Translate this page into:
Bacterial spectrum from diabetic foot Ulcers: A study of antibiotic resistance patterns and phylogenetic diversity
⁎Corresponding author. muhammad.khan@som.umaryland.edu (Muhammad Ajmal Khan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Diabetic foot ulcer (DFU) is one of the most detrimental impacts of diabetes mellitus associated with osteomyelitis and gangrene, accounting for at least two-thirds of non-traumatic amputations with a 5-year survival rate. In this perspective, antimicrobial resistance has been a cause for grave concern for the last 50 years and is among the World Health Organization most pressing “calls to action” for the 21st century. The current study aimed to identify bacterial pathogens present in DFU, their antibiotic resistance profiles, and genetic diversity. A total of 180 samples were collected from DFU patients hospitalized at healthcare institutions in Pakistan. All samples were cultured on three distinct types of media − nutritional agar, McConkey agar, and mannitol salt agar to identify both Gram-negative and Gram-positive bacteria. Biochemical, morphological, and molecular (16 s rRNA) investigations were employed to characterize the bacterial species. Out of the 180 samples collected, Staphylococcus aureus (S. aureus) was isolated from 98 (54 %) samples, Escherichia coli (E. coli) from 75 (41.6 %) samples, S. epidermidis from 20 (11.1 %) samples, and Pseudomonas aeruginosa (P. aeruginosa) from 18 (10 %) samples. Furthermore, PCR amplification confirmed the presence of antibiotic resistance genes in the resistant E. coli and S. aureus isolates. In S. aureus, the most commonly found antibiotic resistance genes were erm(B) and aac(6′) aph (2′) whereas in E. coli the prevalent genes were ampC (tetA) and erm (B). The distributions of many genes associated with drug resistance differed from those documented worldwide. These findings will aid in guiding the empirical use of antibiotics for treating diabetic foot infections, thereby reducing the risk of inappropriate antibiotic use and the development of antibiotic resistance.
Keywords
Diabetic foot ulcer
Staphylococcus aureus
Escherichia coli
Staphylococcus epidermidis and Pseudomonas aeruginosa
Antibiotics
Resistance
1 Introduction
Diabetic foot ulcers (DFUs) are one of the most severe problems in diabetes patients. People with diabetes sometimes develop chronic ulcers that lead to amputation. The DFU pertains to an infection in the lower extremities of individuals. This condition is characterized by ischemia and neuropathy in the affected area, resulting in necrosis (Wang et al., 2022)). It is estimated that 15–25 % of diabetic patients are likely to develop diabetic foot ulcers as disease progression. The mortality risk for those with DFUs is higher than that of diabetic patients. According to the International Diabetes Federation, there is an estimated annual incidence of 9.1–26.1 million cases of DFUs worldwide (Anvarinejad et al., 2015).
Diabetic foot ulcers, often known as DFUs, are severe diabetic complications that significantly affect an individual's social, mental, and financial well-being. The existence of biofilms is one of the primary causes of diabetic foot ulcers' resistance to healing. Biofilms can cause infection development and persistence because they exacerbate wound inflammation and exhibit an apparent absence of response to host defenses or alternative therapies. Foot ulcers are more likely to develop in all of these diabetic problems, and twenty percent of hospital stays among people with diabetes are thought to be the outcome of DFUs. Diabetic foot ulcers can result in the spread of infection, gangrene, amputation, and, in cases where appropriate care is not given, even death. It has been estimated that approximately fifty to seventy percent of all lower limb amputations (LLAs) are caused by diabetes-related foot ulcers. Furthermore, there is an increased risk of amputation once a diabetic foot ulcer develops. The risk of vascular lower limb amputations in people with diabetes is expected to be eight times greater in the entire population (those over 45) than in people without the disease. In people over 85 years of age, the prevalence in men and women is projected to be fifteen and twelve times higher, respectively, compared to the average prevalence rates across all population groups (Afonso et al., 2021). Pathogenic microorganisms have the ability to colonize diabetic foot ulcers, and the immune deficits associated with diabetes promote infections. Aerobic and anaerobic Pathogenic bacterial species such as S. aureus, P. aeruginosa, and Klebsilla, as well as coliform bacteria, play a role in these diseases. The several microbes in diabetic foot ulcers might be either plankton or sessile. When bacteria create biofilms, they enclose themselves in a self-made polymeric matrix that protects them from both antimicrobial agents and the body's immune response. Thus, even with systemic antibiotic therapy, bacterial biofilms in diabetic foot ulcers might be the cause of the infection's slow recovery and subsequent persistence. A DFU is a significant healthcare and socioeconomic issue, affecting 40–60 million individuals worldwide. An older age, a male gender, Type 2 diabetes, a lower BMI, hypertension, diabetes, diabetic retinal degeneration, and a history of smoking are the key risk factors for DFUs. Amputations due to diabetic foot ulcers, particularly severe ulcers, can result in a marked decline in life expectancy and a rise in early death (Pouget et al., 2020).
The antibiotic-resistant bacteria are considered to pose a serious risk to the health of the public. Excessive and improper use of antibiotics is the main contributor to antibiotic resistance. A number of variables, including prolonged wound healing, repeated hospital stays, and inadequate administration of antibiotics, may increase the incidence of multidrug-resistant microorganisms in individuals with diabetes foot ulcers. Additionally, peripheral artery illnesses might make it difficult for antibiotics to penetrate the tissues of the lower limbs, which encourage the development of resistant strains of bacteria. These conditions are frequently prevalent among individuals with DFUs. While S. areus and Streptococcus bacteria typically cause bacterial infections in DFUs, other microbial species or mixed bacteria (enteric bacteria spp., Gram-negative bacillus, Gram-positive anaerobic cocci) may also play a role. The most common type of microbe to be isolated is staphylococci. MRSA has been found in 15–30 % of diabetic foot ulcer infections, according to various investigations. There are multiple factors contributing to antibiotic resistance, but the two most significant ones are improper use of antibiotics and disregard for personal hygiene. While polymicrobial outbreaks are substantially more prevalent, monomicrobial infections can occur occasionally (Kandemir et al., 2007). The E. coli has also the highest prevalence in patients with DFUs in some studies (Sari et al., 2018). In a relevant study, E. coli showed the maximum multidrug resistance (81.81 %). The maximum of the Gram-negative bacteria was resistant to antibiotic ampicillin (Baral et al., 2024).
The effective management of diabetic foot infections requires accurate diagnosis, proper collection of specimens for culture, deliberate selection of antimicrobial therapy, prompt determination of the need for surgical treatment, provision of any additional wound management that may be required, and complete attention to the patient. The management of DFIs through a methodical and evidence-based strategy is likely to yield better results, particularly in terms of illness resolution, and prevent consequences including amputation of the lower extremities. The most effective way to deliver this is through collaborative groups, whose membership should ideally include an expert in infectious disorders or clinical or medical microbiology. Appropriate local wound care (such as cleaning and removing debris), pressure off-loading, vascular evaluation and therapy if necessary, and metabolic (especially glycaemic) regulation should all naturally be prioritized by this team.
There are a number of guidelines available to help clinicians manage diabetic foot infections. Since 2004, the International Working Group on the Diabetic Foot (IWGDF) has gathered a panel of specialists in infectious diseases to issue widely utilized guidelines every four years (Lipsky et al., 2020). More over, the appropriate determination of the causal microorganisms that cause outbreaks is a crucial component in managing diabetes-related foot ulcers. While biopsy specimens, cultures, and swabs are more commonly used traditional diagnosis approaches, new molecular methods are currently investigated for the detection and measurement of bacteria. Understanding antibiotic resistance and the microbiological causes of DFUs is essential for managing and treating these wound infections effectively (Ghotaslou et al., 2018).
The lack of the proper screening facilities and expertise in diagnostic microbiology at the grassroots level further impedes the collection, isolation, and characterization of bacterial isolates from DFU patients. Lastly, the lack of digitalized public health system in Pakistan adds another layer of complexity to addressing and catch up this issue effectively. These all factors contribute to the perceived information gap. The current study aimed to describe the predominant multidrug-resistant bacteria in DFU and to elaborate the molecular mechanisms of antibiotic resistance. Here we showed higest prevalance of S. aureus in DFU followed by E.coli. The findings of the current study highlight the importance of local surveillance and understanding regional patterns of antibiotic resistance. This information will assist healthcare professionals in Pakistan to make informed decisions regarding antibiotic choices, reducing the risk of inappropriate antibiotic use to effectively treat diabetic foot ulcers.
2 Materials and methods
2.1 Samples collection and processing
A total of 180 DFU samples were collected admitted to the Surgery and Medical Department in different hospitals of Pakistan (Table S1). Ethical approval was acquired from the Ethical Committee of the University of Swabi, KP, Pakistan. Patients with DFU were included in the present study if they have had an infected ulcer. The grading system employed in the current study to assess diabetic foot ulcers was the Wagner Classification System (Mehraj and Shah, 2018). The system provides a standardized way to categorize the severity of foot ulcers. It is based on the deepness of the ulcer and the occurrence of infection. The exclusion criteria for the present study were non-diabetic patients with open wound infections or diabetic patients with non-infected open wound. The samples were collected using a standard procedure (Khan et al., 2019). Samples were brought to the Microbiology Laboratory (Biosafety level 2), Department of Microbiology, University of Swabi.
2.2 Culturing
All samples were cultured on three different media types for isolating Gram-negative and Gram-positive i.e. Mannitol Salt Agar (MSA), Nutrient agar, and MacConkey agar (Oxide, United Kingdom). The subculture of all the samples was done on the MacConkey media and MSA media. For sub culturing, a small portion of inoculum was transferred to a fresh culture medium using a loop to pick up a bacterial colony. After 24 h, many colonies were found on the plates. Hemocytometer was used for colony counting. MacConkey agar promotes Gram-negative bacteria growth, particularly those that ferment lactose while inhibiting the growth of Gram-positive bacteria. MSA media is selective for Gram-positive including S. aureus.
2.3 Biochemical and morphological identification
Microorganisms were identified using biochemical and morphological tests. The choice of specific tests depends on the bacterial species being identified. For identification of Gram-positive bacteria, catalase, coagulase and mannitol fermentation tests were used. Lactose fermentation, indole and oxidase tests were used to identify Gram-negative bacteria. Morphological tests comprised colony morphology, Gram staining, and cell shape which contributed to the identification process. Clinical Laboratory Standards Institute (CLSI, 2020) guidelines were followed to ensure accuracy, and reliability in laboratory practices (Grice et al., 2008).
2.4 Antibiotic susceptibility testing
Testing for antimicrobial resistance was carried out using Mueller-Hinton agar (MHA). In the current study, eight different antibiotics were utilized according to (CLSI, 2020) against E. coli (chloramphenicol (30 μg), sulphamethaxazole (1.25 μg), ceftriaxone (30 μg), tetracycline (30 μg), streptomycin (10 μg), erythromycin (15 μg), ampicillin (10 μg), amoxicillin clavulanate (20 μg) were evaluated to determine their efficacy against E. coli. Antibiotics used against Pseudomonas aeruginosa were amoxicillin (20 μg) clavulanate (20 μg), ceftraxione (30 μg), imipenem (10 μg), ceftazidime (30 μg), meropenem (10 μg), cefepime (30 μg), amikacin (10 μg) and ofloxacin (5 μg).
Antibiotics against S. aureus and S. epidermidis were sulfamethaxazole (1.25 μg), tetracycline (30 μg), streptomycin (10 μg), erythromycin (15 μg), chloramphenicol (30 μg), vancomycin (30 μg), daptomycin (3 μg), methicillin (10 μg), and penicillin (30 μg). Bacterial resistance to three or more antibiotic classes is referred to as multidrug resistance (MDR) (Magiorakos et al., 2012).
2.5 Extended-spectrum beta-lactamase-producing isolates (ESBLs)
Bacterial isolates were screened for ESBLs production using a double disc method (Jarlier et al., 1998). The disc amoxiclav was placed in the center of the nutrient agar medium containing the petri dish. Ceftriaxone and ceftazidime and were placed at a distance of 15 mm from amoxiclav. The plates were incubated for 24 h at 37 °C. An increase in the inhibition zone around cefotaxime or ceftazidime (>5 mm) toward the disc of amoxicillin-clavulanate) were read as ESBLs positive. A zone of inhibition of 15 mm or more around the cefotaxime disc showed that the bacterium is semsitve to cefotaxime. A zone of inhibition of 15 mm or more around the amoxicillin-clavulanic acid disc showed that the bacterium was susceptible to amoxicillin-clavulanic acid. If the inhibition zone around the cefotaxime of ceftazidime disc was less than 15 mm, but the zone of inhibition around the amoxicillin-clavulanic acid disc was 15 mm or more, then the bacterium was likely producing ESBLs.
2.6 DNA extraction
GeneJET Genomic DNA purification kit (Thermo Scientific, Lithuania, #K0721) was used to extract DNA from E. coli and S. aureus. Pure 2x109 bacterial cells were harvested in a 1.5 mL microcentrifuge tube and centrifugated for 10 min at 10,000 xg 4 °C. The cell pellet was resuspended in 50 μL of lysis buffer. The lysate was incubated at 56 °C for 15 min. Then, added 10 μL of proteinase K to the lysate and incubated at 56 °C for 15 min. Cold ethanol (500 μL) was added to the lysate and mixed well. The lysate was incubated on ice for 15 min followed by centrifugation at 12,000 x g for 15 min at 4 °C. The supernatant was discarded. The DNA pellet was washed with 70 % ethanol and centrifuged at 12,000 x g for 5 min at 4 °C. The supernatant was discarded and air-dried the DNA pellet for 5 min. The DNA pellet was resuspended in 100 μL of elution buffer. The DNA quality was checked using the nanodrop technique and the elution buffer containing DNA was preserved at −20 ˚C.
2.7 Molecular identification and phylogenetic network analysis
2.7.1 16S rRNA gene amplification
Molecular identification of isolated species was performed by amplifying the 16S rRNA gene using universal primers obtained from Macrogen Universal primer 785F —5′- GGATTAGATACCCTGGTA −3′ and 907R– 5′-: CCGTCAATTCMTTTRAGTTT-3′. There were selected 30 isolates on random basis for amplification to examine antibiotic-resistant genes. The polymerase chain reaction (PCR) profiles were set as suggested by the manufacturer (Solis BioDyne-5X FIREPol® Master mix).
2.7.2 Antibiotic resistance genes amplification
The DNA (5 µL) was used in PCR. The most prevalent E. coli and S. aureus isolates were randomly selected for amplification to examine antibiotic-resistant genes using primers already designed. The PCR parameters and conditions used were followed using standard procedure (Abdelgader et al., 2018; Fawzy et al., 2017, Khan et al., 2023). The PCR products were studied in the GelDoc system, and images were captured. PCR products were purified and sequenced through Macrogen (.
https://www.macrogen.com) using both forward and reverse primers, as shown in Table 1.
Bacterial Isolate
Antibiotic
Gene
Primer Sequence
Product size
S. aureus
Penicillin
blaZ
(F)ACTTCAACACCTGCTGCTTTC
(R)TGACCACTTTTATCAGCAACC
173
Tetracycline
tet(K)
(F)GTAGCGACAATAGGTAATAGT
(R)GTAGTGACAATAAACCTCCTA
360
Erythromycin
msr(A)
erm(C)
erm(B)
(F)GCAAATGGTGTAGGTAAGACAACT
(R)ATCATGTGATGTAAACAAAAT
(F)ATCTTTGAAATCGGCTCAGG
(R)CAAACCCGTATTCCACGATT
(F)CATTTAACGACGAAACTGGC
(R)GGAACATCTGTGGTATGGCG
400
295
425
Aminoglycoside
aac (6′) aph (2′)
(F)GAAGTACGCAGAAGAGA
(R)ACATGGCAAGCTCTAGGA
491
Escherichia coli
Ampicillin
ampC
(F)AATGGGTTTTCTACGGTCTG
(R)GGGCAGCAAATGTGGAGCAA
191
Tetracycline
tet(A)
(F)GGTTCACTCGAACGACGTCA
(R)CTGTCCGACAAGTTGCATGA577
Erythromycin
erm(B)
erm(A)
erm(C)
(F)GAAAAAGTACTCAACCAAATA
(R)AATTTAAGTACCGTTAC
(F)TCTAAAAAGCATGTAAAAGAAA
(R)CGATACTTTTTGTAGTCCTTC
(F)TCAAAACATAATATAGATAAA
(R)GCTAATATTGTTTAAATCGTCAAT642
533
642
Streptomycin
aadA1
(F) TATCCAGCTAAGCGCGAACT
(R) ATTTGCCGACTACCTTGGTC286
2.7.3 Phylogenetic network analysis
16S rRNA sequencing was achieved for the molecular identification of the isolates. The chromatograms received from Macrogen were refined by removing the redundant reads by employing software (Chromas) 2.6.6 (http://technelysium.com.au/wp/chromas/ (accessed on 10 January 2023). The refined sequences were used for similarity to 16S reference sequences by using Basic Local Alignment Search Tool from a National Center for Biotechnology Information database. The sequences were submitted to GenBank, and the allotted accession numbers were summarized in Table S2. The Maximum Likelihood and Tamura-Nei Model Gamma distributed with invariant sites (G + I) were used in Molecular Evolutionary Genetics Analysis software 7 (http://www.megasoftware.net (retrieved on 30 January 2023) to conduct the phylogenetic analysis, and the precision of the results was assessed using bootstrap values obtained from 1000 repeats (Saitou and Nei, 1987, Felsenstein, 1985, Tamura et al., 2004, Kumar et al., 2016).
3 Results
3.1 Microbiological assessment of samples
According to morphology, Gram staining, and biochemical tests, bacterial species were determined from the DFUs patients. in the total samples (1 8 0), the frequency distribution of S. aureus, E. coli, S. epidermidis and P. aeruginosa were reported 98 (54 %), 75 (41.6 %), 20 (11.1 %) and 18 (10 %) respectively.
3.2 Antimicrobial susceptibility
Antimicrobial sensitivity testing was performed on all bacterial isolates. The overall antibiotic resistance patterns of the bacterial isolates from patients with DFUs are shown in Figs. 1-4.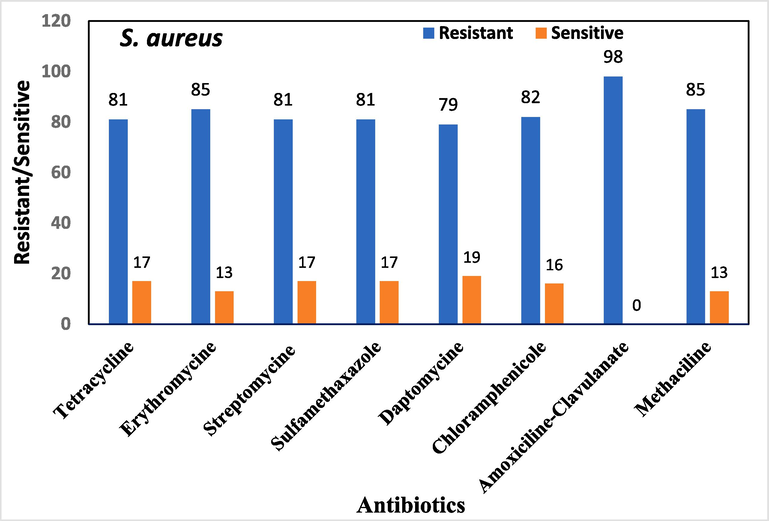
Overall antibiotic resistance patterns of S. aureus isolated from DFUs patients.
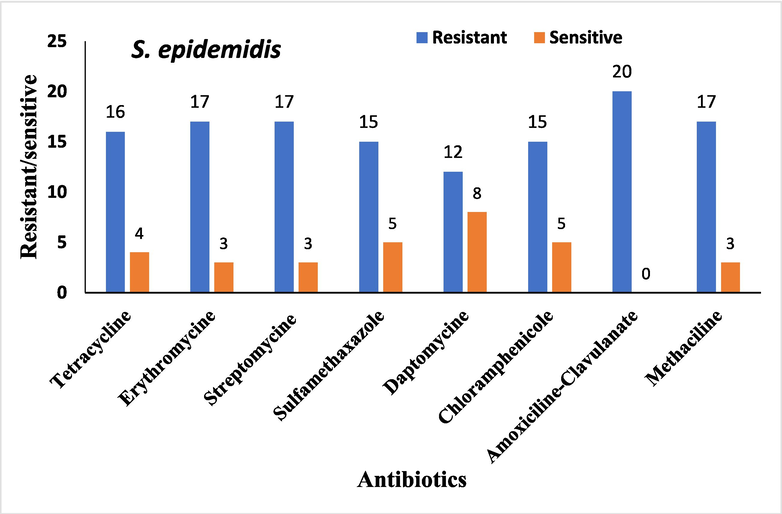
Overall antibiotic resistance patterns of S. epidemidis isolated from DFUs patients.
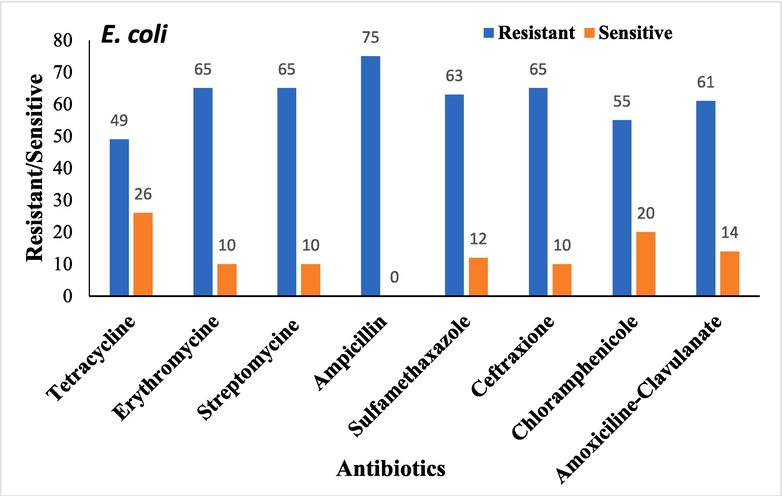
Overall antibiotic resistance patterns of Escherichia coli Isolated from DFUs patients.
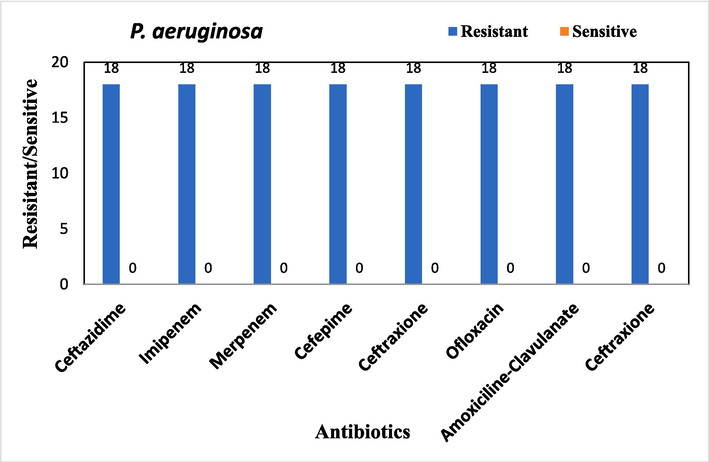
Overall antibiotic resistance patterns of P. aeruginosa Isolated from DFUs patients.
3.3 Phenotypic detection of extended-spectrum β-lactamases
Gram-negative bacterial species for ESBL activity were evaluated. Out of 18P. aeruginosa, 22.2 % (n = 4) were ESBL-positive phenotypically, and 20 % isolates of E. coli were ESBL-positive, as shown in Figs. 5 and 6.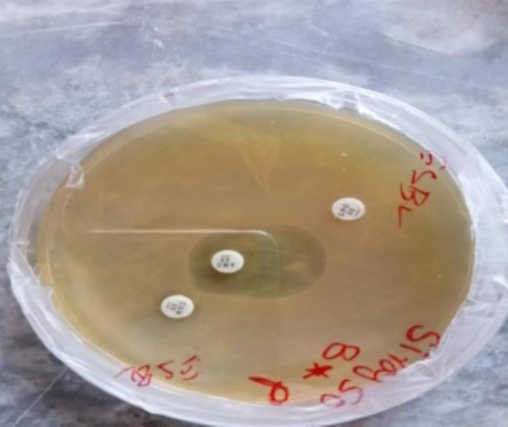
ESBL activity against P. aeruginosa.
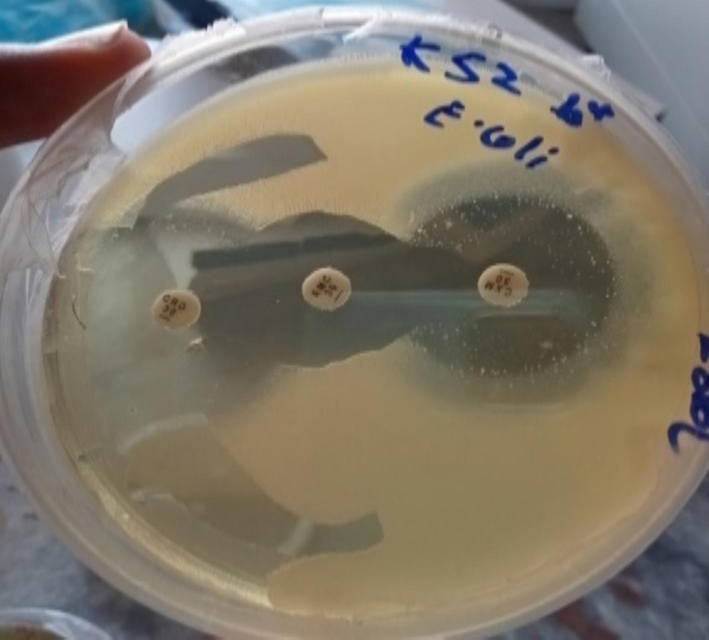
ESBL activity against E. coli.
3.4 Molecular identification and phylogenetic network analysis
Molecular identification of isolated species was performed by amplifying the 16S rRNA gene using universal primers, i.e., 785F and 907R. Based upon the sequencing data, the phylogenetic tree for E. coli, P. aeruginosa, and S. aureus from the current study gathered with each other and with reference sequences showing their high similarity based on 16S rRNA (Figs. 7-9). The sequencing results further validate bacterial identification based on sequence BLAST.
Evolutionary relationships of E. coli isolates based on 16 S rRNA gene sequences with reference sequences. The analysis included 21 GenBank sequences to construct phylogenetic tree by using MEGA. 7. Dendrograms were constructed and genetic diversity was observed in the E. coli. It can be concluded that high genetic diversity is observed in the isolated strains.

Evolutionary relationships of P. aeruginosa based on 16 S rRNA gene sequences with reference sequences. The analysis included 15 GenBank sequences. Dendrograms were constructed and genetic diversity was observed in P. aeruginosa isolates. It can be concluded that high genetic diversity is observed in the isolated P. aeruginosa strains as compared to E. coli.
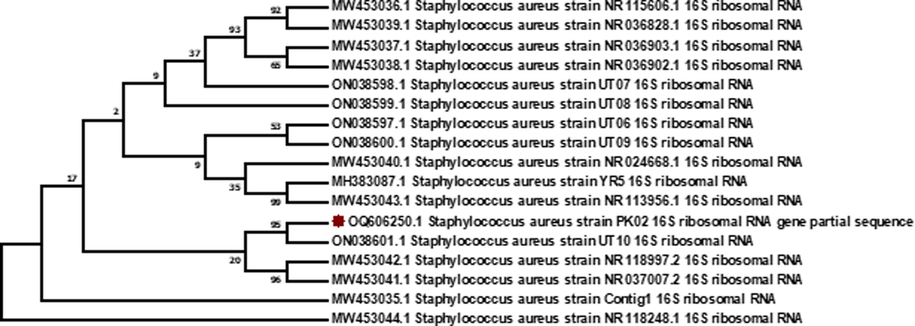
Evolutionary relationships of S. aureus based on 16 S rRNA gene sequences with reference sequences. The analysis included 16 GenBank sequences. Dendrograms were constructed and genetic diversity was observed in S. aureus isolates. It can be concluded that high genetic diversity is observed in the isolated S. aureus strains as compared to E. coli and P. aeruginosa.
The distribution of different antibiotic-resistant genes was reported by polymerase chain reaction, as shown in Table 2. The most commonly detected antibiotic resistance genes (erythromycin and aminoglycoside) in S. aureus were erm(B) and aac (6′) aph (2′). The results revealed that aac (6′) aph (2 ') was detected in 18 isolates (60 %), and erm(B) was detected in 14 isolates (46.6 %) of 30 isolates. blaZ, tet (K), msr (A), and erm (C) were not found in any isolates. In E. coli, the most common antibiotic resistance genes (ampicillin, tetracycline, and erythromycin) were ampC, tet (A), and erm(B). The results revealed that the ampC was detected in twenty-four isolates (80 %), and tet(A) and erm(B) were detected in sixteen isolates (53.3 %) out of thirty isolates. erm(A), erm(C), and aadA1 genes were not found in any isolates. The results showed high antibiotic résistance in E. coli and S. aureus strains. The distributions of genes associated with drug resistance differed from those reported worldwide. The phylogenetic tree for E. coli, P. aeruginosa, and S. aureus from the current study clustered with each other and with reference sequences showing their close similarity based on 16S rRNA (Figs. 8,9).
Antibiotic
Gene
Distribution/Percentage
Total Isolates
S. aureus
Erythromycin
erm(B)
18 (60 %)
30
Aminoglycoside
aac(6′)aph (2′’)
14 (46.6 %)
30
Penicillin
blaZ,
0
30
Tetracycline
tet(K)
0
30
Erythromycin
msr(A),
0
30
Erythromycin
erm(C)
0
30
E. coli
Ampicillin,
ampC
24 (80 %)
30
Tetracycline
tet(A)
(53.3 %)
30
Erythromycin
erm(B)
(53.3 %)
30
Erythromycin
erm(A)
0
30
Erythromycin
erm(C)
0
30
Streptomycin
aadA1
0
30
4 Discussion
Diabetic foot ulcer infection is a serious complication commonly observed in elderly diabetic individuals and is difficult to treat. Pakistan is a high-burden diabetes zone of South Asia; however little evidence is obtainable about the molecular characteristics of the bacterial strains dominant in the region. Current study reports on the molecular characterization of multidrug resistance among bacterial isolates from Pakistan. Unfortunately, diabetic foot ulcers have been largely overlooked in healthcare research and planning. Therefore, clinical practice is often guided more by personal opinion than scientific evidence. Moreover, understanding of the underlying pathological mechanisms is limited and communication between the various specialties involved is often disjointed (Khan et al., 2019).
In a study conducted by Ramakant et al. (2011), a global estimate of the prevalence of DFUs was determined through a meta-analysis of 67 published articles. The reported prevalence rate ranged from 1.5 % to 16.6 %. The prevalence rate of 1.5 % was observed in the Australian population, while the highest rate of 16.6 % was observed in the population in Belgium. The prevalence rate observed in the Indian population was 11.6 %. The present study observed that out of 180 samples, the most commonly isolated pathogenic bacteria based on differential media, morphological and biochemical tests were S. aureus 98 (54 %) and E. coli 75 (41.6 %). S. epidermidis 20 (11.1 %) and P. aeruginosa 18 (10 %) were lowest among all isolates. In the current study, we also employed 16S rRNA sequencing to validate bacterial identification. As here, we see most of current study sequences cluster with the sequences reported from Pakistan previously.
In some relevant studies, Gram-negative infection is predominant (Ali et al., 2019). In the relevant study, the main organisms isolated were S. aureus (16 %), E. coli (15 %), Klebsiella pneumoniae (7 %), Proteus mirabilis (11 %), and P. aeruginosa (7 %) (Mutonga, D. M., et al.,2019). Our study showed high resistance to antibiotics against S. aureus and S. epidermidis, including tetracycline, erythromycin, streptomycin, sulfamethoxazole, daptomycin, chloramphenicol, amoxicillin-clavulanate, methicillin, and tetracycline. High resistance was reported against antibiotics (erythromycin, streptomycin, ampicillin, sulfamethoxazole, ceftriaxone, chloramphenicol, and amoxicillin-clavulanate used to treat E. coli infections including. High resistance was also shown against antibiotics used to treat P. aeruginosa, i.e., ceftazidime, imipenem, meropenem, cefepime, amikacin, ceftriaxone, ofloxacin, and amoxicillin-clavulanate. In a comparable study, isolated bacteria showed resistance to antibiotics such as ceftazidime, amoxicillin, tetracycline, ampicillin, piperacillin-tazobactam, cefuroxime, cefepime, erythromycin, clindamycin, and trimethoprim-sulfamethoxazole (Mutonga et al., 2019). Previous research has identified S. aureus, S. epidermidis, and P. aeruginosa as common bacteria found in diabetic foot ulcer (DFU) wound fluids. Some studies have suggested that delayed wound healing may be attributed to the involvement of particular pathogenic microorganisms. The presence of polymicrobial organisms in the wound site might lead to delays in wound healing. Although the bacterial load may significantly affect the wound healing process, the antibiotic resistance pattern found in wound fluid could also play a significant role. Despite the limited effectiveness of most β-lactams against staphylococci, enterobacteria, and acinetobacter spp, piperacillin proved to be the most potent antibiotic against P. aeruginosa (Khan et al., 2019). In contrast, Paterson et al. (2005) found amikacin and piperacillin/tazobactam effective against Pseudomonas, and ciprofloxacin was identified as the most effective drug for Pseudomonas aeruginosa infections. However, 46 % of strains from diabetic wounds in this study were resistant to ciprofloxacin. The resistance against most β-lactams is well-documented for Pseudomonas aeruginosa, the resistance to fourth generation cephalosporins poses major concerns. Gales et al. (2001) reported similar findings with Pseudomonas aeruginosa strains showing higher susceptibility to ceftazidime than cefepime in the Asia-Pacific region. In a study by Gadepalli et al. (2006), enterococci exhibited high levels of resistance to ciprofloxacin, erythromycin, and tetracycline, while showing low levels of resistance to high levels of aminoglycosides. Despite being commonly referred to as commensals, enterococci can act as opportunistic pathogens in diabetic individuals, as noted by Citron et al. (2007). Various studies have demonstrated the presence of biofilm-forming microorganisms in chronic wounds, as reported by James et al. (2008). Multispecies communities in biofilms contribute a critical role in the wound-healing process (Gupta et al., 2023; Tiwari et al., 2012).
In the current investigation, the antibiotic resistance genes frequently detected in S. aureus were erm(B) and aac (6 ') aph (2′). The results revealed that out of thirty isolates, aac (6′) aph (2′) was detected in 18 isolates (60 %), and erm(B) was detected in fourteen (46.6 %) isolates. The erm(B) gene encodes a protein that makes S. aureus resistant particularly to erythromycin. The aac(6′) aph (2′) gene encodes an enzyme that modifies aminoglycoside antibiotics. Erythromycin is not as commonly used as methicillin to treat S. aureus infections, so the erm(B) gene is not globally as common as the mecA gene which imparts methicillin resistance. blaZ, tet (K), msr(A), erm(C) were not found in any isolates. In E. coli, the most common antibiotic resistance genes (ampicillin, tetracycline, and erythromycin) are ampC, tet (A) and erm(B) in Pakistani population as reported in current investigation. The results revealed that ampC was detected in 24 isolates (80 %), and tet(A) and erm(B) were detected in 16 isolates (53.3 %) of 30 isolates. erm (A), erm(C), and aadA1 genes were not found in any isolates. The most common antibiotic resistance gene found in E. coli isolates from DFUs worldwide is blaCTX-M. It encodes a protein that makes E. coli resistant to extended-spectrum beta-lactam antibiotics, such as cefotaxime and ceftazidime which pose a serious global threat. Other common antibiotic resistance genes found in E. coli isolates from DFUs worldwide include ampC, qnrB, and sul3. The geographical variation in the distribution of antibiotic resistance genes in DFUs is a complex issue. Many factors contribute to this variation, including the use of antibiotics, the environment, and the genetic makeup of bacteria. In a relevant study, PCR was performed to identify 13 virulence genes in E. coli using their specific primers. The distribution of the tetracycline-resistant gene, tetA, was higher in Sudan and China isolates by 54 % and 84 %, respectively, comparable to our study and other studies reported globally (Enne et al., 2008; Abdelgader et al., 2018). In another relevant study, out of 125 samples, 19 S. aureus isolates were identified. All the identified isolates were MDR. The isolates resistant to penicillin, tetracycline, erythromycin, and kanamycin were studied for the resistance genes blaZ (100 %), (msrA(100 %), ermB(0 %), and ermC (100 %), aac (6 ') aph (2′) (62.5 %) and tetK (100 %). The distribution of genes is somehow different from those reported in our study (Fawzy et al., 2017).
The distributions of genes associated with antibiotic resistance in the studied region differ from those reported worldwide (Mutonga et al., 2019).
The findings of the study can help clinicians decide which antibiotics to prescribe as initial empiric therapy. If a patient has a DFU caused by an ESBL-producing E. coli strain, choosing an antibiotic that is not affected by the resistance mechanism like carbapenems may prove more effective. The current study provided valuable local data endorsing the revision of treatment guidelines specific to the region. This data can contribute to national surveillance efforts allowing public health authorities to monitor trends, identify emerging resistance patterns, and implement effective infection control strategies. Current investigation also calls to explore alternative approaches including novel antimicrobial peptides to treat DFUs. Comparative studies between different regions can provide valuable insights into the epidemiology of DFUs.
This study has certain limitations, including a small sample size. It highlights the need for further research involving a larger patient population to validate the findings. Although the results are preliminary, they provide valuable insights for informing treatment decisions for patients with DFU. The high incidence of staphylococcus aureus and extended-spectrum β-lactamase-producing strains highlights the importance of judicious antibiotic use to manage DFUs effectively. The use of antibiotics at an alarming rate in the developing countries such as in Pakistan to treat DFUs causes high resistance and demands for the new antibiotics screenings.
5 Conclusions
The most commonly isolated organisms from DFUs were S. aureus and E. coli. The lowest among all the isolates were S. epidermidis and P. aeruginosa. The antibiotic resistance genes most commonly detected in S. aureus and E. coli were erm(B) and aac(6′) aph (2′) and ampC, tetA, erm(B), respectively. The distributions of genes associated with drug resistance differed from those reported worldwide. These findings will aid in guiding the empirical use of antibiotics for treating diabetic foot infections, thereby reducing the risk of inappropriate antibiotic use and the development of antibiotic resistance. The increase of general awareness programs can help to stop the progression of infection and more importantly, the risk of lower extremity amputation can be decreased with multimodal approaches, improved diagnostic techniques, appropriate antibiotic use, surgical interventions, and routine foot evaluations.
Institutional review board statement
The study was approved by institutional review board, University of Swabi, Pakistan.
Informed consent statement
The patient informed consent was obtained at the time of sample collection.
Consent to participate
All authors consent to participate in the manuscript publication
Consent for publication
All authors approved the manuscript to be published.
Availability of data and material
The datasets generated during the current study are available from the corresponding author on reasonable request.
Ethics approval
Not applicable.
Funding
This research received no external funding.
CRediT authorship contribution statement
Muhammad Idrees: Conceptualization, Investigation, Methodology. Imran Khan: Investigation, Methodology. Amin Ullah: Conceptualization, Investigation, Project administration, Resources, Supervision. Syed Muhammad Mukarram Shah: Formal analysis, Resources, Supervision. Hafiz Ullah: Formal analysis, Software. Muhammad Ajmal Khan: Data curation, Methodology, Software, Writing – review & editing. Rafa Almeer: Funding acquisition, Resources, Writing – review & editing. Zafar Abbass Shah: Formal analysis, Funding acquisition, Writing – review & editing. Tariq Nadeem: Data curation, Writing – review & editing.
Acknowledgements
The authors acknowledge the Surgery and Medical Wards of different tertiary-care hospitals and microbiology laboratories of KP, Pakistan. The authors would like to extend their sincere appreciation to the Researchers Supporting Project Number (RSP2024R96), King Saud University, Riyadh, Saudi Arabia.
References
- Antibiotics resistance genes screening and comparative genomics analysis of commensal Escherichia coli isolated from poultry farms between China and Sudan. BioMed. Res. Int. 2018
- [CrossRef] [Google Scholar]
- Biofilms in diabetic foot ulcers: impact, risk factors and control strategies. Int. J. Mol. Sci.. 2021;22(15):8278-8290.
- [CrossRef] [Google Scholar]
- Microorganism culture and antibiotic sensitivity pattern isolated from diabetic foot infections at tertiary care hospital Mardan. Pak. J. Surg.. 2019;35(3):220-223.
- [Google Scholar]
- Isolation and antibiotic susceptibility of the microorganisms isolated from diabetic foot infections in Nemazee hospital, Southern Iran. J. Pathogens. 2015;1–7
- [CrossRef] [Google Scholar]
- Bacteriological analysis and antibiotic resistance in patients with diabetic foot ulcers in Dhaka. PLoS One. 2024;19(5):e0301767.
- [Google Scholar]
- Bacteriology of moderate-to-severe diabetic foot infections and in vitro activity of antimicrobial agents. J. Clin. Microbiol.. 2007;45(9):2819-2828.
- [CrossRef] [Google Scholar]
- CLSI, (2020): Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2020.
- A high prevalence of antimicrobial resistant Escherichia coli isolated from pigs and a low prevalence of antimicrobial resistant E. coli from cattle and sheep in Great Britain at slaughter. FEMS Microbiol. Lett.. 2008;278, 2:193-199.
- [CrossRef] [Google Scholar]
- Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39(4):783-791.
- [Google Scholar]
- A clinico-microbiological study of diabetic foot ulcers in an Indian tertiary care hospital. Diabetes Care. 2006;29(8):1727-1732.
- [CrossRef] [Google Scholar]
- Characterization of Pseudomonas aeruginosa isolates: occurrence rates, antimicrobial susceptibility patterns, and molecular typing in the global SENTRY Antimicrobial Surveillance Program, 1997–1999. Clinical Infectious Dis.. 2001;32:S146-S155. (Supplement_2)
- [CrossRef] [Google Scholar]
- Classification, microbiology and treatment of diabetic foot infections. J. Wound Care. 2018;27(7):434-441.
- [CrossRef] [Google Scholar]
- A diversity profile of the human skin microbiota. Genome Res.. 2008;18(7):1043-1050.
- [Google Scholar]
- Extended spectrum beta lactamases conferring transferable resistance to newer β-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis.. 1998;10:867-878.
- [CrossRef] [Google Scholar]
- Risk factors for infection of the diabetic foot with multi-antibiotic resistant microorganisms. J. Infect.. 2007;54(5):439-445.
- [CrossRef] [Google Scholar]
- Antimicrobial Susceptibility Pattern of Isolates From Diabetic Foot Ulcers. J. Islam. Int. Med. College (JIIMC). 2019;14(3):105-110.
- [Google Scholar]
- Antibiotic resistance profiling and phylogenicity of uropathogenic bacteria isolated from patients with urinary tract infections. Antibiotics. 2023;12(10):1508.
- [Google Scholar]
- MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol.. 2016;33(7):1870-1874.
- [Google Scholar]
- Lipsky, B. A., Senneville, É., Abbas, Z. G., Aragón‐Sánchez, J., Diggle, M., Embil, J. M., & International Working Group on the Diabetic Foot (IWGDF). (2020). Guidelines on the diagnosis and treatment of foot infection in persons with diabetes (IWGDF 2019 update). Diabetes/metabolism research and reviews, 36(7), 32-80. Doi: 10.1002/dmrr.3280.
- Bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect.. 2012;18:268-281.
- [CrossRef] [Google Scholar]
- A review of Wagner classification and current concepts in management of diabetic foot. Int J Orthop Sci. 2018;4(1):933-935.
- [Google Scholar]
- Bacterial isolation and antibiotic susceptibility from diabetic foot ulcers in Kenya using microbiological tests and comparison with RT-PCR in detection of S. aureus and MRSA. BMC. Res. Notes. 2019;12(1):1-6.
- [Google Scholar]
- Biofilms in diabetic foot ulcers: significance and clinical relevance. Microorganisms. 2020;8(10):15-80.
- [CrossRef] [Google Scholar]
- The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol.. 1987;4(4):406-425.
- [Google Scholar]
- Sensitivity of Escherichia Coli bacteria towards antibiotics in patient with diabetic foot ulcer. Pharmaceutical Sci. Res.. 2018;5(1):19-24.
- [CrossRef] [Google Scholar]
- Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci.. 2004;101(30):11030-11035.
- [Google Scholar]
- Diabetic foot ulcers: classification, risk factors and management. World J. Diabetes. 2022;13(12):1049.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103320.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







