) and energy dispersive x-ray (EDX)
SEM equipped with EDX was used to measure the dispersion, chemical make-up and structure of the produced AgNPs. By splattering the glass coverslips, a thin coating of nanoparticles was produced. Using gold coating sputtering, samples were gold-coated. After that, the films were examined at 20 kV voltages.
2.6 Transmission electron microscopy (TEM)
TEM (Jeol) 2100 was used to determine the size and shape of the nanoparticles. The sample (nanoparticle particles) was immobilized on a copper grid and irradiated in vacuum. An electron beam was used to analyse the material.
2.7 Bacterial isolates
The clinical isolates were obtained from patient’s samples sent to the Microbiology Department of JN Medical College & Hospital, Aligarh, for routine diagnosis. Samples such as Pus, urine, fluids, and blood, etc were cultured on conventional media as per the laboratory policy and protocols. The growth of the culture was routinely biochemically analysed and identified by the Vitek-2 automated system (Biomerieux, France) using the VITEK 2 ID card. In addition, Mueller Hinton agar was used for antibiotic susceptibility using the Kirby-Bauer disc diffusion method and the Vitek-2 automated system. Clinical bacterial isolates; Enterococcus faecalis, Streptococcus spp, methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococci (VRE), Escherichia coli, Enterobacter cloacae, Citrobacter freundii, Burkholderia cepacia complex, Salmonella typhi, and carbapenem-resistant Enterobacterales were used in the present investigation. In addition to the above bacteria, sensitive strains such as E. coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 were also used for the antibacterial screening of Pn-AgNPs (Kumar et al., 2020).
2.8 Antibacterial activity of Pn-AgNPs
2.8.1 Well diffusion assay
Agar well diffusion method was used to assess the antimicrobial action using respective bacterial isolates that were incubated overnight at 37 °C (Shahid et al., 2007). Bacterial culture (about1–2 × 108CFU/mL) was lawn-cultured on to Muller-Hinton agar petri dish making use of a sterile cotton swab. Using a sterile borer, 6 mm wells were created on an agar plate and 100 µL of biosynthesized Pn-AgNPs were added to each well (Mathew et al., 2018). The controls used were AgNO3, distilled water, and native plant extract (without synthesized AgNPs). A criterion for determining the effectiveness of the NPs was the inhibition zone around the well (in mm). Tests were performed in duplicate and average results were used.
2.8.2 Minimum inhibitory concentration (MIC)
MIC is the lowest dose value/the least concentration of nanoparticles needed to kill the bacterial growth. The microtiter broth dilution technique was used to calculate the MIC, as reported previously (Alyousef et al., 2021). 96-well ELISA plates were used to perform the experiments, and Triphenlytetrazolium chloride (TTC-3 mg/mL) was used as a cell viability indicator.
2.8.3 Interaction of Pn-AgNPs with bacterial cells examined HR-TEM and SEM
The interaction of Pn-AgNPs with E. coli and MRSA was observed using 20 kV high resolution transmission electron microscopy (Jeol 2100)). MRSA and E. coli cells were grown at 37 °C in nutrient broth (NB) for 24 h. Cells were centrifuged after incubation, pellet from the culture was dispersed in 1X PBS (phosphate buffer saline). After being dissolved in PBS, the pellet was tested with respective MICx2 of Pn-AgNPs and incubated for 24 h at 37 °C. Subsequently, the treated pellets were centrifuged, the supernatant was removed, pellet was rinsed thrice and fixed in 2.5% glutaraldehyde at 4 °C for 4 h followed by three washes with scaled series of alcohols (30, 50, 70, and 90%). Further, the cells were immersed in white resin overnight for bonding (Ali et al., 2018). Following this, drops of Pn-AgNPs-treated bacterial cells and a control were placed separately on the coverslip for the final examination under TEM and SEM.
2.8.4 Protein leakage assay
Assay to determine the Protein leakage was done employing method described previously (Dharmaraj et al., 2021). Briefly, treated and untreated MRSA and E. coli cells were grown in NB at 37 °C. The supernatant was taken at time 0 and after 3 h after the cells were centrifuged at 6200 g for 10 min. Protein concentration was estimated using Bradford method. Briefly, 800 µL of Bradford reagent were added to 200 µL of the supernatant from each sample solution and incubated for 10 min. At 595 nm after incubation, optical density was measured. As a positive control, bovine serum albumin (BSA) was utilized.
2.9 Effect on biofilm formation
2.9.1 Detection of biofilm by tube method
The Pn-AgNP’s antibiofilm activity was measured qualitatively using the tube method (Kalishwaralal et al., 2010). The turbidity of a 50 µL overnight grown bacterial culture of the selected bacteria was corrected to 0.5 McFarland standards in Luria Bertani broth. AgNPs (MIC/2) were mixed in separate tubes; then the mixture was placed into tubes with 2 mL sterile LB broth and incubated for 24 h at 37 °C. The experiment also comprised of an untreated control. The insides of the tubes were stained for half an hour using crystal violet (CV) dye (0.1%), surplus dye was eliminated, gently cleaned with distilled water, and then the inhibition of biofilms was assessed.
2.9.2 Quantitative evaluation of inhibition of biofilm formation
The quantitative efficacy of Pn-AgNPs in biofilm prevention was determined using a microtiter plate assay (Qais et al., 2020). Overnight grown bacteria were seeded in 96-well microtiter plate wells amended with or without Pn-AgNPs. Post 24 h incubation, cells were removed, well were washed three times with phosphate buffer solution and CV (0.1%) was used to stain the wells. A microtiter plate reader was used to quantify the optical density of dyed biofilms at 620 nm after excess stain was discarded and dye that stained the biofilm was dissolved in 90% ethanol.
2.9.3 Antibiofilm activity of the Pn-AgNPs by SEM
Biofilms were developed on glass coverslips with or without the addition of nanoparticles. Coverslips were given a 2.5% glutaraldehyde treatment after being rinsed with sterile phosphate buffer. The adhering biofilm cells were dehydrated for 10 min in a gradient of ethanol (20, 40, 60, 80, and 100%) solutions. Dried coverslips were coated with gold particles before SEM examination (JEOLJSM 6510 LV) at 20 kV.
3 Results and discussion
Synthesis of nanoparticles from plants is a quick, economical, and environmentally benign process. Characterization of biogenic Pn-AgNPs were done using various spectroscopic and microscopic techniques and assessed for their antibacterial potential against drug resistant pathogenic bacteria. The reaction mixture's noticeable color change is a sign that AgNPs have been synthesized. Addition of Phyllanthus niruri extract to AgNO3 solution tends to start the nanoparticle synthesis within few minutes which is denoted by change of colour in the reaction mixture (Fig. 1). The reduction reaction from Ag + to Ag0 is shown by turning a colorless solution of silver nitrate into a dark brown color. This rapid change in colour of the solution indicates speedy AgNP synthesis (Fig. 1).
3.1 Uv–vis analysis
Pn-AgNPs' UV–Vis spectra were measured within 200 and 800 nm. (Aadil et al., 2019). The Peak region of Pn-AgNPs was observed between 350 nm and 480 nm. The absorbance peak corresponding to Pn-AgNPs was ascribed at 425 nm (Fig. 2A). The absorption maxima for AgNPs was observed in a broad region of 425 nm which suggested that the synthesized particles are visible light active. Previous reports have suggested that the spherical form of AgNPs generally exhibit absorption maxima in the range of 400–440 which is also in support of our study. Further, the appearance of the broad region from 425 to 520 nm suggests that the AgNPs have been successfully capped by the plant extract (Hasan et al., 2020).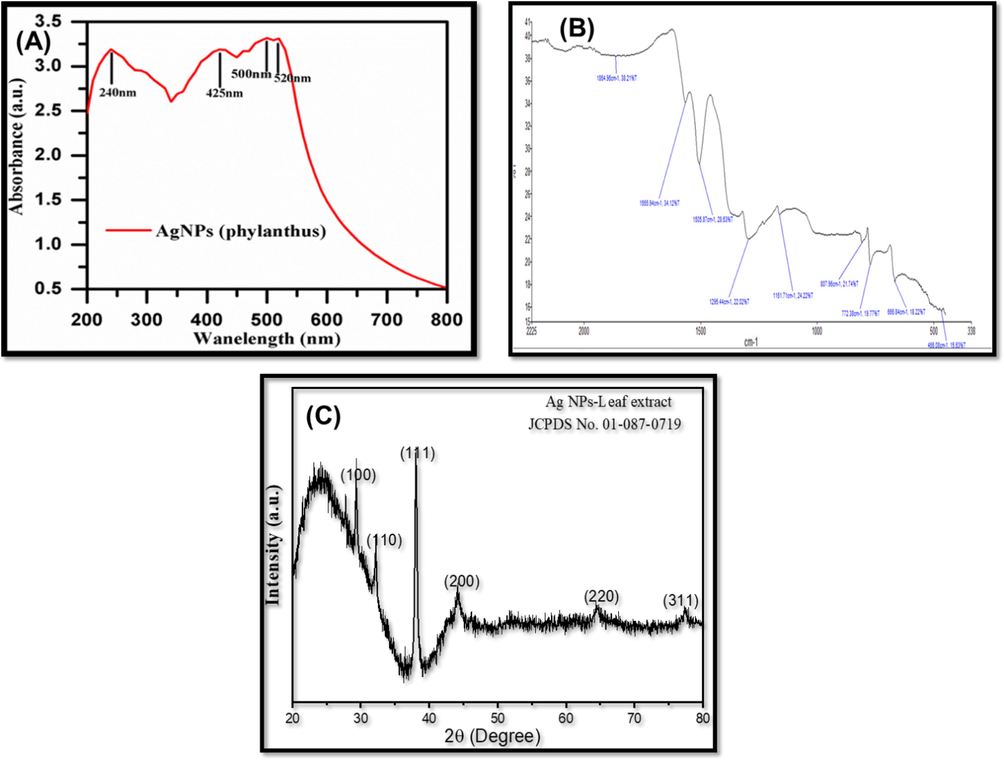
(A). UV–Vis spectrum analysis of silver NPs; (B). FTIR analysis; (C). XRD pattern of Pn-AgNPs.
3.2 FTIR analysis of nanoparticles
Fig. 2B depicts the FTIR spectra of the biosynthesized Pn-AgNPs to identify the presence of functionality of P. niruri leaf extract, that cause the bio-reduction of silver ions (Ag + ) and increase the stability of AgNPs. The FTIR spectra exhibited several absorption bands at 687 cm−1 associated with Ag coordinated with –OH and -C = O groups, 1630 cm−1 (-C = O stretching vibrations), 2066 cm−1 due to C = N stretching vibrations and 3437 cm−1 (–OH stretching vibrations) present in the extract of P. niruri. Findings of the obtained spectra suggested that the AgNPs were capped through the key functional groups of plant extract which also act as stabilizing agent. These results are supported by the findings reported earlier with porous Ag nanoparticles (Venkatesan et al., 2017).
3.3 XRD
The XRD spectra of biosynthesized Silver NPs, showing distinctive peaks at 29.31, 32.29, 38.08, 44.13, 64.48 and 77.36 corresponding to Miller indices values (1 0 0), (1 1 0), (1 1 1), (2 0 0), (2 2 0) and (3 1 1) simulated with JCPDS No. 01–087-0719 are shown in Fig. 2C. The spectra suggested the NPs with some amorphous nature which evaluated due to stabilization and functionalization through the polyphenolic groups of plant extract. Using the Debye-Scherer formula which is D = 0.9λ/βCosθ; where D is crystalline size in nm, λ is wavelength, β is the intensity of half width of maximum intensity line, 18 nm was the observed average crystalline size which was further validated by TEM analysis.
3.4 SEM and EDX analysis
The distribution of nanoparticles was demonstrated using scanning electron microscopic (SEM) images and further their elemental makeup was determined by EDX as depicted in Fig. 3. Micrograph showed the particles to be highly agglomerated and this could be accredited to the centrifugation and drying process applied for the preparation of sample of SEM visualization (Fig. 3A) (Moodley et al., 2018). Due to their surface plasmon resonance and tiny peaks that belong to carbon and oxygen that likely represent the capping structures (surface biomolecules) present in the plant extract, Pn-AgNPs exhibit a specific signal for silver at 3 keV in their EDX examination. (Fig. 3B and C) (Moodley et al., 2018). In the produced nanomaterial, Ag's weight percentage and atomic percentage were, respectively, 74.20% and 28.20%, whereas O's weight percentage and atomic percentage were, respectively, 19.18% and 49.18%. The weight and atomic percentages of C were 6.62% and 22.60%, respectively.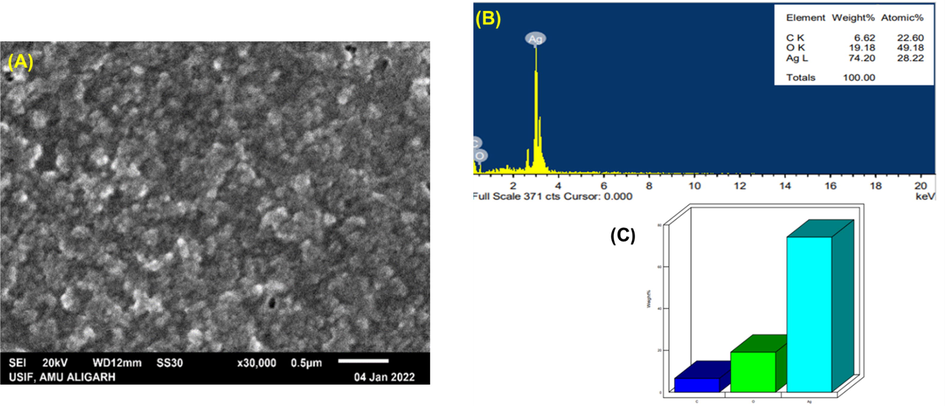
SEM and EDAX analysis of AgNPs biosynthesized from P. niruri. (A). SEM images of Pn-AgNPs; (B). Weight (%) of atoms present in Pn-AgNPs; (C). EDAX spectrum of Pn-AgNPs.
3.5 TEM analysis
The transmission electron microscope (TEM) was also used to characterize biogenic Pn-AgNPs, which revealed spherical-shaped NPs with ∼ 20 nm being the mean particle size (Fig. 4). Most of nanoparticles were evenly dispersed but few were observed to form small clumps plausibly due to improper capping or agglomeration. TEM micrograph shows some particles of different shape and size and these variations in nanoparticles produced using plant extracts have been reported previously [2, 39].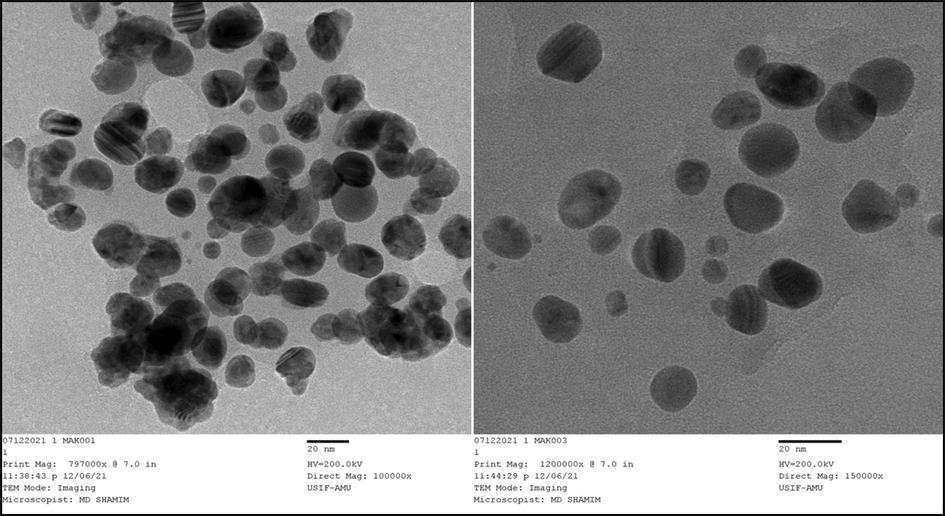
TEM image of Pn-AgNPs.
3.6 Antibacterial activity
Fig. 5 shows that Pn-AgNPs demonstrate bactericidal potential against both standard (ATCC) strains as well as drug resistant human bacterial isolates like MRSA, VRE, and CRE. Zone of inhibition ranged from 10 to 30 mm against all the test bacteria. Comparable inhibition zones were reported with AgNPs produced from M. koengii leaf extract against drug-resistant strains (MRSA and E. coli producing ESβL) (Qais et al., 2019).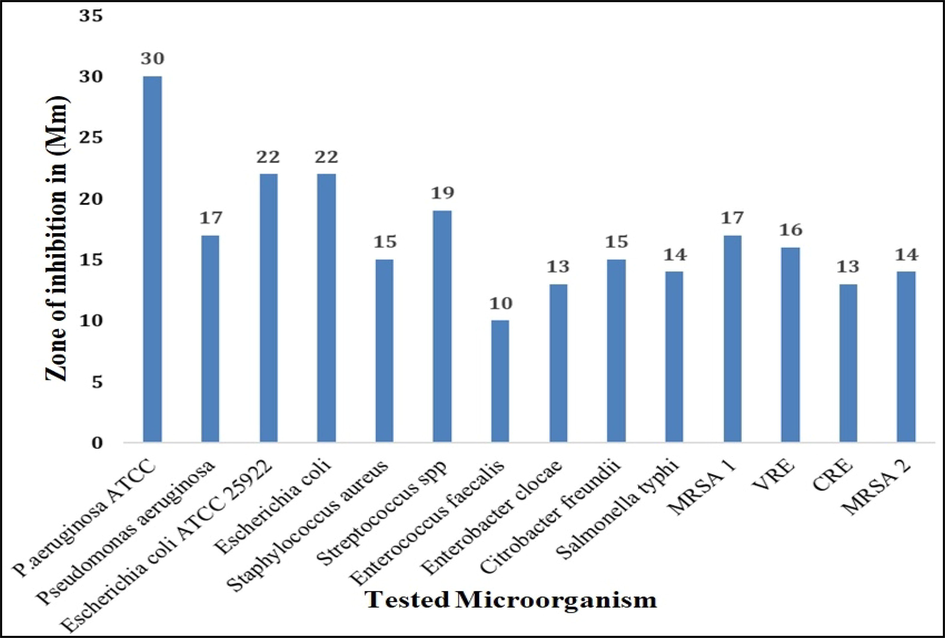
Antibacterial activity (zone of inhibition) of biosynthesized Pn-AgNPs against drug-resistant isolates using well diffusion method.
Additionally, the minimum inhibitory concentration (MIC) of Pn-AgNPs was evaluated in terms of its antibacterial activity. The MIC values of green synthesized Pn-AgNPs ranged from 10 to 40 µg/mL. These nanoparticles demonstrated MIC values of 40 µg/mL against Pseudomonas aeruginosa ATCC, Staphylococcus aureus, Streptococcus spp, Salmonella typhi, MRSA-1, CRE, and MRSA-2; 20 µg/mL against Escherichia coli, Pseudomonas aeruginosa, Citrobacter freundii; and 10 µg/mL against Enterococcus faecalis and Enterobacter clocae, as shown in Table 1. Our research findings agree with those reported with green synthesized AgNPs using Carum copticum extract wherein, MIC values of 10, 20, and 30 µg/mL was observed against C. violaceum, S. marcescens and P. aeruginosa, respectively (Qais et al., 2020). The intrinsic tolerance levels of the strains utilized, shape and size of the nanoparticle, and the MIC measurement method may be responsible for these variations in the measured MICs. It is hypothesized that Pn-AgNPs' antibacterial activity results from Ag's strong adhesion to and penetration of the bacterial cell wall, which releases intracellular components and ultimately causes cell death.
Bacterial Isolates
MIC value (µg/mL)
Escherichia coli ATCC 25922
20
Pseudomonas aeruginosa ATCC
40
Escherichia coli
20
Pseudomonas aeruginosa
20
Staphylococcus aureus
40
Streptococcus spp
40
Enterococcus faecalis
10
Enterobacter cloacae
10
Citrobacter freundii
20
Salmonella typhi
40
MRSA-1
40
VRE
40
CRE
40
MRSA-2
40
3.7 SEM examination to assess effect of Pn-AgNPs on cell morphology
Scanning electron microscopy (SEM) imaging was performed to decipher as to how Pn-AgNPs interacted with the MRSA and E. coli cells. The cell walls of the bacteria treated with Pn-AgNPs showed increasing signs of damage during the experiment. Most cells had rumples and had distorted morphology (Fig. 6). The fact that untreated cells did not exhibit any symptoms of damage provides compelling evidence that Pn-AgNPs are effective at killing bacteria. The key factor contributing to the morphological defects is the rupturing of the cell membrane brought on by treatment with AgNPs (Fig. 6). Pn-AgNPs can cause damage in a number of ways, such as by destroying parts of bacterial cells, tearing the cell membrane, stopping the mechanism that controls microbial respiration, and getting into the cytoplasm [41, 42].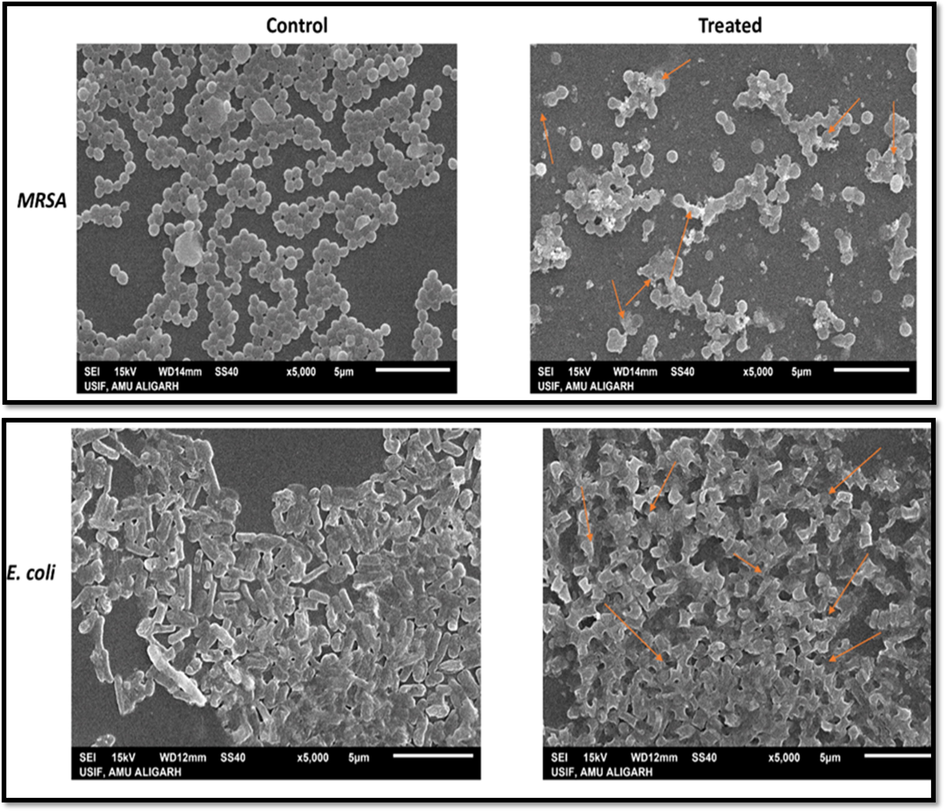
Scanning electron micrograph of MRSA and E. coli grown in the presence and absence of Pn-AgNPs. (Control) Untreated, with intact cell wall, no change in morphology; (Treated) treatment with Pn-AgNPs at 35 µg/ml, cell wall damage, cell distortion and clumping.
3.8 TEM examination to assess effect of Pn-AgNPs on bacterial cells
The internal structures of untreated (control) MRSA and E. coli cells appeared normal, with a cytoplasmic and outer membrane (Fig. 7). After treatment with Pn-AgNPs at respective MICx2, the bacterial cells seem severely injured with the leaking of the cell's interior components is visible in Fig. 7. AgNPs cause cell wall breakage, as well as these nanoparticles enter the cells from various locations, severely damaging the cell and ultimately killing the cells. Our results of the microscopic examination corroborate well with observations made with the in vitro antibacterial activity and MIC determination.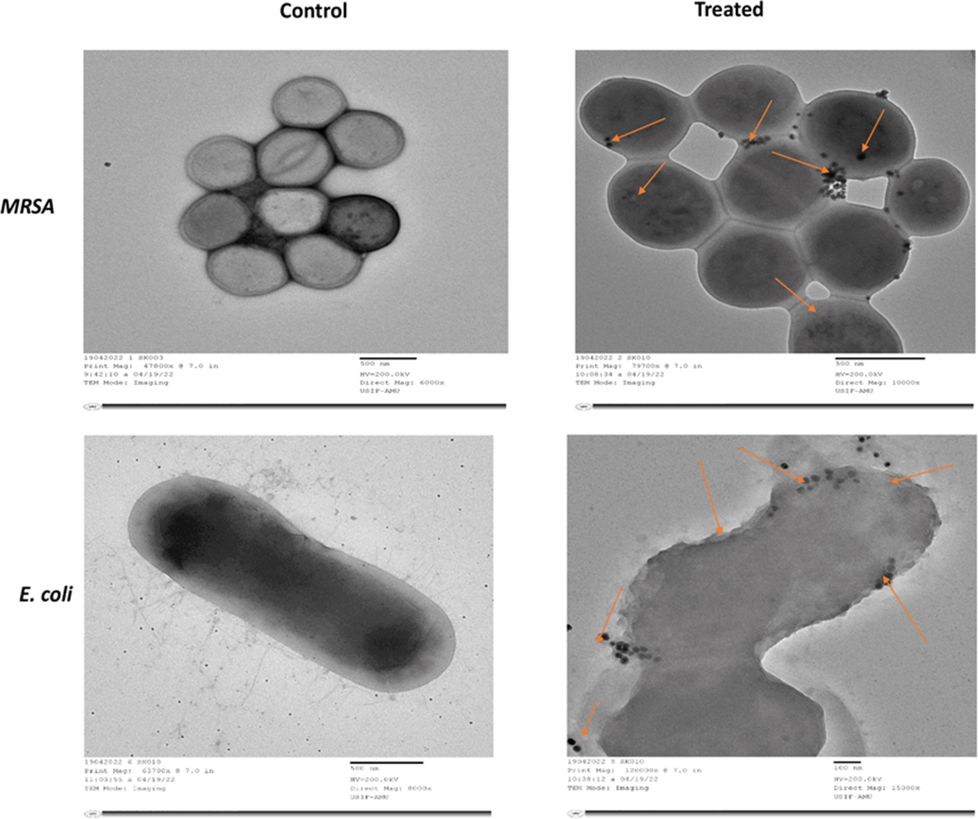
Transmission electron micrograph of MRSA and E. coli grown in the presence and absence of Pn-AgNPs. (Control) Untreated, with intact cell wall and organelles; (Treated) treatment with Pn-AgNPs at 35 µg/ml, cell wall damage, loosening of cell membrane; lysis and shrinkage and penetration of AgNPs.
3.9 Protein leakage assay
Effect of Pn-AgNPs on protein leakage in E. coli and MRSA cells was determined using Bradford method. Experimental results showed that, compared to the corresponding untreated controls, protein leakage in Pn-AgNPs-treated E. coli and MRSA was 4.7 and 5.1 times higher as depicted in Fig. 8. Thus, it is envisaged that the observed antibacterial action of Pn-AgNPs is caused by bacterial cell membrane rupture, which causes Ag to infiltrate the cell wall more frequently, eventually triggering the release of intracellular contents and cell death. (Angamuthu et al., 2023; Dharmaraj et al., 2021).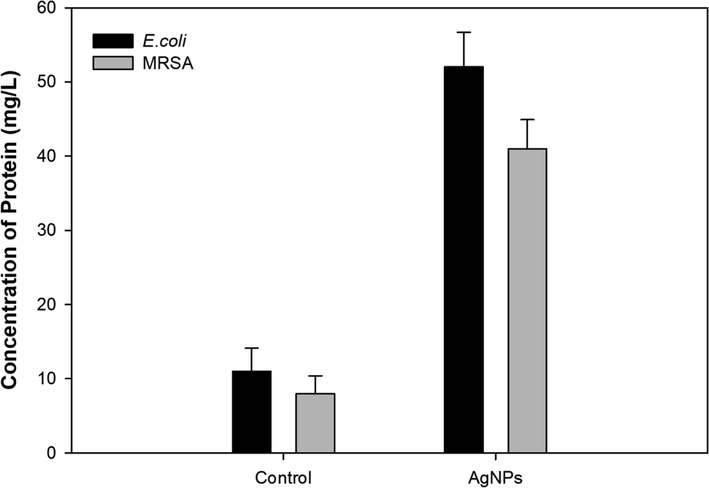
Effect of Pn-AgNPs on protein leakage in test bacterial pathogens.
3.10 Inhibition of biofilm formation
Biofilms have therapeutic importance because they play a role in the bacterial pathogenesis and make the pathogen almost 1000 times more resistant to antibiotics (Hall and Mah, 2017). The persistence of the biofilm is linked to the matrix, which contains the bacteria living in biofilm mode and is mostly made up of EPS, proteins, lipids, and eDNA. [38, 39]. Considering the importance of biofilm in persistent drug-resistant infections, Pn-AgNPs were evaluated for their biofilm inhibitory potential. Biofilm development in the understudy bacteria cultured with and without Pn-AgNPs was evaluated in tubes. Fig. 9A clearly demonstrates the reduced biofilm formation in tubes amended with Pn-AgNPs as compared to the untreated control. Further, the extent of biofilm inhibition was determined using crystal violet assay. Treatment with 10 µg/mL Pn-AgNPs reduced E. coli biofilm formation by 64.70% and P. aeruginosa biofilms by 64.39%. However, a reduction of biofilm by 67.45% and 67.08% was noticed in S. aureus and B. subtilis, respectively, when treated with 20 µg/mL of Pn-AgNPs (Fig. 9B). Findings of CV assay were further confirmed by the SEM visualization of Pn-AgNPs treated and untreated cells of P. aeruginosa (Fig. 9C-D). As evident from the micrograph, untreated cells were observed to form dense, compact, intact biofilm structure embedded in a matrix of EPS (Fig. 9C). On the contrary, reduced colonization, loosely scattered cells and a disturbed biofilm architecture with less EPS was observed in P. aeruginosa cells treated with 10 µg/mL concentration of Pn-AgNPs (Fig. 9D).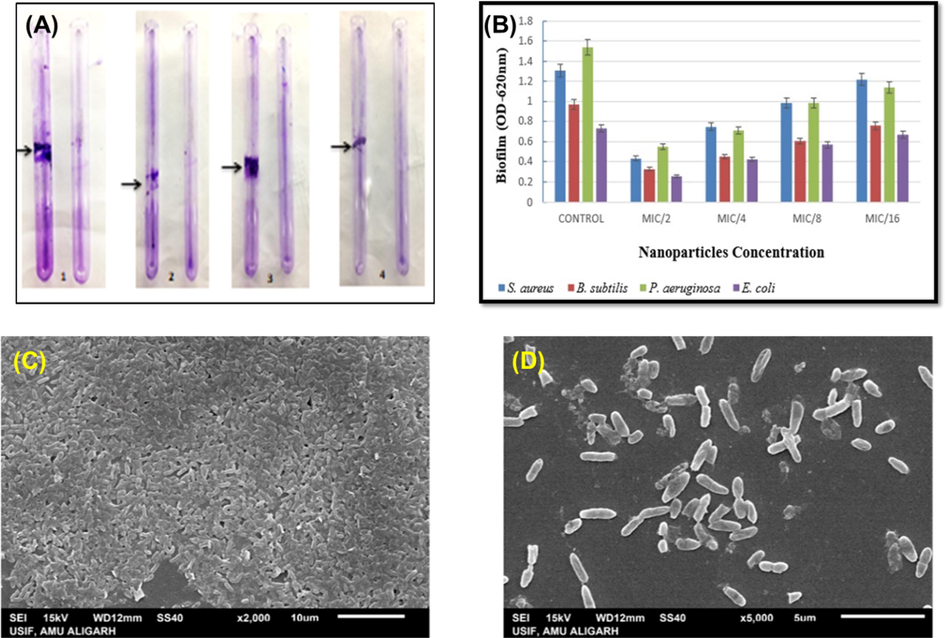
Effect of Pn-AgNPs on biofilm formation (A). qualitative assessment using tube method (B). bar graph showing percent inhibition of biofilm. Scanning electron micrograph (SEM) of P. aeruginosa biofilm (C). Untreated control; (D). 0.5xMIC of Pn-AgNPs.
It is speculated that AgNPs may diffuse and generate antibiofilm activity via water channels in the matrix of biofilms, which are used to transfer nutrients. Nanoparticles have also been shown to bind to and penetrate bacterial membranes, aggregate within bacteria, and kill bacteria (Ansari et al., 2014). AgNPs work by making bacterial membranes more permeable, damaging cells through reactive oxygen species (Al-Shabib et al., 2020), and interacting with the phosphate residues in DNA (Seo et al., 2021), as well as membrane and intracellular proteins (Kim et al., 2007; Morones et al., 2005).
4 Conclusion
These results demonstrate that plant-mediated biosynthesis of AgNPs from the well-known medicinal plant Phyllanthus niruri is a reliable, ecologically friendly, and a safe technique. Biosynthesized AgNPs have especially outstanding optical, morphological, and colloidal characteristics. The green-synthesized Pn-AgNPs demonstrated antibacterial properties against drug-resistant clinical bacterial strains such as MRSA, VRE, and CRE. Moreover, the biogenic AgNPs were also effective in inhibiting the biofilm formed by pathogenic bacteria. It is reasonable to predict that protein leakage from the ruptured cell membranes of the bacterial cells is what is responsible for the antibacterial and antibiofilm activities of the Pn-AgNPs. Due to its broad-spectrum antibiotic potential against clinically significant drug-resistant bacterial strains, the current work provides a novel perspective on the development of biologically produced silver nanoparticles from Phyllanthus niruri as a therapeutic option. Since, Pn-AgNPs also exhibited anti-biofilm properties, it could further be exploited in treatment of biofilm based persistent infections. Further studies are required to determine and unearth the entire molecular pathways that make the synthesized NPs work against bacteria and biofilm.
Acknowledgement
The authors would like to thank the Researchers Supporting Project Number (RSPD2023R729), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Green synthesis of silver nanoparticles using acacia lignin, their cytotoxicity, catalytic, metal ion sensing capability and antibacterial activity. J. Environ. Chem. Eng.. 2019;7:103296
- [CrossRef] [Google Scholar]
- Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037-1050.
- [CrossRef] [Google Scholar]
- Antibacterial and antibiofilm potential of green synthesized silver nanoparticles against imipenem resistant clinical isolates of P. aeruginosa. Bionanoscience. 2018;8:544-553.
- [CrossRef] [Google Scholar]
- Bio-inspired facile fabrication of silver nanoparticles from: in vitro grown shoots of Tamarix nilotica: Explication of its potential in impeding growth and biofilms of Listeria monocytogenes and assessment of wound healing ability. RSC Adv.. 2020;10
- [CrossRef] [Google Scholar]
- Myrtus communis and its bioactive phytoconstituent, linalool, interferes with Quorum sensing regulated virulence functions and biofilm of uropathogenic bacteria: In vitro and in silico insights. J. King Saud Univ. - Sci.. 2021;33:101588
- [CrossRef] [Google Scholar]
- Characterization of silver nanoparticles by green synthesis method using Pedalium murex leaf extract and their antibacterial activity. Appl. Nanosci.. 2016;6:399-408.
- [CrossRef] [Google Scholar]
- Biogenic preparation and characterization of silver nanoparticles from seed kernel of Mangifera indica and their antibacterial potential against Shigella spp. Molecules. 2023;28:2468.
- [CrossRef] [Google Scholar]
- Gum arabic capped-silver nanoparticles inhibit biofilm formation by multi-drug resistant strains of Pseudomonas aeruginosa. J. Basic Microbiol.. 2014;54:688-699.
- [CrossRef] [Google Scholar]
- Green synthesis of metal nanoparticles using microorganisms and their application in the agrifood sector. J. Nanobiotechnology. 2021;19:86.
- [CrossRef] [Google Scholar]
- A review on phytosynthesis, affecting factors and characterization techniques of silver nanoparticles designed by green approach. Int. Nano Lett.. 2020;10:159-176.
- [CrossRef] [Google Scholar]
- Antibacterial activity of green synthesized silver nanoparticles using vasaka (Justicia adhatoda L.) Leaf Extract. Indian J. Microbiol.. 2015;55:163-167.
- [CrossRef] [Google Scholar]
- Biomedical applications of silver nanoparticles: an up-to-date overview. Nanomaterials. 2018;8:1-24.
- [CrossRef] [Google Scholar]
- Protein leakage induced marine antibiofouling activity of biosynthesized zinc oxide nanoparticles. J. Clust. Sci.. 2021;32:643-650.
- [CrossRef] [Google Scholar]
- Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev.. 2017;41:276-301.
- [CrossRef] [Google Scholar]
- Synthesis, characterization and photo-catalytic activity of guar-gum- g -aliginate@silver bionanocomposite material. RSC Adv.. 2020;10:7898-7911.
- [CrossRef] [Google Scholar]
- Silver nanoparticles impede the biofilm formation by Pseudomonas aeruginosa and Staphylococcus epidermidis. Colloids Surfaces B Biointerfaces. 2010;79:340-344.
- [CrossRef] [Google Scholar]
- Alpinia calcarata: potential source for the fabrication of bioactive silver nanoparticles. Nano Converg.. 2018;5:37.
- [CrossRef] [Google Scholar]
- Antimicrobial effects of silver nanoparticles. Nanomedicine. 2007;3:95-101.
- [CrossRef] [Google Scholar]
- Biosynthesis of silver nanoparticles from Phyllanthus niruri leaf extracts and its antibacterial activity against antibiotics-resistant clinical isolates. Pathology. 2020;52:S127.
- [CrossRef] [Google Scholar]
- The pharmacological potential of Phyllanthus niruri. J. Pharm. Pharmacol.. 2016;68:953-969.
- [CrossRef] [Google Scholar]
- Nanoparticles in the treatment of infections caused by multidrug-resistant organisms. Front. Pharmacol.. 2019;10
- [CrossRef] [Google Scholar]
- Multidrug resistance and transferability of bla CTX-M among extended-spectrum β-lactamase-producing enteric bacteria in biofilm. J. Glob. Antimicrob. Resist.. 2016;6:142-149.
- [CrossRef] [Google Scholar]
- Sunlight mediated rapid synthesis of small size range silver nanoparticles using Zingiber officinale rhizome extract and its antibacterial activity analysis. Inorg. Nano-Metal Chem.. 2018;48:139-145.
- [CrossRef] [Google Scholar]
- Green synthesis of silver nanoparticles from Moringa oleifera leaf extracts and its antimicrobial potential. Adv. Nat. Sci. Nanosci. Nanotechnol.. 2018;9
- [CrossRef] [Google Scholar]
- The bactericidal effect of silver nanoparticles. Nanotechnology. 2005;16:2346-2353.
- [CrossRef] [Google Scholar]
- Nanotechnology in sustainable agriculture: recent developments, challenges, and perspectives. Front. Microbiol.. 2017;8
- [CrossRef] [Google Scholar]
- Antibacterial effect of silver nanoparticles synthesized using Murraya koenigii (L.) against multidrug-resistant pathogens. Bioinorg. Chem. Appl.. 2019;2019
- [CrossRef] [Google Scholar]
- Green synthesis of silver nanoparticles using Carum copticum: Assessment of its quorum sensing and biofilm inhibitory potential against gram negative bacterial pathogens. Microb. Pathog.. 2020;144:104172
- [CrossRef] [Google Scholar]
- A review on green synthesis of silver nanoparticles and their applications. Artif. Cells, Nanomed. Biotechnol.. 2017;45:1272-1291.
- [CrossRef] [Google Scholar]
- Antibiofilm activity of silver nanoparticles against biofilm forming Staphylococcus pseudintermedius isolated from dogs with otitis externa. Vet. Med. Sci.. 2021;7:1551-1557.
- [CrossRef] [Google Scholar]
- Antibacterial activity of aerial parts as well as in vitro raised calli of the medicinal plant Saraca asoca (Roxb.) de Wilde. Can. J. Microbiol.. 2007;53:75-81.
- [CrossRef] [Google Scholar]
- In vitro immunomodulatory, antifungal, and antibacterial screening of Phyllanthus niruri against to human pathogenic microorganisms. Environ. Dis.. 2018;3:63.
- [CrossRef] [Google Scholar]
- Antibacterial activity and characteristics of silver nanoparticles biosynthesized from Carduus crispus. Sci. Rep.. 2021;11:21047.
- [CrossRef] [Google Scholar]
- Antimicrobial and anticancer activities of porous chitosan-alginate biosynthesized silver nanoparticles. Int. J. Biol. Macromol.. 2017;98:515-525.
- [CrossRef] [Google Scholar]
- Eco-friendly synthesis of antibacterial zinc nanoparticles using Sesamum indicum L. extract. J. King Saud Univ. - Sci.. 2020;32:1116-1122.
- [CrossRef] [Google Scholar]







