Translate this page into:
Antimicrobial quercetin 3-O-glucoside derivative isolated from Streptomyces antibioticus strain ess_amA8
⁎Corresponding author. essam_92003@yahoo.com (Essam Nageh Sholkamy) elisi@ksu.edu.sa (Essam Nageh Sholkamy)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Streptomyces antibioticus strain ess_amA8 was isolated from the insect Tapinoma simrothi, and was tested for the production of compounds that inhibit the growth of microorganisms. The cultural and physiological descriptions of the strain revealed that it belonged to the Streptomyces genus. The 16S rRNA gene from the Streptomyces isolate was sequenced, and its length was 1156 bp and it was submitted to Genbank (KF996507). According to molecular and morphological studies, the isolated strain was ultimately identified as strain ess_amA8 of Streptomyces antibioticus. The key compound was extracted from the strain ess_amA8, which was active in vitro, against a wide range of microorganisms, such as Staphylococcus aureus ATCC6538, Bacillus subtilis ATCC 6633, Pseudomonas aeruginosa ATCC 9027, Escherichia coli ATCC-7839, Candida albicans, Fusarium moniliforme, Aspergillus niger, and Aspergillus flavus. The physico-chemical property of the purified antimicrobial compound analyzed and its proposed empirical formula was C27H25O13. According to the IUPAC name, this compound was identified as 5,7-diethoxy-2-(3-methoxy-4-methylphenyl)-3-[(2R,3S,4R,5R,6S)-3,4,5,6-tetrahydroxy-2-(hydroxymethyl)tetrahydro-2H-pyran-3-yl] oxy-4H-chromen-4-one, and was a quercetin 3-O-glucoside derivative. In summary, the present study highlights the fact that the purified quercetin 3-O-glucoside antimicrobial derivative, first report to this Streptomyces sp., and it is a flavanone glycoside.
Keywords
Streptomyces antibioticus
Tapinoma simrothi
Quercetin
Flavanone glycoside
1 Introduction
Incurable diseases are becoming increasingly commonly, with the rise in non-sensitivity of microbes to antibiotics; the lack of new alternatives poses a threat to world health (Jones et al., 2008). This serious rise in antibiotic resistance of microbes necessitates research on unconventional methodologies for discovering new drugs (Kumar et al., 2017). Biological diversity is a major resource for exploring new products with specific biological activities; exploring natural, biologically active compounds from insect-microbe symbioses is one of the most unconventional methods of drug discovery (Brachmann and Bode, 2013). Oh et al. (2011) isolated Streptomyces sp. from wasp associated symbiotic microbes and those insects are lactam family members. Poulsen et al. (2011) studied the antagonistic activity between Streptomyces sp. associated with solitary wasps, to find whether they produced compounds with antimicrobial activities against fungi and bacteria. Their results support the fact that isolated actinomycetes are associated with insect and they are the completely new source for biologically active novel compounds. A study by Barke et al. (2010) showed the potential role of the symbiotic relationship in the production of antifungal metabolites from Streptomyces sp. associated with leaf-cutting ants; these species could be a valuable source of novel antimicrobial agents. Candicidin macrolides, which are extracted from antagonistic actinomycetes, are effectively kill the fungal pathogen Escovopsis, but not against the ant’s garden fungus Leucoagaricus gongylophorus (Haeder et al., 2009). Previous studies showed that the actinomycetes isolated from termite gut samples were selectively active against some gram-negative and gram-positive pathogenic bacteria, and pathogenic yeast (Arango et al., 2016). Our study aimed to isolate a new strain of Streptomyces species competent to produce antimicrobial compounds combat pathogenic bacteria viz., P. aeruginosa, B. subtilis, S. aureus and E. coli, and pathogenic fungi such as Fusarium moniliforme, A. niger, A. flavus, C. albicans. Further, the biologically active compound was purified and identified.
2 Materials and methods
2.1 Tested pathogenic microorganisms
The tested pathogens used for preliminary screening were as follows: gram-positive bacteria (B. subtilis ATCC-6633 and S. aureus ATCC-6538); gram-negative bacteria (P. aeruginosa ATCC 9027 and E. coli ATCC-7839); and unicellular and filamentous fungi (C. albicans, F. moniliforme, A. niger and A. flavus).
2.2 Isolation of actinomycete strain
The actinomycete isolates isoalted from the insect Tapinoma simrothi and it was captured from the botanical garden of the King Saud University, Riyadh, Saudi Arabia. The isolate was prepared by crushing the insect in sterilized distilled water. This was followed by the pour plate technique, for which starch casein agar was used and composition as follow; (g/L), Starch-10.00, Casein-0.30, KNO3-2.00, MgSO4·7H2O-0.05, K2HPO4-2.00, NaCl-2.00, CaCO3-0.02, FeSO4·7H2O-0.01, Agar-18.00 and pH 7.0 ± 0.1. All the plates were incubated for 7 days at 28 °C. The colonies grown on the agar plates were noticed as having a leathery texture; a single colony of different morphologies were aseptically selected and streaked onto a separated starch casein agar plate and incubated for 3 days at 28 ± 2 °C.
2.3 Preliminary screening of actinomycetes for antagonistic activity
Actinomycete isolates were introduced into starch casein agar plates (SCA) and incubated at 28 ± 2 °C for 3 days. The sterile Mueller Hinton (MH) agar plates were inoculated with the respective pathogenic bacteria and the well were made in the center of the plates using sterile cork borer (6 mm diameter). Discs containing the well-grown actinomycete culture were placed in these wells. The inoculated Petri plates were incubated at 37 °C for 24 h for bacteria. In case of fungi, the medium used to measure the antagonistic activity was potato dextrose agar (PDA). The antimicrobial activities of the actinomycetes were determined by the size of the inhibition zone.
2.4 Cultural and biochemical characterization of the actinomycete isolate
The actinomycete isolate was morphologically and biochemically characterized according to Nonomura (1974) and Goodfellow et al. (2012).
2.5 Physiological characterization of actinomycete isolate
The isolate was physiologically characterized (temperature of 4, 28, 37 and 45 °C, salinity of 1, 4, 7, 10 and 14%, and pH of 5.0, 7.0 and 10.0). The enzymatic activity assay was performed as per methodology to Hopwood (1957), including tests for starch and casein hydrolysis, and cellulase, protease, and lipase activity. These tests were performed in triplicate.
2.6 DNA extraction from the actinomycete isolate
Total genomic DNA of actinomycete isolate was extracted by the method stated by Barsotti et al. (1987).
2.7 Amplification of 16S ribosomal RNA and sequencing
The 16S ribosomal DNA gene was amplified by PCR using the universal primer pair Star-F 5′-AGAGTTTGATCGTGGCTCAG-3′ and 1387- R 5′-CGGGCGGTGTGTACAAGG-3′ (2001). The amplified products were analyzed by GATC Biotech, European Custom Sequencing Center, D-51105 Cologne, Germany. The DNA sequence analysis was performed with BLAST multiple sequences analysis from NCBI.
2.8 Production of extracellular bioactive secondary metabolites by liquid state fermentation
For the biosynthesis of the active substance using submerged culture, an agar seven days old slant culture of the actinomycete isolate was inoculated into starch nitrate broth (pH 7.0), and it was incubated in shaker with 200 rpm for 3 days at 30° C. Fifty ml of the mother culture was inoculated in the fermentation flasks, each containing 1000 ml of the fermentation medium. Further, the inoculated flasks were incubated at 30° C in shaker with 250 rpm. After the incubation period, the cultures were collected and filtered with cotton wool. The clear filtrate was collected and subjected to extraction using different organic solvents, and then vaporized until dry in a rotary evaporator at 40° C (1976).
2.9 Extractions of the bioactive fractions from the fermented broth
After fermentation, the filtrate from each flask was separated from the mycelium by filtration. Whole broth was concentrated to the least volume by evaporation under vacuum, and then subjected to sequential solvent extraction in a separating funnel using solvents of various polarities namely; n-hexane, ethyl acetate, diethyl ether, chloroform and n-butanol. The extracts were then dried to remove the solvents. The extracted compounds were reconstituted with dimethyl sulfoxide (DMSO). Their biological activity against the test organisms was then analyzed.
2.10 Separtation of crude extract by thin layer chromatography (TLC)
The active fractionations of the crude extract were separated by TLC (Stahl, 2013) on precoated plates using Chloroform: Methanol (9:1 V/V) solvent systems. The TLC plates were air-dried, and examined under UV spectrum.
2.11 Determination of minimum inhibitory concentration (MIC)
The antimicrobial activity of the purified compound was determined by the agar disc diffusion method (Grammer, 1976). Different concentrations of purified compound, which were 5, 10, 40, and 80 μg/ml, were used against the pathogenic test bacteria. Discs soaked with the compound were air-dried and placed on petri plates inoculated with the different microbes using a pair of sterilized forceps. The plates were incubated at 37° C for 24 h for bacterial pathogens. Paper discs soaked in methanol were used as control samples in each test. All the experiments were performed in triplicate.
2.12 Physicochemical characteristics of purified active compound
The physicochemical properties of the purged substance, viz., solubility, color, acid and base behavior and melting point were also studied.
2.13 Spectral analysis
Elucidation of the structure of the isolated compounds includes the use of the following physical methods. The UV spectrum of the purified active substance, which was developed on a TLC plate (silica gel G-60 aluminum sheet, Merck, Germany) was recorded. The Infra-Red absorption spectrum was assessed with FTIR, Perkin-Elmer 1650 spectrophotometer, at the Microanalytical Center of Cairo University, Giza, Egypt. Proton nuclear magnetic resonance spectrum was measured on EM-500 MHZ (NMR) spectrometer using chloroform (CHCl3) as a solvent at the National Research Center (NRC), El-Dokii, Egypt.
3 Results
The actinomycete isolate was identified to the species level according to its specific characteristics. Microscopically, it was observed that the morphology of the spore chains of aerial mycelium is retinaculum apertum (Fig. 1). Furthermore, for adequate identification, melanin production is one of the important physiological characters in Streptomyces taxonomy. The finding showed (Table 1) the isolate did not produce melanin pigments on the media used, whereas the aerial mycelium is with different colors accoriding to growth medium. The isolate was inoculated on 9 different media and the growths were observed after 48 h. The results were interpreted as the following values: weak, moderate, good, very good. Good Growth of the isolate was observed on ISP2, ISP4, ISP5, ISP7, Starch nitrate agar, Nutreint agar and Czapek's agar while was weak on ISP3 and ISP6 (Table 1). The Streptomyces sp. utilized protein and starch, but not lipids and no producing H2S as shown in Table 2. It liquefied gelatin. The actinomycete isolate was selected for an analysis of the enzyme activities of the various species, to identify the isolates exhibiting the highest enzyme potential. The results showed that the amylase, protease, catalase, DNase, and urease tests yielded positive results, while the chitinase and lipase tests yielded negative results; this is shown in Table 2. After 7 days of incubation under different conditions, the growth of isolates was observed and the results were recorded, as shown in Table 2. The results show that the prime temperature for growth of this isolate is 30 °C; it did not grow at 45 °C. Excellent growth of the Streptomyces isolate could be seen within a temperature range of 26–30 °C (Table 2). Good growth was observed within an NaCl range of 1–4 %, and at a pH of 7.0 (Table 2). The results in Table 3 showed that the isolate can utilize rhaffinose, fructose, sucrose, arabinose, lactose, D-galactose, D-xylose, Meso-inositol, cellobiose and maltose, but could not utilize salicine, and cellulose as a carbon source. Good Growth of the Streptomyces sp., as shown in Table 3, was observed when L-valine, lycine, L-methionine, L-arginine, L-aspargine, L-orinthine and L-histidine were used as nitrogen sources; weak growth was seen in case of L-Phenyl alanine, and no growth was observed in case of L-Cysteine. The Streptomyces sp. was sensitive to erythromycin (15 µg), gentamicin (10 µg), kanamycin (1000 µg), colistin sulfate (10 µg), ceftazidime (30 µg), and ciprofloxacin (1 µg), but not to tetracycline (30 µg) and chloramphenicol (30 µg). The isolate was tested using conditions of environmental stress to identify the isolates with a maximum growth range and resistance to stress. Finally, according to biochemical and molecular characterization, the actinomycete isolate was identified as Streptomyces antibioticus strain ess_amA8 with the accession number KF996507.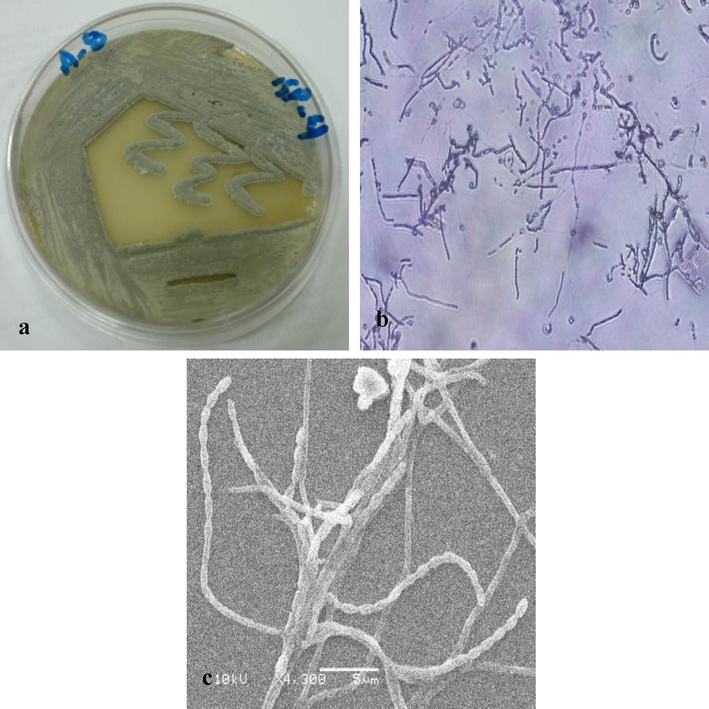
Morphological characterization of strain ess_amA8. a) Culture of S. antibioticus strain ess_amA8 on ISP 4 agar. b) Shape of the spore chain under light microscope. c) Shape of the spore chain under scanning electron microscope.
Types of media
Growth
Aerial mycelium
Substrate mycelium
Diffusible pigments
Yeast – malt extract agar (ISP 2)
Good
Light Gray
(ISCC-NBS 264), Vilvilitly pink colonies with edge grayishgreenish
(ISCC-NBS 264)Very deep red (SCC-NBS 243)
Oat meal agar (ISP 3)
Weak
Light Gray
(ISCC-NBS 264)Light Gray
(ISCC-NBS 264)Very deep red
(SCC-NBS 239)
Inorganic-trace salt- starch agar (ISP 4)
Good
Light Gray
(ISCC-NBS 264)greenish
(ISCC-NBS 264)Gray red
(ISCC-NBS 245)
Glycerol asparagine agar (ISP 5)
Good
Hygroscopic grayish
Light Gray
(ISCC-NBS 264)Light yellow brown
(SCC-NBS 76)
Peptone yeast extract iron agar (ISP 6)
Weak
Hygroscopic growth
colorless
None
Tyrosine agar (ISP 7)
Good
Hygroscopic growth
colorless
None
Starch-nitrate agar
Good
Light Gray
(ISCC-NBS 264)Greenish
(ISCC-NBS 264)Light yellow brown
(SCC-NBS 76)
Nutrient agar
Good
Hygroscopic growth
colorless
None
Czapek’s agar
Good
White
(ISCC-NBS 263)greenish
(ISCC-NBS 264)Slight red
(SCC-NBS 237
Enzyme activity
Amylase
+
Protease
+
Chitinase
–
Catalase
+
DNase
+
Hydrolysis of esculin
+
Lecithin hydrolysis
–
H2S production
–
Nitrate reduction
+
Urea hydrolysis
+
Lipid hydrolysis
–
Growth at different NaCl concentrations
1%
++
4%
+
7%
–
10%
–
14%
–
Growth at different pH
5
–
7
+
10
–
Growth at different temperatures
4
–
26
++
30
+++
37
+
45
–
Carbon sources
Rhaffinose
+
Fructose
+
Sucrose
+
Arabinose
+
lactose
+
D-galactose
+
D-xylose
+
Meso-inositol
+
Cellobiose
+
Maltose
+
Salicine
–
cellulose
–
Nitrogen sources
L-Cysteine
–
L-Valine
+
lycine
+
L-Methionine
+
L-Arginine
+
L-aspargine
+
L-Orinthine
+
L-Phenylalanine
±
L-Histidine
+
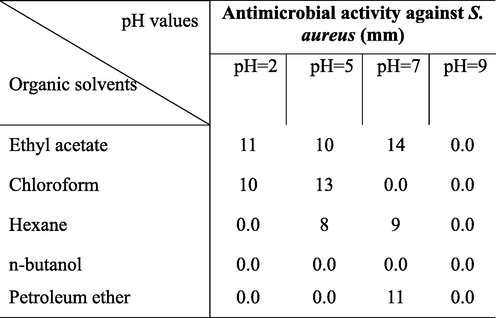
In this study, the antagonistic activity of strain ess_amA8 was tested against F. moniliforme, A. niger, A. flavus, C. albicans, P. aeruginosa, B. subtilis, S. aureus, and E. coli. The results showed that the strain has the ability to inhibit the growth of all the tested pathogenic microbes except A. flavus, as shown in Table 4. The most potent species from the Actinomycetes isolate was isolated on the basis of the wide spectrum of its antimicrobial activity. The results recorded in Table 5 indicated that the maximum extraction of the bioactive substance occurred at a pH range of 2–7 (acidic range) using ethyl acetate, chloroform, and petroleum ether, while the extraction decreased or failed when pH values rose to the alkaline range (>7.0). Additionally, n-butanol failed to extract the active substance at different pH values. Hence, ethyl acetate was chosen as the most suitable organic solvent to extract the active substance, especially when the active filtrate was adjusted to a pH of 7.0. The best solvent system was ethanol: Chloroform: acetic acid (6: 2: 2 v/v/v), and it was utilized at room temperature. This solvent system separated the crude extract into six different spots, and each spot was bioassayed with S. aureus. The active spot was at Rf 0.85. The pure, bioactive compound was also tested with other pathogenic organisms. Finally, the pure active compound had a melting point of 232° C. The purified compound was isolated as an amorphous pale yellow powder with no characteristic odor; it appeared yellow in acidic and alkaline solutions. The isolated compound was soluble in chloroform, ethyl alcohol, ethyl acetate and methanol, and it was insoluble in petroleum ether, hexane, benzene and water. The absorption peaks of compound showed at 252 and 494 nm that is features of highly conjugated systems. Structural elucidation of the compound could be achieved by analysis of its UV, NMR, and MS spectral data (Fig. 2 and Fig. 3). According to physicochemical characterizations, the purified compound produced by strain ess_amA8 was identified as a quercetin 3-O-glucoside derivative (Fig. 2 and Fig. 3).
Test organisms
Inhibition zone diameter (mm)
Gram positive bacteria
B. Subtilis
15.00
S. aureus
22.00
Gram negative bacteria
P. aeruginosa
20.00
E. coli
19.00
Unicellular fungi
C. albican
14.00
Filamentous fungi
A. niger
13.00
A. flavus
0.00
F. moniliforme
15.00
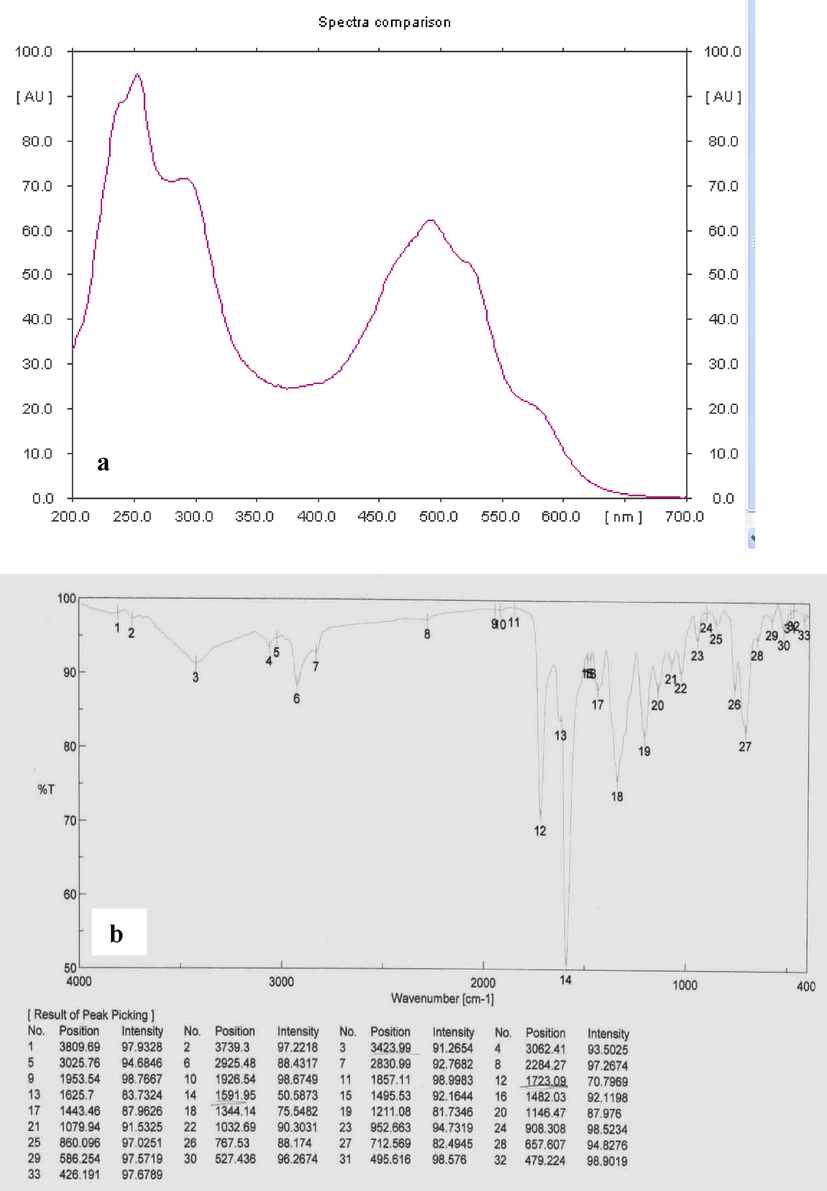
a) UV spectrum; b) FTIR spectrum of active compound.
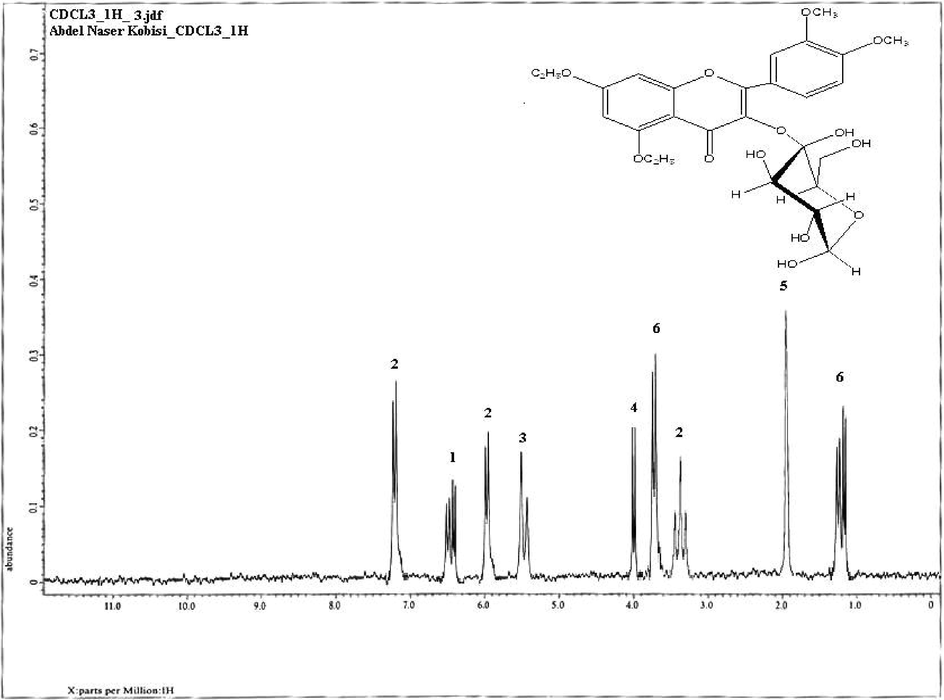
NMR Spectrum of the bioactive compound produced by S. antibioticus strain ess_amA8.
4 Discussion
The isolated actinomycete was identified to the species level according to its specific characteristics. Spore masses were matched against the seven color wheels of Tresner and Backus (1963) as used in the ISP (Shirling and Gottlieb, 1966). Microscopically, it was observed that spore chains of its aerial mycelium showed Retinaculum-Apertum morphology (Pridham et al., 1958). The strain wasidentified as a member of S. antibioticus by 16S rRNA gene sequencing and the sequence has been deposited in GenBank (Accession Number: KF996507). The sequenced 16S rRNA gene has revealed that the strain ess_am8 showed 99.0% similarity with S. antibioticus. Classification of the isolate was initially established, based on its morphological and physiological characterizations. Nowadays, molecular approaches by 16S rRNA gene analysis were commonly used for identification of actinomycetes (Nimaichand et al., 2013). The foremost intention of this research was to discover novel, biologically active secondary metabolites. Insect bodies are the most common habitats of Streptomyces; symbiotic associations occur between the insects and the actinomycetes. Our research was centered on actinomycetes and more particularly on genus Streptomyces, the species of which have documented with their remarkable antimicrobial activities (Thakur et al., 2003). As on day the genus Streptomyces has been assessed at least 1,00,000 novel biocompounds with medically interest (Watve et al., 2001). In an earlier report, the strain PE7 of Streptomyces was found to produce antimicrobial compounds (Gopikrishnan et al., 2013). In our current study, the active antimicrobial compound was identified as the quercetin and this chemical name was 3-O-glucoside derivative, 2- (3,4 - dihydroxyphenyl) − 3,5,7 –trihydroxy-4H-chromen-4-one. Quercetin is a kind of flavonoid commonly present in herbal medicines and plant foods (Materska, 2008). In addition, it also possessed antiviral, anticarcinogenic, and anti-inflammatory effects (Lee et al., 2013).
Acknowledgment
The authors thank the Deanship of Scientific Research at King Saud University for funding this work through research group RG-1439-025 and also thank the RSSU at King Saud University for its technical support.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Antimicrobial activity of actinobacteria isolated from the guts of subterranean termites. Environ. Entomol.. 2016;45(6):1415-1423.
- [Google Scholar]
- A mixedcommunity of actinomycetes produce multiple antibiotics for the fungus farming ant Acromyrmex octospinosus. BMC Biol.. 2010;8:109-112.
- [Google Scholar]
- Rapid isolation of DNA from actinomyces. Ann Inst. Pasteur Microbiol.. 1987;138(5):529-536.
- [Google Scholar]
- Identification and bioanalysis of natural products from insect symbionts and pathogens. Adv. Biochem. Eng. Biotechnol.. 2013;135:123-155.
- [Google Scholar]
- Goodfellow M., Kämpfer P., Busse H.J., eds. Bergey’s Manual® of Systematic Bacteriology. New York, NY: Springer New York; 2012.
- Bioprospecting of actinobacteria from mangrove and estuarine sediments for antifouling compounds. International Journal of Innovation in Science. Eng. Technol.. 2013;2(7):2726-2735.
- [Google Scholar]
- Antibiotic sensitivity and assay test. In: Collins C.H., Lyne P.M., eds. Microbiological Methods. London: Butterworth and Co.; 1976. p. :235.
- [Google Scholar]
- Candicidin-producing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. P. Nat. Acad. Sci.. 2009;106:4742-4746.
- [Google Scholar]
- Jones, K.E., Patel, N.G., Levy, M.A., 2008. Global trends in emerging infectious diseases. Nat. 451 (7181), 990–993.
- Molecular insights into antimicrobial resistance traits of multidrug resistant enteric pathogens isolated from India. Sci. Rep.. 2017;7:144-168.
- [Google Scholar]
- Anti-inflammatory effect of fucoidan extracted from Ecklonia cava in zebrafish model. Carbohydr. Polym.. 2013;92:84-89.
- [Google Scholar]
- Quercetin and its derivatives: chemical structure and bioactivity – a review. Pol. J. Food Nutr. Sci.. 2008;58(4):407-413.
- [Google Scholar]
- Streptomyces manipurensis sp. nov., a novel actinomycete isolated from a limestone deposit site in Manipur, India. Anton. Leeuw. Int. J. G.. 2013;102:133-139.
- [Google Scholar]
- Key for classification and identification of 458 species of the Streptomycetes included in ISP. J. Ferment. Technol.. 1974;52(2):78-92.
- [Google Scholar]
- Oh, D.C., Poulsen, M., Currie, C.R., 2011. Sceliphrolactam, a polyene macrocyclic lactam from a wasp-associated Streptomyces sp. Org. Lett. 13(4), 752–755.
- Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS One. 2011;6(2):e16763
- [Google Scholar]
- A selection of media for maintenance and taxonomic study of Streptomyces. Antibiot. Annu. 1958:947-953.
- [Google Scholar]
- Methods for characterization of Streptomycetes species. Int. J. Syst. Bacteriol.. 1966;16:313-340.
- [Google Scholar]
- Stahl, E., (Ed.), 2013. Thin-Layer Chromatography: A Laboratory Handbook. Springer Science and Business Media.
- Thakur, D., Sharbani, M., Gogoi, D.K., 2003. The Streptomyces flora of Indo-Burma hot spot: isolation and screening for biologically active metabolites. In Bioprospecting of Commercially Important Plants. Proceedings of the National Symposium on “Biochemical Approaches for Utilization and Exploitation of Commercially Important Plants”, Jorhat, India. Indian Soc. Agri. Biochem. 202–206.
- System of color wheels for streptomycete taxonomy. Appl. Microbiol.. 1963;11(4):335-338.
- [Google Scholar]
- How many antibiotics are produced by the genus Streptomyces? Arch. Microbiol.. 2001;176(5):386-390.
- [Google Scholar]







