Translate this page into:
Biochemical and molecular characterization of lactase producing bacterium isolated from dairy effluent
⁎Corresponding author. vidyakodali1017@gmail.com (K. Vidya Prabhakar)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
In the present study, the microbial source for potent lactase producers was explored to supplement the lactase intolerant individuals. Dairy effluent was screened for lactase producing bacteria by conventional microbiological methods. Among the positive isolates, one isolate VUVD001 was found to be a strong producer of lactase enzyme and this strain identified by 16S rDNA analysis. The lactase producing bacterium is identified as Bacillus subtilis by biochemical and 16S rDNA analysis. The VUVD001 strain found to survive in a temperature range of 20–55 °C, pH range of 5–8 and a salt concentration up to 8%. Further, the partially purified enzyme preparation was tested by zymogram analysis for activity testing using x-gal and the enzyme activity of cell free supernatant was estimated to be 15.10 U/ml at 37 °C, 4% NaCl and pH of 7.0. Our finding reports a new isolate Bacillus subtilis VUVD001 strain which can be used as potential strain for commercial production of lactase.
Keywords
Lactase
Bacillus subtilis
ortho-nitrophenyl-β-galactoside (ONPG)
X-gal
16S rDNA
1 Introduction
Lactose intolerance is the inability to hydrolyze dietary lactose due to the deficiency of lactase (Schrezenmeir and deVrese, 2001; Jost et al., 1998). About, 75 percent of population in the world was suffered from lactose intolerance, which significantly decreases their quality of life (Pribila et al., 2000; Venkateswarulu et al., 2017a; Peele et al., 2018). The lactase deficiency could be resolved by supplementary probiotics. The microbial production of lactase enzyme was found in yeast, fungi and bacteria (Rosenberg, 2006; He et al., 2008). Currently, the dairy industry uses probiotic strains namely, Lactobacillus and Bifidobacterium for the production of lactase (Ganeva et al., 2001). However, Kluyveromyceslactis is the most important strain for lactase production due to their dairy environmental habitat. But, its major drawback is lower thermostability (Chen et al., 2008). The bacterium Lactobacillus acidophilus isolated from fermented Ragi (finger millet) produces thermostable lactase enzyme and this enzyme prevents microbial contamination in milk processing (Akolkar et al., 2006). Thus, the bacterial strains have considerable industrial potential for large scale production. The bacterial sources for production of lactase were extensively used for hydrolysis of lactose because of easy fermentation, high activity and good stability of enzyme (Picard et al., 2005; Natarajan et al., 2012; Venkateswarulu et al., 2017b). Lactase producing bacterial strains was used in treatment of milk based products in dairy industry (Sen and Srinivasa Babu, 2005). The enzyme activity was affected by different process variables like type of strain, cultivation conditions and the C:N ratio in the medium (Jurado et al., 2004). In the present study, isolation of lactase producing bacteria from a commercial dairy farm was attempted and also focused on lactase enzyme production from gram positive organism. The main aim of the present work is to screen the dairy effluent for isolation of potential microbial strain for bulk production of lactase such that the produced lactase from this isolate could be used as biotherapeutic agent for treating the patients with lactose intolerance.
2 Materials and methods
2.1 Screening and isolation of lactase producing bacteria
The bacterial strain, isolated from Sangam dairy industry effluent, Vadlamudi Village, Guntur (District), Andhra Pradesh, India. The serial dilutions of dairy effluent were done in sterile saline followed by pour plating on nutrient agar medium containing 1 mM IPTG and X-gal for the isolation of microorganisms. The solidified agar plates were incubated in inverted position at 37 °C for 36 h. Isolated colonies that are blue in color were sub-cultured in nutrient broth and incubated at 37 °C. The lactase producing ability of these strains was confirmed by using ONPG discs (Kalogridou-Vassiliadou, 1992; Sharma and Singh, 2014).
2.2 Morphological and biochemical characterization of lactase producing isolate
The lactase positive isolate was tested for biochemical characterization. The cell morphology and motility of isolate were examined by light microscopy (Bartholomew et al., 1965). Spore-staining was done on the basis of Schaeffer-Fulton method. Smears were made with 24 h old cultures and fixed with gentle heat. The smear was immersed in malachite green stain for 3 to 6 min at 37 °C in dark followed by wash with running tap water and counter stained with Schaeffer & Fulton spore stain-B for 30 s. The smear was then air dried and viewed under a light microscope with 40× resolution.
2.3 Temperature, pH and salt tolerance tests
The isolate was tested for growth tolerance at different temperatures ranging from 20 °C to 55 °C and at different pH values i.e. 5 to 9. The isolate was also tested to determine stability of the bacterium at different sodium chloride concentrations at 2%, 4%, 6%, 8% and 10%.
2.4 Zymogram analysis for lactase activity
Zymogram assay for cell free extract of isolated strain was performed using PAGE method with 10% polyacrylamide gel. After electrophoresis, gel was incubated in 0.25 mM X-gal solution at room temperature for 10 min and the hydrolysis of X-gal is confirmed by formation of blue band on the gel (Trimbur et al., 1994).
2.5 Lactase enzyme assay
The assay was performed with chromomeric substrate namely: ortho-nitrophenyl-β-galactoside (ONPG). The assay was initiated with mixing of 0.5 mL of enzyme source and 2.0 mL of substrate, followed by incubation for 30 min. Then, 0.5 mL of 1 M Na2CO3 was added to stop the reaction and absorbance measured at 420 nm. Lactase enzyme activity, determined from ONP standard graph (Ghosh et al., 2012; Venkateswarulu et al., 2017c).
2.6 Molecular characterization of the lactase producing isolate
2.6.1 Bacterial DNA isolation
The standard protocol of Hoffman and Winston was used for bacterial DNA extraction (Hoffman and Winston, 1987). Lactase producing bacterium grown in broth medium for 24 h at 37 °C. The cells were collected from 5 mL of the broth culture and then, lysozyme (100 μl) was added and incubated at room temperature for 30 min, followed by addition of 800 μl cell lysis buffer (1x TE buffer (pH-8.4); 1% SDS and 100 µg proteinase K). The nucleic acids separated with two rounds of extraction with phenol: chloroform: isoamylalcohol (25:24:1) mixture. Aqueous layer containing genomic DNA was separated and then 800 μl of isopropanol was added on top of the solution. The two layers were mixed gently to precipitate the genomic DNA and then, genomic DNA was harvested by centrifugation at 12,000 RPM for 10 min at 4 °C. The pellet was washed with 70% ethylalcohol and then, DNA pellet air dried, further dissolved in 50 µl of 1X TE buffer (Sen et al., 2010).
2.6.2 PCR amplification
16S rDNA fragment amplification, done in a gradient thermal cycler (Eppendorf, Germany) with the primers: 27F and 1429R (Frank et al. 2008). The reaction mixture of 100 ng DNA, 2.5 Units Taq DNA polymerase, 5 μl of 10X PCR amplification buffer, 200 μM of dNTPs, 10 pico moles each of the two universal primers and 1.5 mM MgCl2. Amplification was performed under the conditions of initial denaturation for 3 min at 94 °C and then subjected to 40 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and elongation at 72 °C for 2 min and a final extension conducted at 72 °C period of 10 min. The amplicon of 16S rDNA was confirmed by agarose gel electrophoresis.
2.6.3 DNA sequencing
DNA sequence is performed at Helini Biomelecules Pvt Ltd at Chennai, India. Obtained DNA sequence subjected to BLAST analysis through PubMed database (http:// www.ncbi.nlm.nih.gov/blast) using BLASTN and then, compared with other sequences for taxonomical identity of bacterial strain through phylogeny.
3 Results and discussion
3.1 Characterization of lactase producing isolate
Lactase producing bacterium was isolated and morphology of isolate was identified as gram-positive and spore forming by microscopic analysis. The lactase producing activity was detected by ONPG disc method. It was observed that the reaction mixture turned deep yellow color after incubation, which indicates that our bacterial strain has the ability to hydrolyze ONPG into ONP (Fig. 1). Further, the fermentation ability of isolate was also tested with different sugars. The isolate is capable of utilizing various carbon sources namely glucose, lactose, sucrose, starch and fructose and the results were compared with previous studies. Sharma et al., (2014) reported similar results for metabolization of various sugars like dextrose, maltose, lactose, sucrose and starch as carbon sources by Lactobacillus delbrueckii for production of lactase. The isolate was able to survive in temperature ranging from 20 to 55 °C with best growth achieved at 37 °C. The growth efficiency of isolate in salt rich environment was studied by salt tolerance test. The isolate showed positive response to different concentrations of sodium chloride such as 2%, 4%, 6% and 8% and the maximum growth was observed with 4% NaCl. In previous studies, reported that the lactic acid bacterium is able to tolerate up to 4% NaCl concentrations (Hudaaneetoo et al., 2015). But our newly isolated strain was showed significant growth up to the concentration of 8% NaCl. Similarly, in the earlier studies the salts like NaCl and MgCl2 were proved as growth influencing factors (Pulicherla et al., 2013). The isolate was able to survive at pH values ranging from 5.0 to 8.0 and optimum proliferation of isolate was found at pH 7. The maximum growth found at pH 6.8 for lactase producing bacterial species namely L. delbrueckii subsp. bulgaricus ATCC11842 and B. animalis subsp. lactis Bp12 (Prasad et al., 2013). The lactase activity 7.76 U/mL was reported from Streptococcus thermophilus strain (Princely et al., 2013). Similarly, Mozumder et al., (2012) reported the production of lactase, 0.85 U/mL from Lactobacillus species isolated from the yogurt. However, VUVD001 strain isolated dairy effluent produced higher lactase activity (15.10 U/mL) in comparison with other microbial sources and also this strain is not reported earlier for production of lactase.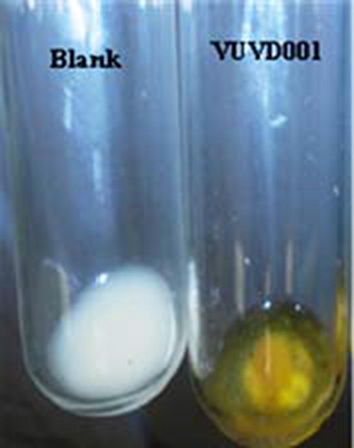
ONPG disc method for screening of lactase producing bacteria.
3.2 Zymogram analysis
Lactase activity in the crude cell free extracts of B. subtilis VUVD001 strain was confirmed through zymogram assay. The native PAGE gel stained with chromogenic X-gal and the enzyme performed hydrolysis of X-gal which was observed in gel with distinct blue color band (Fig. 2). Similar method was reported by Favier et al. (2011) in the biochemical screening of bifidobacterial strain for lactase activity. Previous reports have been revealed that the X-gal hydrolysis through zymogram assay was used for confirmation of lactase producing nature of a cold-adapted bacterium named as Rahnella aquatilis KNOUC 601 (Nam and Ahn, 2011).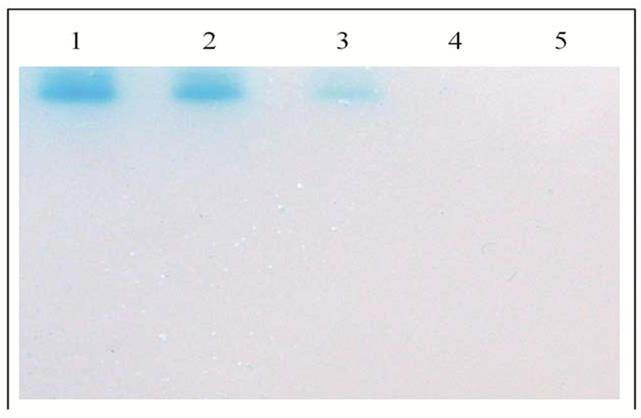
Zymogram analysis of concentrated cell free extracts of B. subtilis VUVD001 isolate. Lane 1 & 2 represents X-gal hydrolyzing enzyme from B. subtilis VUVD001 grown at different temperatures; Lane 3 – positive control (Lactobacillus sp. ATCC-8008); Lane 4 – Negative control (B. cereus ATCC-10876); Lane 5-blank.
3.3 Identification of the lactase producing isolate
The PCR amplification of ribosomal DNA was performed with universal primers of rDNA which produced amplicon of 1400 bp (Fig. 3). Since DNA sequencing was performed directly on PCR amplicon, a 1000 bp DNA was obtained after merging forward and reverse sequencing reads. This highly conserved region of 16S rDNA was identified by blast search and multiple sequence alignment was carried out with other Bacillus species sequences obtained from Genbank. Evolutionary analysis was conducted using MEGA5 and the evolutionary history was inferred by using maximum likelihood method. Bootstrap score indicates the reliability of branches, in the tree lactase producing bacterial isolate was represented as VUVD001. The phylogenetic tree clearly showed that the VUVD001 is 77% similar to Bacillus subtilis strain DBT13 from Genbank database (Fig. 4). The sequencing data of the strain was submitted to National Centre for Biotechnology Information (NCBI) and it was accepted as new strain of Bacillus subtilis strainVUVD001, NCBI-Genbank No: KT894158.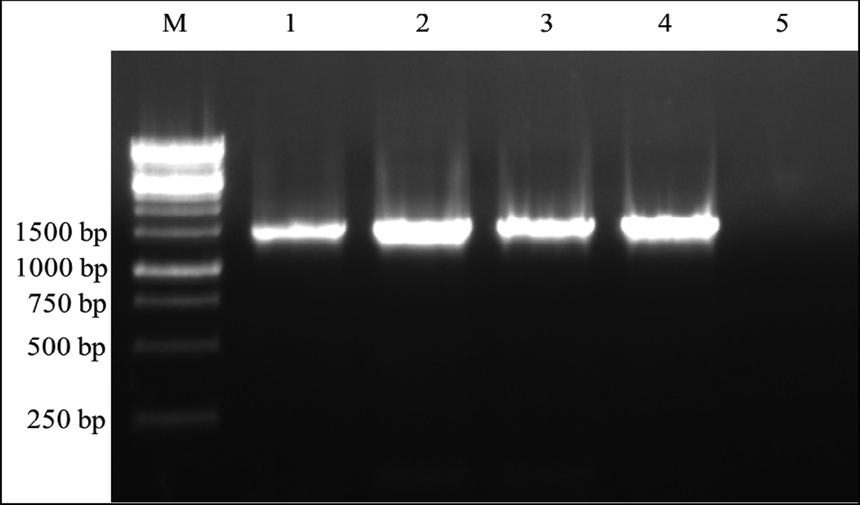
Agarose gel analysis of PCR amplified 16srDNA gene sequences. Lane M-1 KB ladder; Lane 1-16srDNA of B. subtilis VUVD001; lane 2- B. cereus ATCC-10876; lane 3-Staphylococcus aureus NCIM-2122; Lane 4-Escherichia coli K-12.
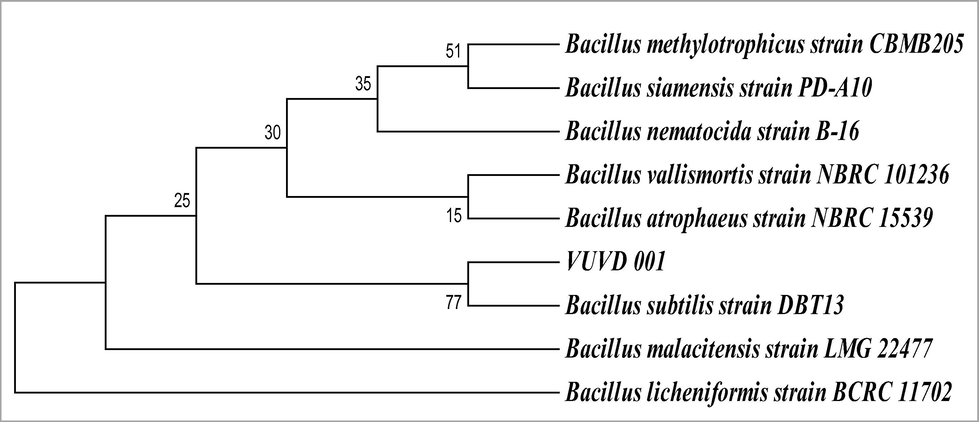
The phylogenetic tree constructed by maximum-likelihood method and is showing the position of strain VUVD 001.
4 Conclusion
The native isolate of B. subtilis VUVD001 strain possessing prominent lactase activity. Since B. subtilis is considered as generally recognized and safe (GRAS) by FDA there is a possibility for utilizing this strain for industrial level cultivation for harvesting lactase enzyme. The bacterial species present in dairy industry effluents have probiotic properties. The newly isolated bacterium could be used as promising strain for commercial production of lactase.
Acknowledgements
Dr. KVP acknowledges Science and Engineering Research Board (SERB), Government of India for the financial support (ECR/2016/000062). Authors also acknowledge DST-FIST (2015-20) support to the Department of Biotechnology, VFSTR.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- β-galactosidase from Lactobacillus acidophilu isolated from fermented ragi (Eleusine coracan) Indian J. Biotechnol.. 2006;5:184-188.
- [Google Scholar]
- Differential spore & lipid staining at room temperature by using fluorescent dye. J. Bacteriol.. 1965;90:1146-1147.
- [Google Scholar]
- Production, purification, and characterization of a potential thermostable galactosidase for milk lactose hydrolysis from Bacillus stearothermophilus. J. Dairy Sci.. 2008;91:1751-1758.
- [Google Scholar]
- Fecal β-D-galactosidase production and Bifidobacteria are decreased in crohn's disease. J. Microbiol. Methods. 2011;279:25-31.
- [Google Scholar]
- Critical evaluation of two primers commonly used for amplification of bacterial 16S rRNA genes. Appl. Environ. Microbiol.. 2008;74:2461-2470.
- [Google Scholar]
- Electroinduced extraction of β-galactosidase from Kluyveromyces lactis. Appl. Microbiol. Biotechnol.. 2001;56:411-413.
- [Google Scholar]
- Cold active β-galactosidase from Thalassospira sp. 3SC-21 to use in milk lactose hydrolysis: a novel source from deep waters of bay-of-bengal. World J. Microbiol. Biotechnol.. 2012;28:2859-2869.
- [Google Scholar]
- Effects of yogurt and Bifidobacteria supplementation on the colonic microbiota in lactose-intolerant subjects. J. Appl. Microbiol.. 2008;104:595-604.
- [Google Scholar]
- A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267-272.
- [Google Scholar]
- Isolation, characterization and determination of probiotic properties of lactic acid bacteria from effluents of a dairy plant in Mauritius. Int. J. Biol. Pharm. All.-Sci.. 2015;4:320-333.
- [Google Scholar]
- Lactase is unchanged in suckling mice fed with lactose-free milk. Gastroentérologie Clinique et Biologique. 1998;22:863-867.
- [Google Scholar]
- Kinetic models of activity for β-galactosidases influence of pH, ionic concentration and temperature. Enzyme Microb. Technol.. 2004;34:33-40.
- [Google Scholar]
- Biochemical activities of Bacillus species isolated from flat sour evaporated milk. J. Dairy Sci.. 1992;75:2681-2686.
- [Google Scholar]
- Study on isolation and partial purification of lactase (β-galactosidase) enzyme from Lactobacillus bacteria isolated from yogurt. J. Sci. Res.. 2012;4 239–239
- [Google Scholar]
- Isolation and characterization of cold-adapted bacteria producing lactose hydrolyzing enzyme isolated from soils of nome area in Alaska. Int. Res. J. Microbiol.. 2011;2:348-355.
- [Google Scholar]
- Isolation and charac/terization of β-galactosidase producing Bacillus sp. from dairy effluent. World Appl. Sci. J.. 2012;17:1466-1474.
- [Google Scholar]
- Plackett-Burman design for screening of process components and their effects on production of lactase by Newly Isolated Bacillus sp. VUVD101 strain from dairy effluent. Beni-Suef Univer. J. Basic Appl. Sci.. 2018;7:543-546.
- [Google Scholar]
- Bifidobacteria as probiotic agents – physiological effects and clinical benefits. Aliment. Pharmacol. Ther.. 2005;22:495-512.
- [Google Scholar]
- Extraction and characterisation of β-galactosidase produced by Bifidobacterium animalis spp. lactis Bb12 and Lactobacillus delbrueckii spp. bulgaricus ATCC 11842 grown in whey. Int. Food Res. J.. 2013;20:487-494.
- [Google Scholar]
- Improved lactose digestion and intolerance among african-american adolescent girls fed a dairy-rich diet. J. Am. Diet. Assoc.. 2000;100:524-528.
- [Google Scholar]
- Biochemical characterization, partial purification, and production of an intracellular beta-galactosidase from Streptococcus thermophilus grown in whey. Eur. J. Exp. Biol.. 2013;3:242-251.
- [Google Scholar]
- Statistical approach for the enhanced production of cold-active β-galactosidase from Thalassospira frigidphilosprofundus a novel marine psychrophile from deep waters of Bay of Bengal. Prep. Biochem. Biotech.. 2013;43:766-780.
- [Google Scholar]
- Current trends of β-galactosidase application in food technology. J. Food Nutri. Res.. 2006;45:47-54.
- [Google Scholar]
- Probiotics, prebiotics and synbiotics-approaching a definition. Am. J. Clin. Nutr.. 2001;73:361-364.
- [Google Scholar]
- Modeling and optimization of the process conditions for biomass production and sporulation of a probiotic culture. Process Biochem.. 2005;40:2531-2538.
- [Google Scholar]
- Molecular characterization and in vitro analyses of a sporogenous bacterium with potential probiotic properties. Probiotics Antimicrob. Proteins. 2010;2:152-161.
- [Google Scholar]
- Isolation and characterization of galactosidase enzyme producing microbe and optimization of its enzyme activity under different culture condition. Int. J. Curr. Microbiol. Appl. Sci.. 2014;3:148-155.
- [Google Scholar]
- Characterization of a psychrotrophic Arthrobacter gene and its coldactive beta-galactosidase. Appl. Environ. Microbiol.. 1994;60:4554-14552.
- [Google Scholar]
- Optimization of nutritional components of medium by response surface methodology for enhanced production of lactase. 3 Biotech. 2017;7:1-9.
- [Google Scholar]
- Modeling and optimization of fermentation variables for enhanced production of lactase by isolated Bacillus subtilis strainVUVD001 using artificial neural networking and response surface methodology. 3 Biotech.. 2017;7:1-7.
- [Google Scholar]
- Optimization of variables for lactase production from isolated Bacillus subtilis strainVUVD001 through submerged fermentation. Curr. Trend. Biotechnol. Pharm.. 2017;11:371-375.
- [Google Scholar]







