Translate this page into:
Bacterioplankton diversity in the estuarine regions of two peninsular rivers: A metagenomic approach
⁎Corresponding author. jishajacob@devagiricollege.org (Jisha Jacob)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Objective
In this present study we evaluated the bacterioplankton diversity employing a culture independent high throughput 16srRNA pyro-sequencing technique in two selected estuarine regions that belongs to two peninsular rivers on which the people living in the area heavily depend for their water needs. In spite of several advantages to metagenome based bacterioplankton diversity analysis, no such studies have been carried out in the region so far.
Methods
Samples were collected from three different sites for each estuaries. A comparative study on physico-chemical analysis was done on water samples from the estuaries using standard analytical methods. High-throughput sequencing reads obtained from metagenomic sequencing were classified into a taxonomic category using the NCBI taxonomy reference database.
Result
In the study we observed higher percentage composition of bacterioplanktons appertaining to the actinobacteria, cyanobacteria, and proteobacteria groups which can be attributed to higher levels of water nutrients such as nitrates, nitrites and phosphates. Firmicutes identified in the samples indicates the faecal contamination in the water. Apart from these numerous nitrogen-metabolizing bacteria, such as Roseobacter, Nitratiruptor, and Sulfitobacter, were present, as observed. One of the most common bacterioplankton species that is responsible for the breakdown of different types of organic matter is the Roseobacter. Similarly, nitratiruptor is a type of bacteria that reduces nitrite and can break down nitrate in water. Sulfitobacter and Pseudoruegeria are other two strains observed in both the Chaliyam and Dharmadam estuarine regions. It is reported that the Sulfitobacter strains are predominantly distributed in the areas of oil polluted marine environments;it has been associated with hydrocarbon removal from seawater.
Conclusion
Most of the diverse bacterioplanktons reported are capable of utilizing the dissolved organic matter for their growth and functioning. Isolation of such bacterial communities is having great significance in the aspects of waste water treatment and pollution abatement. We did not observe any major difference in diversity of particle associated bacterioplanktons in the two estuaries investigated. However, there was a significant variation observed in the diversity of free living bacterioplankton attributed to changes in physio-chemical parameters. Overall, metagenomics is a more effective and insightful way to examine microbial populations than standard culture techniques.
Keywords
Bacterioplankton
Metagenomics
Free-living
Particle-associated
Chaliyar river
Anjarakkandi river
1 Introduction
Diverse aquatic systems, comprising little streams, rivers, lakes, sea waters, and estuaries, are found throughout the Indian subcontinent. Among these, the Western Ghats, a significant supply of water for southern India, are the source of several significant rivers. (Khadse et al., 2010; Khadse et al., 2012; Ramachandra et al., 2018). As they regulate the nitrogen cycle and the disposal of trash, rivers and estuarine environments are essential to the maintenance of biodiversity. These rivers' estuarine regions are extremely productive, have a variety of habitats, and have distinct microbial populations.(Duarte et al., 2017; Julian and Osborne, 2018). Bacterioplanktons are free floating and play a significant part in functioning of global ecosystem through biogeochemical cycle, Primary producton and food web dynamics.These microbial communities are important in the regulation of particulate and dissolved organic carbon and thereby regulating the various mineral and nutrient cycling (Lønborg et al., 2019; Molina and Fernández, 2020, Mou et al., 2013).. Applications in industry and commerce include the targeted isolation of specific bacterial strains and the diversity study of both particle-associated and free-living bacterioplankton (Soares et al., 2018; Mou et al., 2015).
Analysing the distribution pattern of both free-living and particle-associated bacterioplankton is more important because the way these bacteria function continues to be a major factor in determining nutrient fluxes and concentrations because they fix carbon and nitrogen and assimilate and remineralize particulate and dissolved organic matter in the ecosystem. (Gómez-Consarnau et al., 2019; Mou et al., 2013). Consequently, the variety of bacterioplankton seen in aquatic environments can serve as an indication for the water's quality and degree of contamination in the body of water. (Salloto et al., 2012; Zhao et al., 2020). A variety of methods are used to analyse bacterioplankton, with culture-independent techniques being the most often used. (Li et al., 2019; Salloto et al., 2012, Robbins et al., 2011).
Microbial ecologists have identified more efficient techniques for analyzing microbial populations in recent times. The application of advanced sequencing technologies has enabled the examination of microbial group structures through amplicon sequencing and metagenomics. (Abia et al., 2018). The current study assessed the variety of bacterioplankton in the estuarine areas of two adjacent rivers, Chaliyar and Anjarakkandy, which originate in the Western Ghats of southern India, Kerala. Little was known about bacterioplankton diversity of the proposed water bodies, including those that are particle associated or free-living, despite their importance in maintaining the ecosystems. In order to analyse the bacterial makeup, the current work used high throughput pyro-sequencing techniques of the 16 s rRNA gene. To obtain a better understanding of how environmental circumstances affect the taxonomic composition and diversity of bacteria, a thorough physio-chemical examination of the water samples was conducted. It is crucial to study the diversity of bacterioplankton because it provides the answers to many questions about the health, resilience, and sustainability of our planet's essential water supplies as well as the intricate web of interactions that shape aquatic ecosystems.
2 Methods
2.1 Site and sampling
Water samples (Post monsoon) were collected from the esturine regions (Chaliyum and Darmadom) of Chaliyar and Anjarakandi River (Fig. 5). The sites were split up into three stations, and samples were collected from 0.5 m below the air–water interface using a depth sampler. As soon as the samples were collected, they were placed in a dark carboy and preserved at 4 °C. In the site Physico-chemical parameters like Temperature (T), pH, salinity, Total Dissolved Solids (TDS), Conductivity were measured (EUTECH PC 450). Using accepted techniques, the dissolved oxygen (DO), biochemical oxygen demand (BOD), and chemical oxygen demand (COD) of each station's samples were determined. (APHA 2005).
After the samples from three stations were combined, they were filtered via membrane filters with pore sizes of 3.0 μm and 0.2 μm to separate the particle-associated (PA) and free-living (FL) bacterioplankton, respectively. The filtered samples were then kept in a deep freezer at −20 °C. For the nutrient analysis, the residual water sample from each of the three stations was utilised. The filters were subjected to DNA extraction.
2.2 Nutrient analysis
Conventional methods have been employed to analyse water's organic and inorganic nutrients. Using the EDTA titration method, total hardness, calcium, and magnesium were assessed. Ash method was used to determine the particulate organic carbon (POC) and dissolved organic carbon (DOC). Stannous chloride method was used for phosphate, ammonium was determined by Nesseler's method, sulphate by Turbidity method, nitrate was done by Phenol-disulphonic acid method, iron by Phenanthroline method, finally Chloride by Argentometric method. (APHA 2005).
2.3 DNA extraction and PCR amplification
DNA isolation from frozen filter was done using the EXpure Microbial DNA isolation kit (BogarBio Bee stores Pvt Ltd) by following manufactures instructions. The cells grown as monolayer were lysed by suspending 1–3 colonies in0.45 mL of lysis buffer and subsequent pipetting; RNAase and 0.25 mL of neutralization buffer were and incubated at 65 °C for 30 min.The tubes were centrifuged for 20 min (12,000 g)in a refrigerated centrifuge. The viscous supernatant was collected and 0.6 mL of binding buffer was added and again incubated at 25 °C (5 min). The contents were transferred in to a spin column and centrifuged at 12,000 g (2 min). The bound DNA material was washed using washing buffer II and the DNA was collected by elution buffer.
DNA with 16S rRNA genes were obtained via the DNA extraction procedure, and these were then amplified by PCR using primers designed especially for 454 high-throughput pyrosequencing. 27F (5′AGAGTTTGATCMTGGCTCAG3′) was the forward primer, while 1492R (5′AAGGAGGTGATCCAGCCGCA3′) was the reverse primer.. The Taq Master Mix is made up of 0.02 % bromophenol blue, 3.2 mM MgCl2, 0.4 mM dNTPs, and TaqDNA polymerase in 2X Taq buffer. A 25 μL reaction solution is made up of 12 μL of Taq Master Mix, 5 μL of deionized water, 1.5 μL of forward and reverse primers, and 5 μL of extracted DNA..The 25 cycles of this PCR amplification include a 2-minute denaturation step at 95 °C and a 10-minute extension step at 72 °C. Denaturation (95 °C) and Annealing (60 °C) and Extension (72 °C) were the three steps in each cycle. Each cycle had Denaturation (95 °C, 30 s) Annealing (60 °C, 30 s) Extension (72 °C 2 min) (Kikani et al., 2017). The Montage PCR Clean-Up kit was employed to eliminate unincorporated PCR primers and dNTPs from the PCR products. Subsequently, the PCR product was assessed for quality and quantity, yielding an approximate concentration of 100 ng/μl, and its formulation was verified using the Qubit Fluorometer 3.0.
2.4 Sequencing and bioinformatics analysis
The sequencing process was carried out utilising a 1 μg DNA template. AMPure XP bead binding was used to purify the sequenced sample. For priming and loading, an Oxford Nanopore SpotON flow cell was utilised.
2.5 Sequencing annotation and library coverage calculation
The EPI2ME 16 s analysis workflow empowers users to attain genus-level identification from individual reads, providing access to base-called files for thorough examinations at both the species and sub-species levels. After obtaining blast results, a phylogenetic analysis was conducted, comparing the query sequence with closely related sequences, followed by multiple sequence alignment. The process's goal is to match base call sequences with the NCBI 16 s bacterial database, which include 16 s sequences belonging to various species. Based on identification and percentage coverage, each read is categorised. The 16S process is useful for identifying pathogens in a mixture sample or learning more about the makeup of a microbial community.
2.5.1 Sequence analysis
The raw sequence of Fastq format was uploaded to Kaiju webserver. Kaiju is a software designed for precise taxonomic classification of high-throughput sequencing reads originating from metagenomic whole-genome sequencing or metatranscriptomics experiments. It accomplishes this by matching each sequencing read to a taxonomic category within the NCBI taxonomy through comparisons with a reference database that includes microbial and viral protein sequences. Krona chart was prepared for further analysis.
2.6 Statistical analysis
The data were presented as the mean ± standard deviation from three distinct sample locations in each estuary zones. When comparing the nutritional quality of water samples from both locations, the statistical analysis employed a paired t-test, where a difference of p < 0.05 is deemed statistically significant.
3 Results
3.1 Nutrient composition of the estuaries
Table 1 lists the changes in the different physical and chemical parameters as well as the biological elements of the water quality from the Chaliyar River and Anjarakkandi estuary zones. The data show that the temperature of the water in both estuary zones varied minimally, while the dissolved oxygen content of the Anjarakkandi river was significantly lower. Nitrate, ammonium, phosphate, and sulphate levels were found to be elevated together. Furthermore, these samples had considerably greater BOD and COD values than expected (Table 1).
Parameter
Chaliyar
Anjarakkandi
Temperature(°c)
31.0 ± 0.0
31.0 ± 0.00
pH
7.57 ± 0.02
7.72 ± 0.02*
Salinity(ppt)
10.60 ± 0.00
13.67 ± 0.06***
TDS(g/l)
9.94 ± 0.00
12.63 ± 0.06***
Conductivity(ms/cm)
13.95 ± 0.06
22.06 ± 0.07**
Total Hardness(mg/ml)
1548.33 ± 2.89
2150.33 ± 0.058***
DO(mg/l)
6.35 ± 0.03
3.82 ± 0.04**
BOD(mg/l)
2.07 ± 0.01
8.87 ± 0.21**
COD(mg/l)
39.10 ± 0.17
77.83 ± 0.81***
Calcium(mg/l)
100.00 ± 0.00
158.33 ± 1.53*
Magnesium(mg/l)
320.80 ± 3.82
426.00 ± 1.73**
Chloride(mg/l)
5998.44 ± 0.49
8463.41 ± 0.13***
Iron(ppm)
1.42 ± 0.10
0.50 ± 0.00*
Nitrate(ppm)
0.29 ± 0.03
0.46 ± 0.00*
Ammonium(ppm)
0.31 ± 0.01
0.43 ± 0.00*
Sulfate(ppm)
139.62 ± 0.06
186.62 ± 0.04*
Phosphate(ppm)
0.03 ± 0.01
0.02 ± 0.00
POC(%)
0.21 ± 0.02
2.12 ± 0.00***
DOC(%)
0.14 ± 0.01
0.39 ± 0.01**
3.2 Bacterioplankton diversity of the estuaries
Tables 2 and 3 list the diversity of free-living and particle-associated bacterioplanktons found in the samples. The estuarine regions of Anjarakkandi river exhibited a notably greater diversity of bacteria in comparison to Chaliyar River's estuarine parts. Additionally, the diversity of firmicutes (17.24 %) and proteobacteria (67.07 %) in the Anjarakkandi River estuaries was revealed by the phylum level investigation. In addition, the Chaliyar river has less abundant proteobacteria and more firmicutes than Anjarakkandi river, with 46.35 % and 31.51 %, respectively. Bacillus the subtilis, a faecal bacterium, was identified at the species level in estuaries of the Anjarakkandi and Chaliyar rivers at significantly higher percentages 16.19 % and 10.07 %, respectively (Table 3). PA- Particle-associated; FL- Free living.
Kingdom level
Phylum level
Bacteria
Archaea
Eukaryota
Proteobacteria
Firmicutes
Euryarchaeota
Anjarakkandi- PA
1768
217
157
1067
572
181
Anjarakkandi- FL
1027
78
77
778
200
71
Chaliyar- PA
744
84
65
492
186
73
Chaliyar- FL
321
61
21
178
121
13
Anjarakkandi
Chaliyar
Phylum
Proteobacteria
67.07
46.35
Firmicutes
17.24
31.51
Euryarchaeota
6.12
10.68
Chordata
3.45
3.39
Species
Marivivens sp. JLT3646
11.97
10.53
Sulfitobacter sp. AM1-D1
11.97
3.2
Bacillus subtilis
16.19
10.07
3.3 Bacterial diversity and composition of Chaliyar river
Particle associated bacteria:11015 out of 36701 reads were classified. Shown are taxa that comprise at least 0.1 % of classified reads. Bubble plot shows some major phyla of bacteria. Actinobacteria dominates over other phyla of bacteria in the sample, second dominating phyla is proteobacteria. Plot shows the presence of Cyanobacteria in the particle associated bacteria community (Fig. 1b). Krona chart shows the presence some distinctive class like Alphaproteobacteria, Gammaproteobacteria in major share of 30 % of classified reads (Fig. 1a).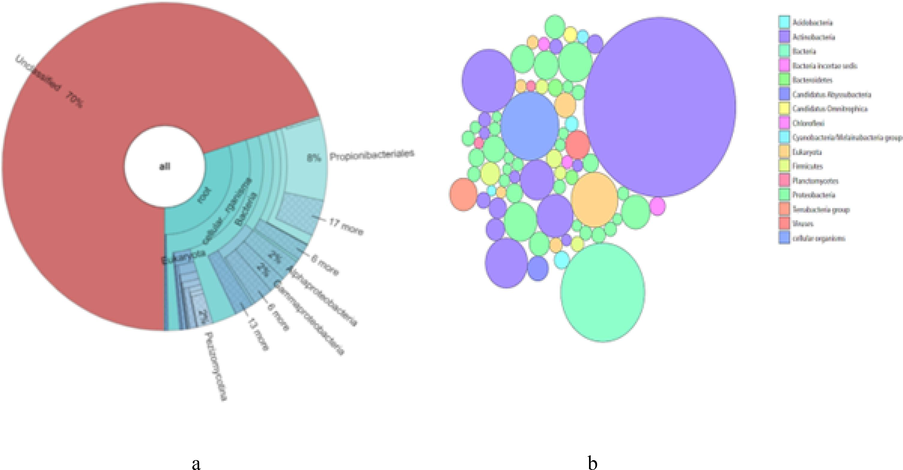
(a) Krona pie chart showing particle- associated Bacteria present in estuary of Chaliyar. (b) Bubble plot showing particle-associated Bacteria present in estuary of Chaliyar.
Free living bacteria:2367 out of 10658 reads were classified. Shown are taxa that comprise at least 0.1 % of classified reads.Bubble plot shows some major phyla of bacteria. Proteobacteria dominates over other phyla of bacteria in the sample. Second dominating phyla is Actinobacteria. Plot shows the presence of large amount of Cyanobacteria in the Particle associated bacteria community than the other samples (Fig. 2b). Krona chart shows 78 % of unclassified reads and within the 22 % classified reads, predominant strains are Phenylobacteriumzucineum, Rhizobiales,Actinomycetia and Bacteroidetes(Fig. 2a).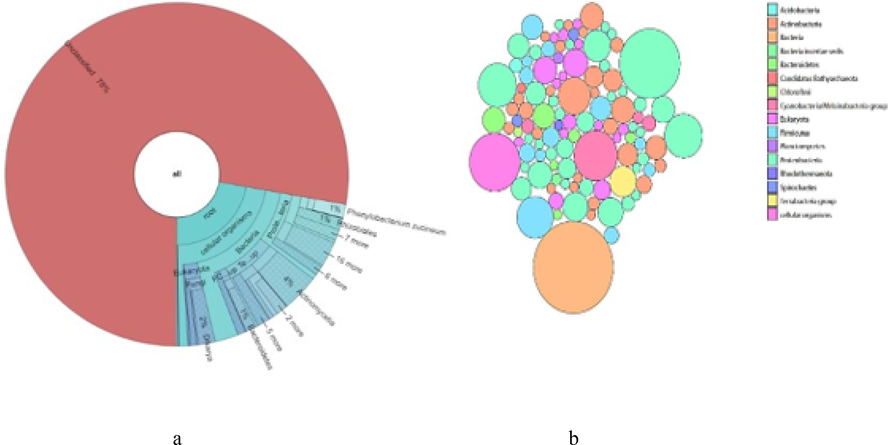
(a) Krona pie chart showing free-living Bacteria present in estuary of Chaliyar. (b) Bubble plot showing free-living Bacteria present in estuary of Chaliyar.
3.4 Bacterial diversity and composition of Dharmadom
Particle associated bacteria:35090 out of 89214 reads were classified. Shown are taxa that comprise at least 0.1 % of classified reads. The bubble plot shows some major phyla of bacteria. Actinobacteria dominates over other phyla of bacteria in the sample, second dominating phyla is Proteobacteria. Plot shows least abundance of Cyanobacteria in the particle associated bacterial community (Fig. 3b). Krona chart shows 61 % of unclassified reads,and within 39 % of classified reads, Propionibacteriales, Micrococcales, Alphaproteobacteia, Gammaproteobacteia and Pezizomycotinawere found to be prominent (Fig. 3a).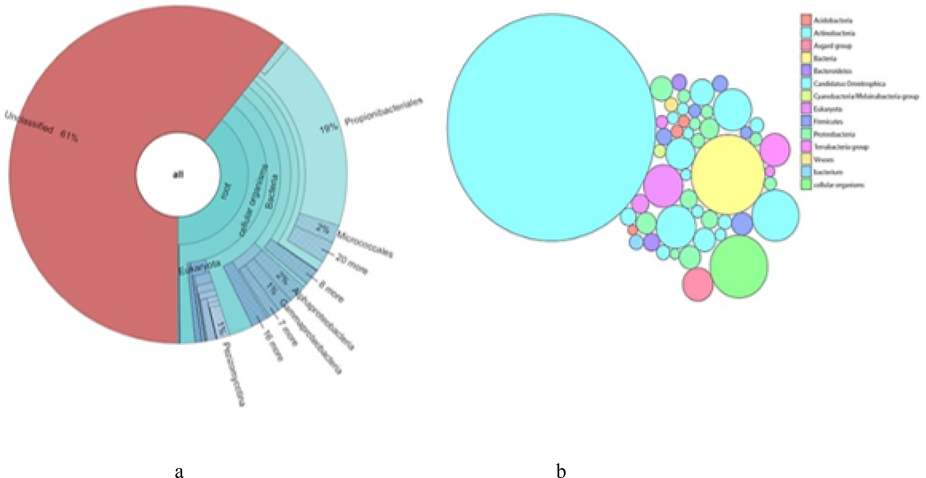
(a) Krona pie chart showing particle- associated Bacteria present in Darmadam estuary. (b) Bubble plot showing particle- − associated Bacteria present in Darmadam estuary.
Free-living bacteria: 12348 out of 40998 reads were classified. Shown are taxa that comprise at least 0.1 % of classified reads. Bubble plot shows the dominance of Actinobacteria, Proteobacteria, Acidobacteria and Firmicutes (Fig. 4b). Krona chart shows prominent presence of Propionibacteriale sand Gammaproteobacteria (Fig. 4a).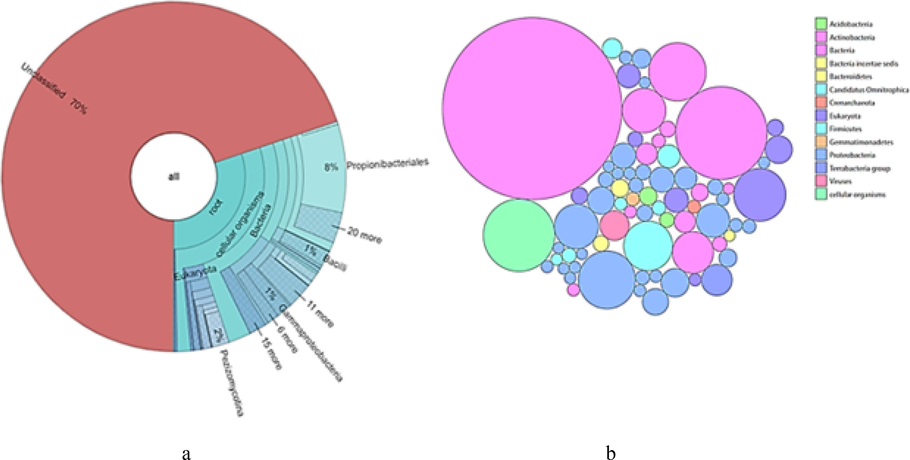
(a) Krona pie chart showing free-living Bacteria present in Darmadam estuary. (b) Bubble plot showing free-living Bacteria present in Darmadam estuary.
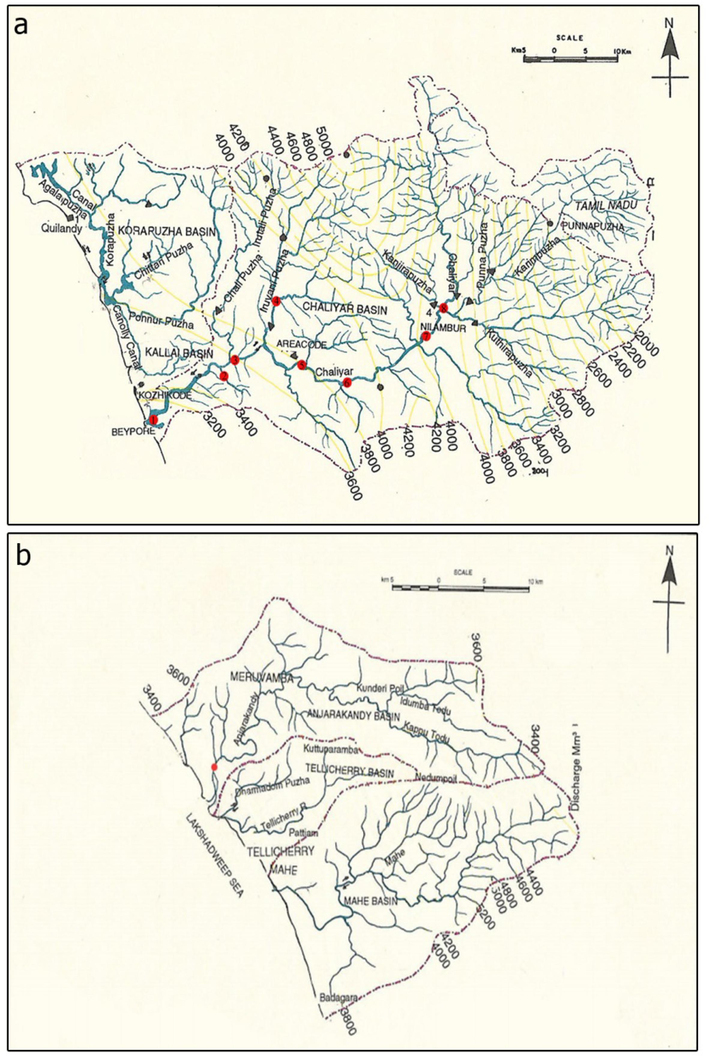
The site map of estuarine regions of Chaliyar river (a) and Anjarakkandi river (b).
4 Discussion
Bacterioplanktons are important in an aquatic ecosystem due to their roles in mineral cycling, improving overall productivity of the system as well as in pollution control (Pommier, 2011; Grossart, 2010). The current study discovered a notably increased quantity of Proteobacteria, Actinobacteria, and Cyanobacteria in the estuary waters of the Anjarakkandi River. Elevated concentrations of water nutrients, including nitrates, nitrites, and phosphates, correlate with a higher proportion of bacterioplankton taxa, predominantly consisting of Actinobacteria, Cyanobacteria, and Proteobacteria. (Kiersztyn et al., 2019; Mou et al.,2013a). Firmicutes identified in the samples indicates the faecal contamination in the water (Paruch et al., 2019a; Paruch et al., 2019b). We also discovered that a range of nitrogen-breaking bacteria, including Sulfitobacter, Nitratiruptor, and Roseobacter, were present. One common bacterioplankton that is well-known for breaking down different types of organic materials is the Roseobacter.(Pohlner et al., 2019). Likewise, Nitratiruptor is a nitrite reducing bacteria, which is capable of metabolising the nitrate in the water (Nakagawa et al., 2005). Sulfitobacter and Pseudoruegeria are other two strains observed in both the Chaliyam and Dharmadom estuarine regions. It is reported that the Sulfitobacter strains are predominantly distributed in the areas of oil polluted marine environments;it has been associated with hydrocarbon removal from seawater (Neethu et al., 2019). Pseudoruegeria is associated with the metabolic pathways for vitamin Bcomplexes (such as B1, B7, B12)(Cho et al., 2020).Further isolation and culturing of these bacteria may yield a source for vitamin B complex biosynthesis. Identifying characteristic strains from particle -associated and free-living communities is vital for their specific applications. Most of the diverse bacterioplankton reported here are capable of utilizing the dissolved organic matter for their growth and functioning. Isolation of such bacterial communities is having great significance in the aspects of wastewater treatment and pollution abatement(Dinasquet et al., 2018; Watanabe et al., 2009). Abundance of members like Actinobacteria, Cynobacteria and Proteobacteria(Table 3) act as biological elements of water quality assessment (Pinto et al., 2021). There are reports of how water contaminations has changed the community compositions of bacteria. However, certain organisms are adapting to the pollutants. Bacterioplanktons are actively participating in pollution control. A few species of Betaproteobacteria are involved in degradation of of microcystin, which is a cyanobacteria induced pollutant (Mou et al. 2013). Some Species belongs to Gammaproteobacteria are involved in hydrocarbon degradation and Rodococcus like species are capable of degrading organic pollutants and they form a biofilm which reduce the mobility of pollutants (Gontikaki et al., 2018; Martínková et al., 2009).
Estuaries exhibit variations not only in bacterial abundance but also in nutrient parameters, with the Anjarakandy River estuary displaying higher levels of physiochemical parameters compared to the Chaliyar River estuary. Specifically, nutrients such as calcium, magnesium, chloride, iron, nitrate, ammonium, sulfate, and phosphate are elevated in the Anjarakandy estuary, along with a higher percentage of organic carbon content.
Various scientific studies have investigated how nutrient parameters impact the bacterial composition of water bodies, rivers, and similar aquatic ecosystems. These studies aim to understand how factors such as nutrient levels, including nitrates, phosphates, and organic carbon, influence the types and abundance of bacteria present in these environments. (Yang et al., 2020; Mou et al., 2013b). Certain investigations have focused on seasonal variations, revealing shifts in bacterioplankton composition and richness influenced by changing environmental conditions. Additionally, these studies explore the role of bacterioplankton in mediating the bioavailability of organic matter, crucial for nutrient cycling within aquatic ecosystems (Xiao et al., 2021,Stephens et al., 2020) We did not observe any major difference in diversity of particle associated bacterioplanktons in the two estuaries investigated. However, there was a significant variation observed in the diversity of free living bactrioplankton attributed to changes in physio-chemical parameters.
5 Conclusions
In conclusion, the study conducted in the Anjarakkandi River estuary in Dharmadom emphasises the critical functions that bacterioplankton play in aquatic environments. These waters' high Proteobacteria, Actinobacteria, and Cyanobacteria abundances indicate that the surrounding environment has an abundance of nutrients. The detection of Firmicutes hints at the possibility of fecal contamination, prompting concerns regarding the water's quality. When comparing bacterioplankton abundance and nutrient parameters between the Chaliyar and Anjarakandy river estuaries, we observed a higher abundance of bacteria and elevated nutrient levels in the Anjarakandy river estuary. These findings suggest a potential link between nutrient availability and bacterioplankton abundance. The diversity of bacterioplankton in natural environments is impacted by various physio-chemical factors, underscoring the importance of ongoing research into these microbial communities for their unique environmental and industrial potential. Overall, the comparison has been easy to draw because metagenomics has yielded a comprehensive understanding of the composition of the communities in both estuaries.
Funding
Researchers Supporting Project Number (RSP2024R11), King Saud University, Riyadh, Saudi Arabia.
CRediT authorship contribution statement
D. Nikhitha: . Arunaksharan Narayanankutty: Writing – review & editing. Manoj Mathews: Writing – review & editing, Methodology, Funding acquisition, Conceptualization. Deepa Sathee: Writing – review & editing, Formal analysis. Ahmed Alfarhan: Software. Rajakrishnan Rajagopal: Software. Jisha Jacob: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Acknowledgement
The authors acknowledge the funding support from Researchers Supporting Project Number (RSP2024R11), King Saud University, Riyadh, Saudi Arabia. The DBT-STAR (project number: BT/HRD/11/09/2020) scheme supported infrastructural development in St. Joseph’s College (Autonomous), Devagiri, Calicut.
Availability of data and material
Upon proper request, the data will be made available.
Ethics approval
There are neither human nor animal subjects in this study.
Consent for publication
The research does not incorporate any external materials or third-party content.
NCBI Accession Number
The data have been deposited with links to BioProject accession number PRJNA750037 in the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Metagenomic analysis of the bacterial communities and their functional profiles in water and sediments of the Apies River, South Africa, as a function of land use. Sci. Total. Environ.. 2018;616–617:326-334.
- [CrossRef] [Google Scholar]
- Elucidation of the biosynthetic pathway of Vitamin B groups and potential secondary metabolite gene clusters via genome analysis of a marine bacterium Pseudoruegeria Sp. M32A2M. J. Microbiol. Biotechnol.. 2020;30:505-514.
- [Google Scholar]
- Enrichment of Bacterioplankton Able to Utilize One-Carbon and Methylated Compounds in the Coastal Pacific Ocean. Front. Mar. Sci.. 2018;5
- [CrossRef] [Google Scholar]
- Biomarker responses to environmental contamination in estuaries: A comparative multi-taxa approach. Aquat. Toxicol.. 2017;189:31-41.
- [Google Scholar]
- Influence of light on particulate organic matter utilization by attached and free-living marine bacteria. Front. Microbiol.. 2019;10
- [CrossRef] [Google Scholar]
- Gontikaki, E., et al. 2018.Hydrocarbon-Degrading Bacteria in Deep-Water Subarctic Sediments (Faroe-Shetland Channel. Journal of Applied Microbiology. 125. 4,1040–53. 10.1111/jam.14030.
- Ecological consequences of bacterioplankton lifestyles: changes in concepts are needed. Environ. Microbiol. Rep.. 2010;2:706-714.
- [Google Scholar]
- From lake to estuary, the tale of two waters: a study of aquatic continuum biogeochemistry. Environ. Monit. Assess.. 2018;190:017-6455.
- [Google Scholar]
- Water resources development and management: an experience in rural hilly area. J. Environ. Sci. Eng.. 2010;52:67-74.
- [Google Scholar]
- Water resource management: an Indian perspective. J. Environ. Sci. Eng.. 2012;54:577-591.
- [Google Scholar]
- Structural and functional microbial diversity along a eutrophication gradient of interconnected lakes undergoing anthropopressure. Sci. Rep.. 2019;9:11144.
- [CrossRef] [Google Scholar]
- Metagenomic and culture-dependent analysis of the bacterial diversity of a hot spring reservoir as a function of the seasonal variation. Int. J. Environ. Res.. 2017;11. 1:25-38.
- [CrossRef] [Google Scholar]
- Dissolved organic carbon source influences tropical coastal heterotrophic bacterioplankton response to experimental warming. Front. Microbiol.. 2019;10
- [CrossRef] [Google Scholar]
- Biodegradation Potential of the Genus Rhodococcus. Environ. Int.. 2009;35. 1:162-177.
- [CrossRef] [Google Scholar]
- Bacterioplankton response to nitrogen and dissolved organic matter produced from salmon mucus. MicrobiologyOpen.. 2020;9:e1132.
- [Google Scholar]
- Metagenomic Identification of Bacterioplankton Taxa and Pathways Involved in Microcystin Degradation in Lake Erie. PLoS. One. 2013;8:e61890.
- [Google Scholar]
- Diversity and distribution of free-living and particle-associated bacterioplankton in Sandusky Bay and adjacent waters of Lake Erie Western Basin. J. Great. Lakes. Res.. 2013;39:352-357.
- [CrossRef] [Google Scholar]
- BrdU-labeling and Fluorescence Activated Cell Sorting of Polyamine-Transforming Bacterioplankton in Coastal Seawater. Environ. Microbiol.. 2015;17:876-888.
- [CrossRef] [Google Scholar]
- Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the epsilon-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough Int. J. Syst. Evol. Microbiol.. 2005;55:925-933.
- [Google Scholar]
- Oil-spill triggered shift in indigenous microbial structure and functional dynamics in different marine environmental matrices. Sci. Rep.. 2019;9:1354.
- [CrossRef] [Google Scholar]
- Aquatic microbial diversity associated with faecal pollution of Norwegian waterbodies characterized by 16S rRNA gene amplicon deep sequencing. J. Microbial. Biotechnol.. 2019;12:1487-1491.
- [CrossRef] [Google Scholar]
- Faecal pollution affects abundance and diversity of aquatic microbial community in anthropo-zoogenically influenced lotic ecosystems. Sci. Rep.. 2019;9:19469.
- [CrossRef] [Google Scholar]
- Bacterioplankton community as a biological element for reservoirs water quality assessment. Water.. 2021;13. 20:2836.
- [CrossRef] [Google Scholar]
- The majority of active rhodobacteraceae in marine sediments belong to uncultured genera: a molecular approach to link their distribution to environmental conditions. Front. Microbiol.. 2019;10
- [CrossRef] [Google Scholar]
- Pommier, T., 2011. Bacterioplankton. In: Reitner J, Thiel V (eds) Encyclopedia of Geobiology. Springer Netherlands, Dordrecht, pp 89-92. doi:10.1007/978-1-4020-9212-1_18.
- Eco-Hydrological Footprint of a River Basin in Western Ghats Yale. J. Biol. Med.. 2018;91:431-444.
- [Google Scholar]
- Bromodeoxyuridine (BrdU) labeling and Subsequent Fluorescence Activated Cell Sorting for Culture-independent Identification of Dissolved Organic Carbon- degrading Bacterioplankton. J. Vis. Exp.. 2011;55:e2855.
- [Google Scholar]
- Pollution impacts on bacterioplankton diversity in a tropical urban coastal lagoon system. PLoS One. 2012;7:e51175-e.
- [CrossRef] [Google Scholar]
- Bacterioplankton Responses to Increased Organic Carbon and Nutrient Loading in a Boreal Estuary—Separate and Interactive Effects on Growth and Respiration. Microb. Ecol.. 2018;76:144-155.
- [Google Scholar]
- Organic Matter Composition at Ocean Station Papa Affects Its Bioavailability, Bacterioplankton Growth Efficiency and the Responding Taxa. Front. Mar. Sci.. 2020;7:590273
- [CrossRef] [Google Scholar]
- Effective isolation of bacterioplankton genus Polynucleobacter from freshwater environments grown on photochemically degraded dissolved organic matter. FEMS. Microbiol. Ecol.. 2009;6:57-68.
- [CrossRef] [Google Scholar]
- Bacterioplankton Richness and Composition in a Seasonal Urban River. Front. Environ. Sci.. 2021;9:731227
- [CrossRef] [Google Scholar]
- Compositional variations and the environmental responses of bacterioplankton in the Pengxi River of the Three Gorges Reservoir, China. J. Freshwater. Ecol.. 2020;35:449-467.
- [CrossRef] [Google Scholar]







