Translate this page into:
Biochemical characterization of human exonuclease1 (hExo1)
⁎Corresponding author at: Department of Biochemistry, Kebbi State University of Science and Technology, Aliero, Nigeria. auargungu96@gmail.com (Aminu A. Umar)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Human exonuclease1 (hExo1) function in different processes of DNA metabolism which includes DNA replication and repair. Characterization of Exo1 has been most focused on the yeast homologue. The full-length human exonuclease1 has only been partially characterized. We report here, the biochemical studies on the full-length hExo1. We show that the stability and resection activity of hExo1 are affected by factors such as ionic strength, pH, and temperature. We determined the substrate preference of hExo1 by assaying its resection activity on different types of DNA substrates, our result show that hExo1 preferably degrades 3’ overhang dsDNA substrate, suggesting the preference of hExo1 resection activity to 3’ overhang dsDNA substrate.
Keywords
hExo1
Resection
Double strand break
Characterization
- ATM
-
Ataxia Telangiectasia Mutated
- ATR
-
Ataxia Telangiectasia and Rad3-related protein
- CtIP
-
C-terminal Binding Protein-Interacting protein
- DSB
-
Double Strand Break
- DDR
-
DNA Damage Response
- FEN1
-
Flap Endonuclease1
- GEN1
-
Gap Endonuclease 1
- hExo1
-
Human Exonuclease1
- HR
-
Homologous Recombination
- MMR
-
Mismatch Repair
- MRX/MRN
-
Mre11–Rad50–Xrs2/Mre11–Rad50–Nbs1
- NHEJ
-
Non-homologous End Joining
- PCNA
-
Proliferating Cell Nuclear Antigen
- PCR
-
Polymerase Chain Reaction
- STR
-
Sgs1–Top3–Rmi1
- XPG
-
Xeroderma Pigmentosum Complementation Group
Abbreviations
1 Introduction
Human exonuclease1 (hExo1) is involved in various processes of DNA metabolism such as DNA replication, homologous recombination (HR), mismatch repair (MMR), and micro-mediated end-joining (Szankasi and Smith, 1995; Schmutte et al., 1998; Jäger et al., 2001; Tran et al., 2004; Bardwell et al., 2004; Morin et al., 2008). Human Exo1 is a member of Rad2/XPG, FEN1 and GEN1 nucleases found in eukaryotes (Lee et al., 2002; Orans et al., 2011). Human Exo1 can process double stranded DNA (dsDNA) (Lee and Wilson, 1999; Genschel and Modrich, 2003). DNA double strand break (DSB) is one of the most cytotoxic classes of DNA damage (Aguilera and Gomez-Gonzalez, 2008). Prompt detection and precise repair of DSB is requisite to preserving the integrity of the genome. The basic step in DSB repair is DNA end resection. The proposed two-step DSB resection model (Zhu et al., 2008; Gravel et al., 2008; Mimitou and Symington, 2008) involves an initial short resection to generate 3′ overhang followed by extended resection that activates HR, ATR checkpoint and subsequently DSB repair. MRX/MRN and Sae2/CtIP executes the initial short resection at the point of DNA break using their endonucleolytic activity to cleave the 5′ strand. Although, DSB with ‘clean’ ends do not necessarily require the initial short resection step, it is however, necessary to resect DSB with covalently bound proteins and chemical adducts at their 5′ ends. Two nucleases that carries out an extended long-range resection are Exo1 and Dna2 (Huertas, 2010; Mimitou and Symington, 2008; Mimitou and Symington, 2009; Zhu et al., 2008). The long range DNA end resection is achieved through two pathways that are parallel to one another. One pathway depend on the 5′→3′ Exo1 activity while the other pathway rely on Dna2 endonuclease coupled with Sgs1–Top3–Rmi1 (STR) complex (Mimitou and Symington, 2008; Zhu et al., 2008; Gravel et al., 2008). Insufficient resection of broken double stranded DNA hinders HR and ATR checkpoint activations, hence truncate DSB repair which can cause deleterious consequences. Whereas, uncontrolled resection can cause negative effects due to unstable nature of single stranded DNA (ssDNA) and vulnerability to degradation. DNA resection is regulated either positively or negatively by factors such as MRN, 14-3-3, PCNA, SOSS1, HELB, Shieldin, and DYNLL1 (Andersen et al., 2012; Chen et al., 2013; He et al., 2018; Mirman et al., 2018; Nimonkar et al., 2011; Noordermeer et al., 2018; Tkác et al., 2016; Yang et al., 2013). Human Exo1 is believed to be the enzyme that generate the required long 3′ single-stranded DNA overhangs for homology-based DNA repair and activation of the ATR-dependent checkpoint (Mimitou and Symington, 2008; Zhu et al., 2008; Gravel et al., 2008). Despite this crucial role of inducing the overall DNA damage response by hExo1, the biochemical characterization (in particular, the thermodynamic factors) of this important protein is still poorly understood.
To date, most biochemical studies of Exo1 concentrate on N-terminal region which is 395 out of the 846 amino acids present in the full-length Exo1 protein (Lee and Wilson, 1999; Lee et al., 2002; Orans et al., 2011). The crystal structure of N-terminal nuclease domain of the bacterial Exo1 protein has been established (Orans et al., 2011). Full-length human Exo1 has not been substantially characterized, although a limited study on its biochemical activity has been reported under different conditions of pH and salt (Bregenhorn and Jiricny, 2014). Substantial biochemical characterization of hExo1 is critical to understanding the essential mechanisms of its exonuclease resection activity (Sun et al., 2002; Schaetzlein et al., 2013; Shao et al., 2014; Bregenhorn and Jiricny, 2014). We present here, our findings on the biochemical characterization of hExo1. Our study focus on factors that influence protein stability. Our results revealed that full-length hExo1 is stable and more active on 3′ dsDNA substrate at temperature ranges between 25 °C and 37 °C distinctly with an increase in resection activity. In addition, we studied the substrate preference of hExo1 in its resection activity by using three different types of DNA substrate. This study report that hExo1 prefers a 3′ overhang dsDNA substrate for its resection activity. It has proven difficult to express and purify the full-length human exonuclease1 protein from bacteria. This work reported an E. coli based expression system to express and purify hExo1 protein.
2 Materials and methods
Plasmid pTXB1 (New England Biolab) was used for expression of hExo1. The coding region of hExo1 (codons 1-846) was amplified by PCR using a full-length clone for the human Exo1 splice variant (hExo1 cDNA) in a pFastBacI plasmid as a template DNA. Two synthetic oligonucleotides, 5′-GGGAATTCCATATGGGGATACAGGGATTGCTA-3′ and 5′-AAGGCCGCTCTTCCGCACATCTGGAATATTGCTCTTTGAACACGG-3′ that contained NdeI and SapI restriction sites were used as forward and reverse primers respectively. PCR product was digested with NdeI and SapI then ligated into pTXB1 plasmid. E. coli BL21 was transformed with the construct (pTXB1-hExo1) and grown in LB medium containing 25 µg/ml chloramphenicol and 100 µg/ml ampicillin. The cell culture was allowed to reach OD600 of 0.3 before protein expression was induced by adding 0.2 mM isopropylthiogalactoside (IPTG). Culture was allowed to grow overnight at 18 °C in a shaking incubator at 250 rpm. Cells were harvested at 18000 rpm for 45 min at 4 °C in JA-20 rotor.
Protein purification was conducted with some modifications to the method described by El-Shemerly, et al. (2005). CH buffer (20 mM Tris pH 8.0, 500 mM NaCl, 0.1% Triton X100, 1 mM EDTA, 10% glycerol) was used to re-suspend bacterial pellets. The bacterial suspension was sonicated and centrifuged at 18000 rpm for 45 min at 4 °C in JA-20 rotor. The cleared supernatant containing the fusion protein of interest was poured into a chitin resin (New England Biolabs) which was pre-equilibrated with CH buffer supplemented with PMSF and allowed to gravity flow (∼0.5 mL/min). The resin was washed using 200 mL CH buffer in the presence of PMSF. Intein affinity tag was removed from the recombinant hExo1 by proteolytic cleavage using 5 mL of CH buffer in the presence of PMSF and 30 mM DTT. The chitin resin containing bound hExo1 were stirred gently with a sterile glass rod and allowed to incubate over night at 4 °C. The fraction containing hExo1 was recovered by gravity flow and further purified by size exclusion with Superose 12 10/300 column (GE Healthcare Life Sciences) using AKTA prime protein purification system. Purified hExo1 was stored at −80 °C until when needed.
Resection assays were carried out with minor modifications to the method described by Nicolette et al, (2010). A 4.8-kb pFastBac-HTA plasmid was linearized with SphI restriction endonuclease, which generated a 4 nucleotides long 3′ overhang dsDNA. A resection reaction containing 10 nM hExo1 protein and 20–50 nM linear dsDNA in a reaction buffer (20 mM HEPES pH 7.0, 100 mM KCl, 5 mM MgCl2, 1 mM ATP, 10% v/v glycerol) was set up to a final volume of 20 µL on ice. Reaction was activated by transferring the tube to 37 °C and incubated for 1 hr. Reaction was terminated by adding stop buffer (2% w/v SDS, 10 mM EDTA) in the presence of 1 µg proteinase K and incubated for 1 h at 37 °C. Reaction products were separated on 1% agarose gels stained with SYBR gold and visualized with Gel Doc XR + system (BioRad).
3 Results
Human Exo1 was purified to near homogeneity (Fig. 1A). The molecular weight of hExo1 is 94 kDa, but in the presence of sodium dodecyl sulfate it migrated similar to proteins larger than its size (about 110 kDa) (Fig. 1A), this migration behaviour of hExo1 was observed in previous studies (Genschel et al., 2002; El-Shemerly et al., 2005; Chen et al., 2013; Bregenhorn and Jiricny, 2014). This could be due to the unusual amount of charged amino acids present in the protein that can affect the amount of SDS bound to the protein during electrophoresis and hence lead to unusual migration. A more detailed observation of hExo1′s substrate preference revealed that this protein preferably degrade a 3′ overhang substrate (Fig. 1B, lane 2) while failing to degrade either of the 5′-overhang (generated using BamHI restriction enzyme) or blunt end DNA substrates (generated using StuI restriction enzyme) as no detectable resection product was observed (Fig. 1B, lanes 3 and 4 respectively).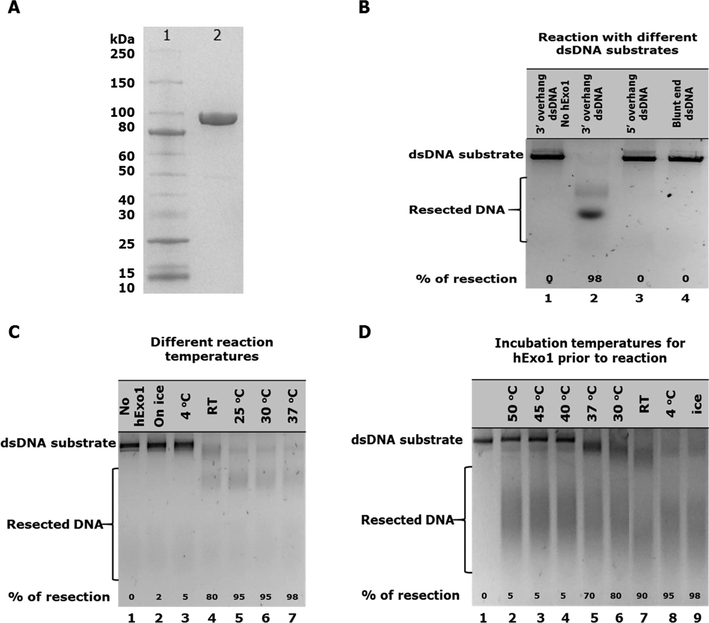
(A) SDS/PAGE analysis of 10 μg purified hExo1 protein lane 2, lane 1 is protein ladder (B) Substrate specificity assay showing hExo1 exonuclease resection activities on different dsDNA substrate types. Lane 1 was a substrate control; no hExo1 enzyme added. Lanes 2–4 contained 10 nM hExo1 and 30 nM dsDNA substrates. Reaction was activated by transferring reaction tubes to 37 °C and incubated for 60 min. (C) Temperature sensitivity assay showing hExo1 exonuclease resection activities on a 3′ overhang dsDNA substrate at different temperatures. Lane 1 was substrate control, no hExo1 enzyme was added. Lanes 2–7 contain 10 nM hExo1. Reaction was activated by transferring reaction tubes to different assay temperatures and incubated for 60 min. (D) Temperature stability assay showing hExo1 exonuclease resection activities on a 3′ overhang dsDNA substrate after incubation of hExo1 at different temperatures prior to reaction set up. Lane 1 was substrate control, no hExo1 enzyme added. Lanes 2–9 contain 10 nM pre-incubated hExo1 and 20 nM dsDNA substrate. Reaction was activated by transferring reaction tubes to 37 °C and incubated for 60 min.
To examine if hExo1 is temperature sensitive, hExo1 activity was tested at different temperatures as follows: ice, 4 °C, room temperature (RT), 25 °C, 30 °C and 37 °C. Maximum resection activity of hExo1 was observed at 37 °C (Fig. 1C, lane 7), although hExo1 resected the DNA substrate almost completely at 25 °C and 30 °C (Fig. 1C, lanes 5 and 6 respectively), activity at these temperatures is less than the one observed at 37 °C. At room temperature, a good resection activity was also observed even though the substrate was not degraded completely (Fig. 1C, lane 4). No degradation of substrate was observed when the reaction was set up on ice and at 4 °C (Fig. 1C, lanes 2 and 3 respectively). These results indicates that hExo1 is enzymatically inactive at low temperature but partially active at room temperature and maximally active at 37 °C. Further to this, we investigated the stability of hExo1 at different temperatures. To determine the stability of hExo1 protein at various temperatures, prior to setting the resection reaction with DNA substrate, hExo1 was incubated at various temperatures as follows: 50 °C, 45 °C, 40 °C, 37 °C, 30 °C, 25 °C, 4 °C and on ice for one hour. After the incubation, resection assays were set up and the products of the reactions were analysed. Little degradation of the DNA substrate was observed from hExo1 that was incubated at 50 °C, 45 °C, and 40 °C (Fig. 1D, lanes 2, 3 and 4 respectively), partial substrate degradation was seen from hExo1 incubated at 37 °C (Fig. 1D, lane 5) while more activity was observed from hExo1 incubated at 30 °C (Fig. 1D, lane 6). When hExo1 was incubated at room temperature prior to setting up reaction, the activity of hExo1 was not drastically affected as more substrate was degraded compared to when hExo1 was incubated at other higher temperatures than room temperature (Fig. 1D, lane 7). From lanes 8 and 9 in Fig. 1D, it is clearly seen that hExo1 activity was maximum as substrate was degraded almost completely. These results revealed that hExo1 is less stable at temperatures higher than 37 °C.
The ionic strength of hExo1 during DNA resection activity was tested. Human Exo1 exhibited maximum resection activity in reaction buffers containing KCl concentration in the range 50 mM to 150 mM (Fig. 2A, lanes 2, 3 and 4). At 200 mM KCl concentration, hExo1 activity decreased slightly (Fig. 2A, lane 5). Resection activity of hExo1 reduced drastically when the reactions were conducted in the presence of between 250 and 500 mM KCl concentrations (Fig. 2A, lanes 6–9). Likewise, at low KCl concentrations (5 and 10 mM KCl) hExo1 resection activity was decreased significantly (Fig. 2A, lanes 10 and 11). Biochemical factors such as pH have been reported to modulate the nuclease activity of both full-length and truncated Exo1 protein (Lee and Wilson, 1999; Bregenhorn and Jiricny, 2014). To determine if full length hExo1 is sensitive to pH, hExo1 resection activity was tested at varying pH conditions of the reaction buffer as follows: pH 5, 6, 7, 7.5, 8, and 9. It was observed that hExo1 resection activity was optimum at pH 7.5 and 8 (Fig. 2B, lanes 4 and 5 respectively). We investigated the role of reaction incubation time to ascertain how long hExo1 is active in the reaction. Six different resection reactions were set up with enough substrate (50 nM) and allowed to run for 1, 5, 10, 15, 30, and 60 min. Reaction products revealed that hExo1 remained active over a period of 60 min and degraded the substrate almost completely after 60 min (Fig. 2D, lane 7). Although most of the substrate was degraded after 30 min (Fig. 2D, lane 6), these data suggests that hExo1 remain active in reaction mixture for up to 60 min at 37 °C.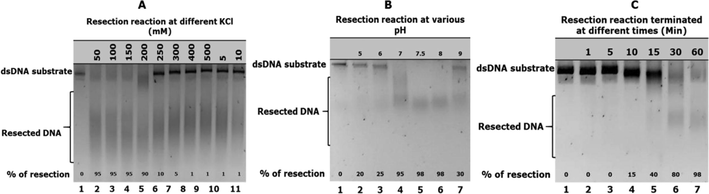
(A) Ionic strength assay showing exonuclease resection activities on a 3′ overhang dsDNA substrate. Lane 1 was substrate control, no hExo1 enzyme added. Lanes 2–11 contain 10 nM of hExo1 and 20 nM dsDNA substrate at different salt concentrations in the reaction buffer, reaction was activated by transferring reaction tubes to 37 °C and incubated for 60 min. (B) pH sensitivity assay showing hExo1 exonuclease resection activities on a 3′ overhang dsDNA substrate. Lane 1 was a substrate control; no hExo1 enzyme added. Lanes 2–7 contained 5 nM hExo1 and 10 nM dsDNA substrate, reaction buffer was set at different pH as indicated. Reaction was activated by transferring reaction tubes to 37 °C and incubated for 60 min. (C) Time course assay showing hExo1 exonuclease resection activities on a 15 nM 3′ overhang 4.8 kb dsDNA substrate at different time course. Lane 1 was a substrate control; no hExo1 enzyme was added. Lanes 2–7 contained 10 nM hExo1 and 50 nM dsDNA substrate. Reactions were incubated at 37 °C and terminated at different times as indicated.
4 Discussion
Understanding the biochemical and thermodynamic properties of a protein is the basis to molecular understanding of its structure and function. There has been limited literature reports describing in clear terms the characterization of full-length hExo1 protein. However, recent biochemical studies on the MMR-deficient phenotype of the Exo1-Glu-109-Lys mouse has highlighted the importance of such studies. These studies reported that the mutant Exo1 and wild-type protein have similar exonuclease activity (Eid et al., 2010; Shao et al., 2014). This is further evidence that, characterization of Exo1 is still incomplete. Shao and colleagues have recently suggested that tagging of Exo1 at the NH2-terminal hindered its nuclease activity (Shao et al., 2014). We, used non-affinity tagged full-length hExo1 in this study.
A more detailed look at the substrate preference of hExo1 revealed that this protein preferably degrade a 3′-overhang duplex substrate (Fig. 1B, lane 2), no substrate degradation was observed when a 5′-overhang dsDNA or blunt end were used as substrates (Fig. 1B, lanes 3 and 4 respectively). Lee and Wilson (1999) reported that the truncated Exo1 (HEX1-N2) degrades both 5′ and 3′ 32P labelled blunt end duplex substrates (Lee and Wilson, 1999). The disagreement between the results found in our study and that of the previously reported (Lee and Wilson, 1999) with regards to substrate specificity of hExo1 may be due to the fact that this study used a full-length protein, and persistently checked its integrity before and after the assay by SDS/PAGE analysis. However, this does not appear to be the case in other assays. There is also a mixture of conditions in the published assays: some were done with protein of unspecified purity but this study used a single band purity. In contrast to what Keijzers and colleagues (2015) reported that the enzymatic activity of Exo1 increased by about 2-fold at 25 °C and 30 °C compared to at 37 °C (Keijzers et al., 2015), we observed that hExo1 exhibited optimal exonuclease resection activity at 37 °C when exonuclease resection activity of hExo1 was tested at different temperature conditions (Fig. 1C). In addition, this study observed that hExo1 protein is not stable at temperature ranges from 40 °C to 50 °C when the protein was pre-incubated at different temperature conditions prior to setting the resection reaction (Fig. 1D). When hExo1 was pre-incubated at 4 °C and on ice it showed high resection activity (Fig. 1D, lanes 8 and 9) compared to when it was pre-incubated at 37 °C (Fig. 1D, lane 5). A drastic decrease in resection activity was observed when hExo1 was pre-incubated at 40 °C, 45 °C or 50 °C prior to resection assay. This is further evidence that hExo1 is sensitive to high temperature and these observations agrees to what was previously reported (Keijzers et al., 2015).
The ionic strength assay for full-length hExo1 was conducted in order to understand the effect of salt in its enzymatic function. In previous studies it was reported that truncated HEX-N2 which represents 1–395 amino acids of the full length Exo1, exhibited optimal activity at 50 mM KCl (Lee and Wilson, 1999). Likewise, in recent studies, full-length Exo1 was reported to show a decreased in nuclease activity at 40 mM and 180 mM KCl (Bregenhorn and Jiricny 2014). Similarly, Keijzers et al. (2015) reported that the ionic strength of full-length is in the range of 50 mM to 150 mM KCl and 200–250 mM KCl decreased the nuclease activity of Exo1. Our study revealed that the ionic strength of the full-length hExo1 is in the range from 50 mM to 200 mM KCl with a slight decreased in resection activity at 200 mM KCl, therefore, hExo1 is active under physiological salt conditions (Fig. 2A). High concentrations of KCl (250–500 mM) reduced hExo1 exonuclease resection activity (Fig. 2A). Likewise, low concentrations of KCl (5 mM and 10 mM) drastically reduced hExo1 enzymatic exonuclease resection activity (Fig. 2). The enzymatic requirements of hExo1 revealed that the pH range for its optimal exonuclease resection activity is in the range 7–8 (Fig. 2B). To determine how long hExo1 protein remain active in the reaction, the role of resection reaction incubation time was investigated in this study. The resection reaction was assayed at 37 °C and found that hExo1 degraded 3′ overhang dsDNA substrate for up to 60 min. Our data shows that hExo1 remain active in resection reaction mixture for up to 60 min at 37 °C (Fig. 2C). These data contradicts what was reported previously that hExo1 became inactive after 10 min of starting the reaction at 37 °C (Keijzers et al., 2015). This may be due to different substrates used in both studies, while a 5′-radioactive labelled 42-mer dsDNA was used as a substrate (Keijzers et al., 2015), here a linearized 4.8 kb non radiolabelled dsDNA was used, which is a much larger target DNA substrate and hence there is more substrate upon which to act (10 nM hExo1 protein against 50 nM dsDNA substrate). These observations are significant in designing in vitro experiments that involve hExo1.
5 Conclusion
We provide more insight into the thermodynamic parameters of hExo1. Our study revealed that hExo1 has strong in vitro DNA resection activity. We show that resection activity of hExo1 is modulated by factors such as pH, temperature and ionic strength. Additionally, our results suggests that hExo1 prefers a 3′ overhang dsDNA substrate. This is very significant to the biochemical characterization studies of hExo1, an enzyme with critical role in the process of maintaining the stability of human genome through the process DNA resection during the repair of DNA lesion.
Author’s contributions
The research was conceived by Aminu A. Umar and Abubakar Abdulhamid. Ahmad Ibrahim Bagudo helped with protein expression studies and Ibrahim Muhammad Magami aided in interpretation of enzymatic assays.
Acknowledgement
We thank Paul Modrich for human Exonuclease1 clone.
Funding
This project received part funding from Nigerian government under Tertiary Education Trust Fund (TetFund) through Kebbi State University of Science and Technology, Aliero as part of staff training and development.
Conflict of interest
We have no conflicts of interest to declare.
References
- Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet.. 2008;9:204-217.
- [Google Scholar]
- 14-3-3 checkpoint regulatory proteins interact specifically with DNA repair protein human exonuclease 1 (hEXO1) via a semi-conserved motif. DNA Repair (Amst.). 2012;11:267-277.
- [Google Scholar]
- Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol.. 2004;5:224-229.
- [Google Scholar]
- Biochemical characterization of a cancer-associated E109K missense variant of human exonuclease 1. Nucleic Acids Res.. 2014;42:7096-7103.
- [Google Scholar]
- PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res.. 2013;41:9325-9338.
- [Google Scholar]
- DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep.. 2010;11:962-968.
- [Google Scholar]
- Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res.. 2005;65:3604-3609.
- [Google Scholar]
- Human exonuclease I is required for 5’ and 3’ mismatch repair. J. Biol. Chem.. 2002;12:13302-13311.
- [Google Scholar]
- Mechanism of 5’-directed excision in human mismatch repair. Mol. Cell. 2003;12:1077-1086.
- [Google Scholar]
- DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev.. 2008;22:2767-2772.
- [Google Scholar]
- DYNLL1 binds to MRE11 to limit DNA end resection in BRCA1-deficient cells. Nature 2018
- [CrossRef] [Google Scholar]
- DNA resection in eukaryotes: deciding how to fix the break. Nat Struct Mol Biol. 2010;17:11-16.
- [Google Scholar]
- HNPCC mutations in the human DNA mismatch repair gene hMLH1 influence assembly of hMutLalpha and hMLH1-hEXO1 complexes. Oncogene. 2001;20:3590-3595.
- [Google Scholar]
- Human exonuclease 1 (EXO1) activity characterization and its function on flap structures. Biosci. Rep.. 2015;35:e00206.
- [CrossRef] [Google Scholar]
- Molecular interactions of human Exo1 with DNA. Nucleic Acids Res.. 2002;30:942-949.
- [Google Scholar]
- The RAD2 domain of human exonuclease 1 exhibits 5’ to 3’ exonuclease and flap structure-specific endonuclease activities. J. Biol. Chem.. 1999;274:37763-37769.
- [Google Scholar]
- Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature. 2008;455:770-774.
- [Google Scholar]
- DNA end resection: many nucleases make light work. DNA Repair (Amst). 2009;8:983-995.
- [Google Scholar]
- 53BP1/Rif1/Shieldin counteract DSB resection through CST/Polα-dependent fill-in. Nature. 2018;560(7716):112-116.
- [Google Scholar]
- Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J.. 2008;27:2400-2410.
- [Google Scholar]
- Mre11-Rad50- Xrs2 and Sae2 promote 5' strand resection of DNA double-strand breaks. Nat. Struct. Mol. Biol.. 2010;17:1478-1485.
- [Google Scholar]
- BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev.. 2011;25:350-362.
- [Google Scholar]
- The Shieldin complex mediates 53BP1-dependent DNA repair. Nature. 2018;560(7716):117-121.
- [Google Scholar]
- Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212-223.
- [Google Scholar]
- Mammalian Exo1 encodes both structural and catalytic functions that play distinct roles in essential biological processes. Proc. Natl. Acad. Sci. U.S.A.. 2013;110:E2470-E2479.
- [Google Scholar]
- Human exonuclease I interacts with the mismatch repair protein hMSH2. Cancer Res.. 1998;58:4537-4542.
- [Google Scholar]
- Hydrolytic function of Exo1 in mammalian mismatch repair. Nucleic Acids Res.. 2014;42:7104-7112.
- [Google Scholar]
- Functional alterations of human exonuclease 1 mutants identified in atypical hereditary nonpolyposis colorectal cancer syndrome. Cancer Res.. 2002;62:6026-6030.
- [Google Scholar]
- A role for exonuclease I from S. pombe in mutation avoidance and mismatch correction. Science. 1995;267:1166-1169.
- [Google Scholar]
- HELB Is a Feedback Inhibitor of DNA End Resection. Molecular Cell. 2016;61:405-418.
- [Google Scholar]
- The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J.. 2013;32:126-139.
- [Google Scholar]
- Sgs1 helicase and two nucleases dna2 and exo1 resect DNA double-strand break ends. Cell. 2008;134:981-994.
- [Google Scholar]







