Translate this page into:
Biodegradation and discoloration of disperse blue-284 textile dye by Klebsiella pneumoniae GM-04 bacterial isolate
⁎Corresponding author. mrafatullah@usm.my (Mohd Rafatullah)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The increase in urbanization and industrialization has led to the discharge of wastes in environment causing more pollution. Textile industries are playing a havoc role in water pollution natural aquatic ecosystems. Bioremediation is the one of the cost effective and ecofriendly achievement of biotechnological novelty. Bioremediation is the key to waste management. It can remove textile dyes that are considered to be heterologous biological compounds and are difficult to degrade. In the present bio-remedial method, Klebsiella pneumoniae GM-04 was used promising microbial tool for the decolorization of disperse blue 284 dye. Klebsiella pneumoniae was isolated from industrial wastewater. Under optimum conditions (37 °C and pH 7) Klebsiella pneumoniae led to 95% decoloration of DB-284 (200 ppm) within 24 hrs. The increase in dye concentration and temperature decreased the bacterial growth results in low decoloration of dye. UV–Vis and FTIR analysis suggested that the parent dye degraded into different products. The data was statistically analyzed.
Keywords
Azo dye
Bioremediation
Disperse Blue 284
Klebsiella pneumonia
Incubation period
1 Introduction
Synthetic dyes are widely used as colorants in various industries as pharmaceutical, cosmetic industries, paper and textile due to high inherent stability, low cost, variety of colors shades as contrasted to natural dyes. There are about 0.1 million ton of synthetic dyes used worldwide with annual production of around 0.28 million tons (Singh et al., 2017). Textile industries consumed 56% of synthetic dyes of the total annually produced worldwide. Textile industries exploit a hefty quantity of water hence generate a bulk of wastewater contaminated with unfixed dyes (Markandeya et al., 2017). Textile industry play important role in the world’s economy but it is largest consumer of portable water, about 200 L water is required to create 1 kg textile product (Barathi et al., 2020), consequently producing significant volume of dyeing wastewater (Wu, 2009). Synthetic dyestuff are mainly divided as: aromatic methane, triphenylmethane, azo, indigo and anthraquinone dyes (Cai et al., 2015), characterized by resistant to alkali, acid and light treatment, carcinogenic and mutagenic to flora and fauna including human (Deng et al., 2018). Azo dyes are the versatile category of dyestuff using in textile industry contributing 60–70% of all dyestuff produced due to ease and low cost (Singh et al., 2015). Azo dyes are distinguish on the basis of azo groups (–N═N–) linked to aromatic rings (Dixit and Garg, 2018). About 10–15% of total dyes released into environment due to ineffectivity in printing and dyeing processes of fabric (Prasad and Aikat, 2014), are a risk to people’s health and natural environment due to their toxic and carcinogenic nature (Han et al., 2012).
Rapid industrialization led to environment pollution and imbalance ecosystem by releasing contaminated wastewater having different toxic organic and inorganic chemicals (Basheer, 2018; Mishra and Maiti, 2019). Textile industrial effluents contain approximately 10–100 mg/L of dye containing carcinogenic amines (Doble and Kumar, 2005) and other chemical pollutants in the form of toxic metals, pentachlorophenol, biocides, chlorine bleaching, free formaldehyde, softeners fire retardants and halogen carriers (Jadhav et al., 2008). Discharge of untreated textile wastewater into water resources lead into reduced sunlight penetration, eutrophication (Eslami et al., 2019), pH alteration, gas solubility and increase in COD, TOC and BOD of water resources posing harmful impacts (Thanavel et al., 2019). It also affect terrestrial ecosystem by reducing plant growth, seed germination, soil fertility and plant productivity (Saranraj, 2013). Use of dye contaminated water for drinking results in cancer, cutaneous stimulation, allergy, gene mutation and dermatitis in human (Kaur and Singh, 2007). Ingestion of dye contaminated water by mammals result in biodegradation of azo bond into aromatic amines by anaerobic bacteria present in lower gastrointestinal tract (Saranraj, 2013). These hazardous aromatic amines are carcinogenic, causing bladder cancer and contracting cancer in kidney and liver (Saha and Rao, 2020). Dye contaminated water is unfit for the perseverance of recreation, irrigation drinking and fishing. So, the scheming of textile wastewater is necessary afore release to the environment.
Physicochemical and biological methods have their own efficiency to eliminate toxins from textile wastewater. However, physical and chemical methods like photocatalysis, flocculation, coagulation, sonication, membrane separations, ion exchange, activated carbon adsorption, ozonation, irradiation, Fenton processes and electrochemical oxidation (Chen et al., 2018) are limited and unfeasible for disposal of textile wastewater due to very expensive, less efficient, inappropriateness to a broad range of dyes and resulting in secondary pollution (Qu et al., 2010). Bioremediation is an environment friendly, cheap, effective and sustainable method to decolorize textile wastewater (Sudha et al., 2014) by converting organic compound into nontoxic products i.e. water and carbon dioxide (Karim et al., 2018). Bacterial genera such as Klebsiella, Pseudomonas, Desulphovibrio, Bacillus, Sphingomonas, Streptococcus, Escherichia, Aeromonas, Proteus, Schewanella, Alcaligenes, and Citrobacter effectively decolorize textile wastewater (Cai et al., 2015; Zabłocka-Godlewska et al., 2015) using dye as carbon and nitrogen source (Senthilkumar et al., 2013). A variety of bacterial enzymes including reductases, laccases and oxygenases have degradation potential via reductive break down of azo bonds by anaerobic degradation and finally biological transformation of aromatic amines in the aerobic conditions (Kumar et al., 2016). The bacterial enzymatic system for dye degradation is well developed and diverse. The biodegradation is followed by aerobic or anaerobic or by sequential method (Khattab et al., 2020). Under anaerobic condition the azo bond breaks down via reductive cleavage and azo dyes are degraded to mutagenic amines, followed aerobic processes for complete breakdown of dye (Eslami et al., 2019). Bacterial oxidoreductases are the significant for the degradation of the textile dyes. Bacteria use xenobiotic compound of dyes as substrate by dynamic metabolism and the dye fragmented to less toxic metabolites. One significance of isolating bacteria from the definite location of effluents is that they are more probably to have the activated enzymes, thereby promoting the breakdown of dyes. Therefore this study was aimed to isolate identify dye degrading potential bacteria and optimize conditions (pH, temperature, and dye concentration) for discoloration and degradation of DB-284 textile dye.
2 Material and methods
2.1 Sampling of textiles wastewater
Textile industrial effluents was sampled from 10 different discharging points of textile industries located in district Shiekhupura. The pH and temperature of wastewater were measured at collection site using pH strips and thermometer respectively. The samples were taken in sterilized screw capped 50 mL falcons and immediately brought to Cell and Molecular Biology Lab Department of Zoology GC University Lahore, Pakistan and stored at 4 °C until further used.
2.2 Chemicals and microbiological media
The widely used textile azo dye (DB-284) was purchased from a local textile dyeing unit, Lahore, Pakistan. The stock solution was made dissolving 1 gm of DB-284 dye in 500 mL distilled water. Dye solutions of desired concentrations (200 ppm, 400 ppm, 600 ppm, 800 ppm and 1000 ppm) were made from stock solution. The solution of dyestuff was filtered using 0.22 µm membrane filter, as azo dyes are unstable to moist–heat sterilization. Bacteria were grown on LB agar and LB broth medium.
2.3 Screening of azo dye degrading bacteria
All the wastewater samples were used for the screening of bacteria using serial dilution method. 100 µL of diluted samples were spread on LB agar plates and incubated overnight. A total of 10 morphologically different colonies named (GM-01, GM-02, GM-03, GM-04, GM-05, GM-06, GM-07, GM-08, GM-09 and GM-10) were selected. All the strains were streaked for pure culture. Inoculum of pure culture was prepared by inoculating a single colony of each strain with sterilized inoculating loop in 50 mL LB broth and then incubated at 37 °C in a shaking incubator (150 rpm) for overnight incubation.
2.4 Bioremediation potential
The bacterial isolates were screened and dye degrading assay was performed. One percent (1%) inoculum of all strains was inoculated in 50 mL of broth containing 200 ppm of DB-284 at 150 rpm and 37 °C for 24 hrs till complete depolarization occurs. 1 mL of sample was taken at every 24 hrs of incubation and then centrifuged at 13,000 rpm for 5 min. Decolorization potential was calculated by measuring the absorbance of the supernatant at 550 nm spectrophotometer. Maximum absorbance of DB-284 was found by UV– Vis spectrophotometer. The discoloration percentage was determined by using given formula (Bekhit et al., 2020). % decolorization = {(A – B) / A} 100 where, A; is an initial absorbance of dyes, B; is final absorbance of dye.
2.5 Molecular characterization of bacterial isolate
The genomic DNA of GM-04 strain was extracted using phenol chloroform method and PCR amplification was carried out of 16S rRNA gene using universal primers 27F 5′ AGAGTTTGATCMTGGCTCAG 3′) and 1541R (5′-AAGGAGGTGATCCAGCCGCA-3′). The amplification was initiated by initial denaturing at 94 °C for 5 min, followed by 35 cycles of 40 sec of denaturing at 94 °C, annealing 50 sec at 51 °C, 1 min and elongation 45 sec at 72 °C with final elongation at 72 °C for 10 min. For the confirmation of amplification the gel electrophoresis was perform and gene (1500 bp) was cleaned using a Genejet Gel extraction Kit (Thermoscientific). The amplicon 16S rRNA gene was sent to First BASE Laboratories Pvt. Ltd., Malaysia, for DNA sequencing. The sequence of 16S rRNA gene was initially analyzed at NCBI server using Basic Local Alignment Search Tool (BLAST) (Kumar et al., 2016).
2.6 Effect of temperature, pH and dye concentration for decolorizing potential of GM-04 strain
The effect of different physicochemical factors like pH, temperature and dye concentration on biodegradation of DB-284 by GM-04 strain was determined by incubating it at various dye concentrations (200–1000 ppm), different temperature (20–65 °C), different pH of medium (4–10) and different incubation period (0 hr to 28 hrs). After incubation 3 mL of sample was drawn after every 4 hrs for the measurement of absorbance (550 nm) for decolorization percentage using spectrophotometer PD 303S (Bekhit et al., 2020).
2.7 Analysis of textile wastewater
The samples were analyzed using different analytical techniques. The culture was centrifuged at 10000 rpm for 10 min and supernatant was separated for analysis. The UV–visible spectra of control and the sample treated with bacterial isolates were recorded in the range between 200 and 900 nm in UV–Visble spectrophotometer (UV 2100, Shimadzu, Japan) in order to observe the changes in the peak values of the degraded products (Kilany, 2017). To identify variation in functional group of DB-284 before and after treatment with K. pneumoniae, fourier transformed infrared spectrometry (FTIR) technique was used. Shimadzu 8400S spectrophotometer was used to record FTIR spectra in the 500–4000 cm−1 IR region (Lade et al., 2012).
2.8 Statistical analysis
Data was statistically analyzed by using analysis of the variance (one-way ANOVA) through Tukey posttest (P ∼ 0.05) using SPSS software.
3 Results and discussion
3.1 Isolation, identification and phylogenetic position of DB-284 decolorizing bacteria
Two isolates, GM-04 and GM-06, collected from textile industrial effluent, showed the ability to decolorize DB-284 in LB broth media. GM-04 strain was selected for further use. Based on the characterization of isolate that refer to Bergey’s manual showed that GM-04 isolate belong to the genus of Klebsiella (Guerrero, 2001). Molecular identification was carried out to verify the identity of this bacterial isolate that was capable of decolorizing DB-284. Polymerase chain reaction (PCR) was used to polymerized 16S rRNA gene of K. pneumonia and confirmed by visualizing a single DNA band of about 1500 bp as shown in the Fig. 1(a, b). The sequence of 16S rRNA gene was blast on NCBI. BLAST showed that GM-04 isolate was 100% identical with K. pneumoniae.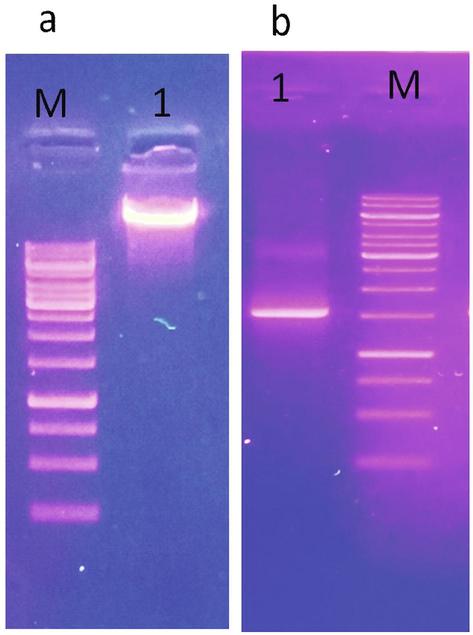
(a) Agarose gele electrophoresis of genomic DNA (1), b) PCR amplicone of 16S rRNA gene of Klebsiella pneumoniae. M, 1 kb DNA ladder mix.
3.2 Decolorization potential and mechanism of DB-284 biodegradation
Decolorization efficiency of K pneumonia was observed by change in color of azo dye DB-284 within 24 hrs. K pneumonia showed different level of decolorization and biodegradation efficiency at various temperature, pH and different dye concentration. Maximum decolorization of 200 ppm DB-284 by 1% (v/v) inoculum of K. pneumonia was 95% at pH 7 and 37 °C within 24 hr. Under optimum condition, 1% inoculum of K. pneumoniae led to 95% decolorization of DB-284 (200 ppm) within 24 hrs as shown in Fig. 2 (a, b). Klebsiella sp. Y3 showed maximum degradation of methyl red at 37 °C and 7 pH (Cui et al., 2014). Whereas K. pneumonia showed 95% discoloration of DB-284 occurred at 200 ppm and dropped drastically to 79%, 75%, 52% and 35% with the increase in concentration of dye as 400, 600, 800, 1000 ppm respectively. The reduction in decolorizing efficiency of K. pneumonia at high concentrations of dye was probably due to toxic effects of dye or because of slow growth rate of bacteria (Mate and Pathade, 2012).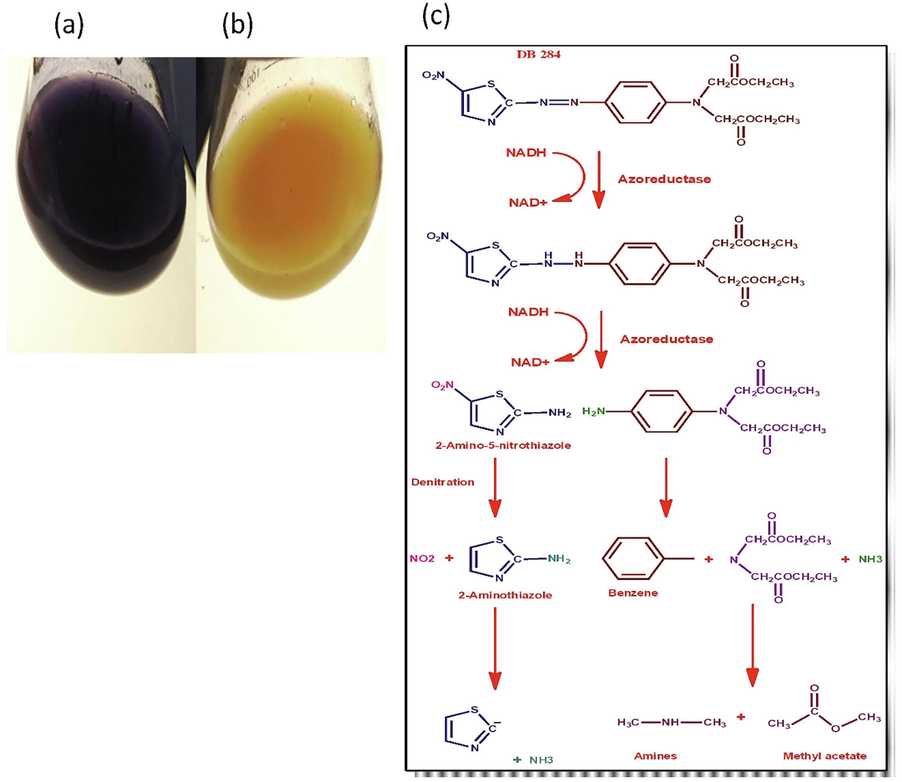
(a) Control of DB-284 (b) after treated with K. pneumoniae (c) Proposed mechanism of DB-284 azo dyes by K. pneumoniae.
Klebsiella sp. has potential to degrade textile azo dyes. The initial stage in breakdown of azo dye, mediated by bacteria, is the reduction of –N = N– bond under both aerobic and anaerobic condition. This reduction can either be carried out by the enzymes, the redox mediators, and physicochemical reduction employing sulfide or combination of reductants as shown in Fig. 2c. Reduction of azo dyestuff via enzyme may be specific or nonspecific to dyestuff. Rafii, et al., (1990) described that the azoreductase present in the Clostridium and Eubacterium sp. for the very first time. Azoreductase isolated from given strains showed sensitivity toward and synthesized and liberated constitutively. Afterward, azoreduction in Clostridium perfringens were found to carry out by FAD dehydrogenase (Rafii and Cerniglia, 1995). Azoreductase gene of C. perfringens has been cloned and expressed in Escherichia coli (Rafii and Coleman, 1999). The other process for degradation of dye is cytosolic flavin-dependent reduction is that involves transfers of electron via flavin to the azo dyes. However, recombinant Sphingononas strain BN6 has been found recently to reduce in vitro by cytosolic flavin-dependent azo reduction of sulfonated azo dyes (Russ et al., 2000).
3.3 Effect of dye concentration on discoloration
Under optimum condition, decolorization rate attained at 200, 400, 600, 800, 1000 ppm of DB-284 was as 95%, 79%, 75%, 52% and 35% respectively, revealed that degradation efficiency of K. pneumonia decreased with increase in dye concentration as shown in Fig. 3a. Cui, et al., 2014 revealed that Klebsiella sp. have great potential to degrade azo dyes at its initial concentrations and potential was decrease due to high concentration of dye, which result low growth rate of bacteria.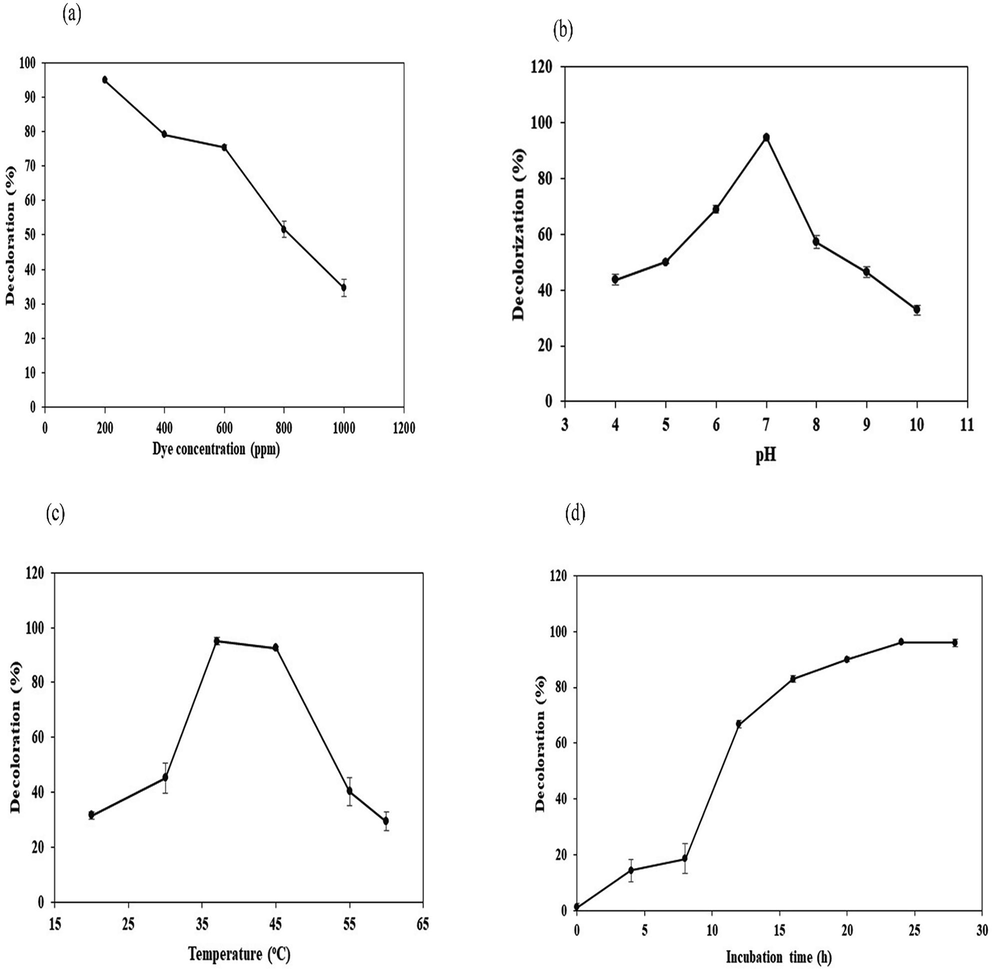
Effect of dye concentration (a), pH (b), temperature (c) and incubation time period (d) on decolorization of DB-284 by K. pneumoniae strain GM-04.
3.4 Effect of pH
The highest biodegradation 95% observed at pH 7, however at pH 4, 5, 6, 8, 9 and 10; decolorization efficiency was reduced to 44%, 50%, 71%, 59%, 48% and 30% respectively as shown in Fig. 3b. K. pneumonia showed maximum potential (95%) at neutral pH 7. With increasing pH from 4 to 7, dye degradation efficiency was increased gradually from 44% to 95% due to increased bacterial biomass. While dye biodegradation potential was decreased gradually from 95% to 30% by incubating GM-04 at pH 7 to 10 due to its lower growth efficiency (Cui et al., 2014; Kurade et al., 2015; Krishnan et al., 2017). Afzal et al., 2017 founded that slightly alkaline pH and neutral for optimum removal of dye while Klebsiella species have optimum pH in the range of 6–8. Hsueh and Chen, 2007 observed that at low pH decreased dye degradation efficiency due to slow growth rate and protonation of azo dyes due to change in their chemical composition, bacteria were unable to degrade the dyes.
3.5 Effect of temperature
Our study showed that K. pneumoniae (GM-04 strain) has statistically significant potential to DB-284 within 24 hrs under aerobic condition. K. pneumonia showed different level of decolorizing potential between 20 °C and 6 °C. Due to increased growth at temperature range 20 °C to 37 °C, the decolorization efficiency increased gradually from 23% to 95% while dye degradation efficiency was decreased from 95% to 23% due to slower growth at 40 oC to 65 °C respectively. K. pneumoniae did not break methyl red at 45 oC due of slow growth rate at high temperature (Elizalde-González et al., 2009; Franciscon et al., 2009; Lin et al., 2010).
The optimum temperature for degradation of DB-284 is 37 °C. With the increase in temperature of incubation from 20 oC to 37 oC, decolorization efficiency was also increased from 44% to 95%. While upon increasing temperature from 37 oC to 60 °C, biodegradation potential decreased from 95% to 33% respectively. These results are supported by Wong and Yuen (1996) describe that K. pneumoniae strain RS-13 isolated from textile waste sludge completely decolorized methyl red 100 mg/L. Biodegradation potential of K. pneumonia at different range of temperature is shown in Fig. 3c.
3.6 Effect of incubation period
The influence of incubation period studied on decoloration of DB-284. The result showed that decolorization potential of DB-224 by K. pneumonia increases with incubation time. With increase in incubation time from 4, 8, 12, 16 and 24 hrs, decoloration of DB-224 increases as 14%, 19% and 95% respectively as shown Fig. 3d. Siddeeg, et al., 2020 reported that the decolorization increased by time increment of incubation period which help to increase the biomass of bacteria.
3.7 UV–visible spectrophotometry
UV visible spectra showed changed in wavelength of absorbance from 340 to 390 nm and 550 to 460 nm after treated with K. pneumoniae showed formation of new metabolites as shown in Fig. 4. The reduction in absorbance was likely due to the degradation of DB-284 azo bond. The biodegradation and decolorization of dye (DB-284) was observed 95% after 24hr of incubation at 200 ppm concentration. After biological treatment of dye, the UV–Visble spectrum showed that major change in the spectrum of maximum absorbance of DB-284 (550 nm) and disappearance of peak suggested that the decolorization of DB-284 disperse azo dye occur by bacterial (Wu, 2009; Bekhit et al., 2020).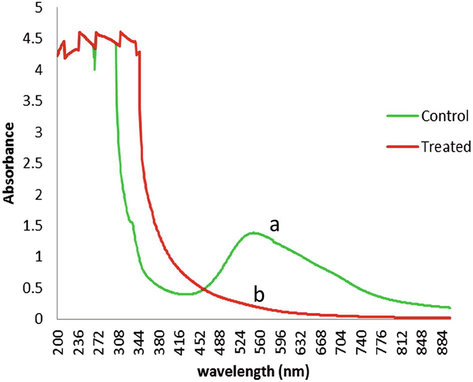
UV spectrum of DB-284 before (line a) and after treated (line b) with K. pneumoniae, the absorbance peak at 550 nm disappeared after 24 hrs of incubation.
3.8 Fourier transformed infrared spectrometry (FTIR) analysis
Characterization of functional group of control DB-284 and its metabolite after treatment with K. pneumoniae Fourier transformed infrared spectrometry (FTIR) further evident biodegradation of DB-284. Comparison of FTIR spectra of control and experimental DB-284 exhibited emergence of new peaks and increases in intensity of peaks at 1000–3500 cm−1 range. The FTIR spectra of control DB-284 showed peaks at 1004, 1238, 1600, 1667, 2310 and 3329 cm−1 representing C–H, C-N, N = N, C = C and N–H respectively as shown in Fig. 5a. A Specific peak of N = N at 1600 cm−1 and 3329 cm−1 for N–H confirmed aromatic structure of DB-284, after treatment with K. pneumoniae, FTIR spectra of experimental DB284 exhibited rise in intensity at 1028, 1242, 1739, 2328, 3383 cm−1 and appearance of specific new peaks at 1363 and 1541 cm−1 showing NO2 evident of biodegradation of DB-284 in the form of new product formation as shown in Fig. 5b. Disappearance of peaks at 1600 cm−1 in experimental group showed breakdown of azo bond of DB-284 led to formation of new metabolites.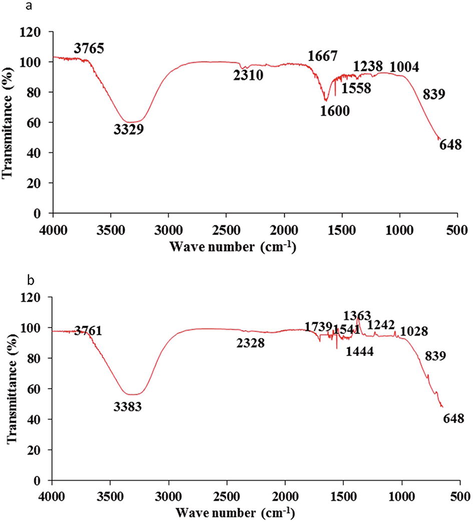
FTIR spectra of control DB-284 (a) and decolorized DB-284 by K. pneumoniae (b).
4 Conclusions
Wastewater from the textile industries reveals intimidations to ecosystem, because the major amounts of dyes are consumed. K. pneumoniae has a promising potential to decolorize and degrade DB-284 (95%) after 24 hrs of incubation at temperature 37 °C, pH 7 and 200 ppm dye concentration. This bacterium completely degraded 200 ppm of DB-284 within 24 hrs. The bacterial fauna showed efficient decolorization potential at various pH ranging from 4 to 9 and temperatures range from 25 to 45 °C. The potential of the strain to tolerate and decolorize azo dyes in a wide range of temperature, pH and dye concentrations gave it an edge to use them in the biodegradation of the textile industrial effluents. The biodegradation of DB-284 was evident by UV–Vis spectrophotometric and FTIR analysis. Bioremediation is an efficient technique for the treatment of textile industrial effluents with the help of bacterial fauna.
Acknowledgements
The authors are grateful to the Universiti Sains Malaysia and financial support through the Fundamental Research Grant Scheme (FRGS) grant number (203/PTEKIND/6711822).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Assessment of heavy metal tolerance and biosorptive potential of Klebsiella variicola isolated from industrial effluents. AMB Express. 2017;7:184.
- [Google Scholar]
- Biodegradation of textile dye Reactive Blue 160 by Bacillus firmus (Bacillaceae: Bacillales) and non-target toxicity screening of their degraded products. Toxicol. Rep.. 2020;7:16-22.
- [Google Scholar]
- Chemical chiral pollution: impact on the society and science and need of the regulations in the 21st century. Chirality. 2018;30:402-406.
- [Google Scholar]
- Decolorization and degradation of the Azo dye by bacterial cells coated with magnetic iron oxide nanoparticles. Environ. Nanotechnol. Monit. Manage.. 2020;14:100376
- [Google Scholar]
- Biodegradation of Azo Dye Disperse Orange S-RL by a Newly Isolated Strain Acinetobacter sp. SRL8. Water Environ. Res.. 2015;87:516-523.
- [Google Scholar]
- Biodegradation and detoxification of Direct Black G textile dye by a newly isolated thermophilic microflora. Bioresour. Technol.. 2018;250:650-657.
- [Google Scholar]
- Decolourization of azo dyes by a newly isolated Klebsiella sp. strain Y3, and effects of various factors on biodegradation. Biotechnol. Biotechnol. Equip.. 2014;28:478-486.
- [Google Scholar]
- Biodegradation of environmentally hazardous azo dyes and aromatic amines using Klebsiella pneumoniae. J. Environ. Eng.. 2018;144:04018035.
- [Google Scholar]
- Biotreatment of Industrial Effluents. Oxford, United Kingdom: Elsevier Butterworth-Heinemann; 2005. p. :19-38.
- Degradation of immobilized azo dyes by Klebsiella sp. UAP-b5 isolated from maize bioadsorbent. J. Hazard. Mater.. 2009;161:769-774.
- [Google Scholar]
- Decolorization and biodegradation of reactive Red 198 Azo dye by a new Enterococcus faecalis–Klebsiella variicola bacterial consortium isolated from textile wastewater sludge. World J. Microbiol. Biotechnol.. 2019;35:38.
- [Google Scholar]
- Microaerophilic–aerobic sequential decolourization/biodegradation of textile azo dyes by a facultative Klebsiella sp. strain VN-31. Process Biochem.. 2009;44:446-452.
- [Google Scholar]
- Bergey's manuals and the classification of prokaryotes. International Microbiology. 2001;4:103-109.
- [Google Scholar]
- Exploring new strains of dye-decolorizing bacteria. J. Biosci. Bioeng.. 2012;113:508-514.
- [Google Scholar]
- Comparative study on reaction selectivity of azo dye decolorization by Pseudomonas luteola. J. Hazard. Mater.. 2007;141:842-849.
- [Google Scholar]
- Biodegradation of methyl red by Galactomyces geotrichum MTCC 1360. Int. Biodeterior. Biodegrad.. 2008;62:135-142.
- [Google Scholar]
- Decolorization of textile reactive dyes by bacterial monoculture and consortium screened from textile dyeing effluent. J. Genet. Eng. Biotechnol.. 2018;16:375-380.
- [Google Scholar]
- TiO2 mediated photocatalytic degradation studies of Reactive Red 198 by UV irradiation. J. Hazard. Mater.. 2007;141:230-236.
- [Google Scholar]
- Textile dyeing industry: environmental impacts and remediation. Environ. Sci. Pollut. Res.. 2020;27:3803-3818.
- [Google Scholar]
- Isolation, screening and molecular identification of novel bacterial strain removing methylene blue from water solutions. Appl. Water Sci.. 2017;7:4091-4098.
- [Google Scholar]
- Effect of pH, inoculum dose and initial dye concentration on the removal of azo dye mixture under aerobic conditions. Int. Biodeterior. Biodegrad.. 2017;119:16-27.
- [Google Scholar]
- Bioinformatics aided microbial approach for bioremediation of wastewater containing textile dyes. Ecol. Inf.. 2016;31:112-121.
- [Google Scholar]
- Bacterial–yeast consortium as an effective biocatalyst for biodegradation of sulphonated azo dye Reactive Red 198. RSC Adv.. 2015;5:23046-23056.
- [Google Scholar]
- Enhanced biodegradation and detoxification of disperse azo dye Rubine GFL and textile industry effluent by defined fungal-bacterial consortium. Int. Biodeterior. Biodegrad.. 2012;72:94-107.
- [Google Scholar]
- Biodegradation of reactive blue 13 in a two-stage anaerobic/aerobic fluidized beds system with a Pseudomonas sp. isolate. Bioresour. Technol.. 2010;101:34-40.
- [Google Scholar]
- Toxicity of disperse dyes and its removal from wastewater using various adsorbents: a review. Research Journal of Environmental Toxicology. 2017;11:72-89.
- [Google Scholar]
- Biodegradation of CI Reactive Red 195 by Enterococcus faecalis strain YZ66. World J. Microbiol. Biotechnol.. 2012;28:815-826.
- [Google Scholar]
- Optimization of process parameters to enhance the bio-decolorization of Reactive Red 21 by Pseudomonas aeruginosa 23N1. Int. J. Environ. Sci. Technol.. 2019;16:6685-6698.
- [Google Scholar]
- Study of bio-degradation and bio-decolourization of azo dye by Enterobacter sp. SXCR. Environmental technology. 2014;35:956-965.
- [Google Scholar]
- Decolorization of reactive dark blue KR by the synergism of fungus and bacterium using response surface methodology. Bioresour. Technol.. 2010;101:8016-8023.
- [Google Scholar]
- Reduction of azo dyes and nitroaromatic compounds by bacterial enzymes from the human intestinal tract. Environ. Health Perspect.. 1995;103:17-19.
- [Google Scholar]
- Cloning and expression in Escherichia coli of an azoreductase gene from Clostridium perfringens and comparison with azoreductase genes from other bacteria. Journal of Basic Microbiology: An International Journal on Biochemistry, Physiology, Genetics, Morphology, and Ecology of Microorganisms. 1999;39:29-35.
- [Google Scholar]
- Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl. Environ. Microbiol.. 1990;56:2146-2151.
- [Google Scholar]
- The function of cytoplasmic flavin reductases in the reduction of azo dyes by bacteria. Appl. Environ. Microbiol.. 2000;66:1429-1434.
- [Google Scholar]
- Biotransformation of Reactive Orange 16 by alkaliphilic bacterium Bacillus flexus VITSP6 and toxicity assessment of biotransformed metabolites. Int. J. Environ. Sci. Technol.. 2020;17:99-114.
- [Google Scholar]
- Bacterial biodegradation and decolourization of toxic textile azo dyes. African Journal of Microbiology Research. 2013;7:3885-3890.
- [Google Scholar]
- Response surface Optimization for Biodegradation of Textile Azo dyes using isolated bacterial strain Pseudomonas sp. Arabian Journal for Science and Engineering. 2013;38:2279-2291.
- [Google Scholar]
- Iron oxide/chitosan magnetic nanocomposite immobilized manganese peroxidase for decolorization of textile wastewater. Processes. 2020;8:5.
- [Google Scholar]
- Enzymatic decolorization and degradation of azo dyes–A review. Int. Biodeterior. Biodegrad.. 2015;104:21-31.
- [Google Scholar]
- Role of azoreductases in bacterial decolorization of azo dyes. Current Trends in Biomedical Engineering & Biosciences. 2017;9:50-52.
- [Google Scholar]
- Microbial degradation of azo dyes: a review. International Journal of Current Microbiology and Applied Sciences. 2014;3:670-690.
- [Google Scholar]
- Combined biological and advanced oxidation process for decolorization of textile dyes. SN Applied Sciences. 2019;1:97.
- [Google Scholar]
- Decolorization and biodegradation of methyl red by Klebsiella pneumoniae RS-13. Water Res.. 1996;30:1736-1744.
- [Google Scholar]
- Photodegradation of CI Reactive Red 2 in UV/TiO2-based systems: Effects of ultrasound irradiation. J. Hazard. Mater.. 2009;167:434-439.
- [Google Scholar]
- Dye decolourisation using two Klebsiella strains. Water Air Soil Pollut.. 2015;226:2249.
- [Google Scholar]







