Translate this page into:
Antimicrobial potential of Chlorella sorokiniana on MRSA – An in vitro study and an in silico analysis on ClpP protease
⁎Corresponding authors at: School of Health Sciences, Swinburne University of Technology, Hawthorn campus, Melbourne, VIC 3122, Australia (Lloyd Charmaine) and Department of Microbiology, Saveetha Dental College and Hospitals, Saveetha Institute of Medical and Technical Sciences (SIMATS), Saveetha University, Chennai 600077 (A.S. Smiline Girija). calloyd@swin.edu.au (Charmaine Lloyd), smilinegirija.sdc@saveetha.com (A.S. Smiline Girija)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objective
Methicillin-resistant Staphylococcus aureus (MRSA) strains are a leading cause of communicable disease in community and nosocomial settings. They are responsible for high morbidity and mortality. Researchers currently pursue novel antimicrobials from natural sources against non-traditional drug targets of staphylococci to ensure a pipeline of potent drugs, in the face of rising drug resistance. The focus of this study was to screen compounds from a freshwater isolate of Chlorella sorokiniana for anti-staphylococcal activity, using traditional microbiology, phytochemical analysis and bioinformatics approaches.
Methods
Chlorella sorokiniana methanol extract was investigated for its antimicrobial potential on Staphylococcus aureus strains (ATCC and MRSA isolates) by Kirby Bauer disc diffusion, broth microdilution, cell cytotoxicity and thin layer chromatography-bioautography (TLC-BA). Two antimicrobial TLC-BA antimicrobial fractions (A and B) were subject to gas chromatography mass spectrometry (GCMS). The structures of 9 compounds representing GCMS peaks were tested in silico, for their pharmacokinetic properties and binding energy efficiency with the target, using Molinspiration tool and Autodock 4.2.
Results
Mean zone diameter of inhibition of growth by CSME (20 mg) was 21 mm, MIC/MBC was 0.31/2.5 mg/L. GCMS analysis of TLC fraction-A revealed 31 phytochemicals, of which 2-pentanone,4-hydroxy-4-methyl- had the highest area % (65.61) and TLC fraction-B revealed 4 peaks of which pentadecanoic acid and 1-(+)-ascorbic acid 2,6-dihexadecanoate had the highest area % (45.57, 48.09).
In silico analysis of 9 peak compounds on the target of interest showed that compound 2: 2-pentanone,4-hydroxy-4-methyl- and compound 7: 1,2 – benzene dicarboxylic acid, mono (2- ethylhexyl) ester, satisfied Lipinski’s rule of 5, and displayed the least binding energies −6.93 and −5.74 with ClpP protease, thus holding pharmaceutical potential, and supporting further investment into in vitro and in vivo studies.
Conclusions
C. sorokiniana, a less studied microalga thus offers a promising natural resource for anti-MRSA phytochemicals, capable of targeting ClpP1 protease.
(290 words)
Keywords
Chlorella sorokiniana
Microalgae
MRSA
Staphylococci
ClpP1
Zone of inhibition of growth
- CSME
-
Chlorella sorokiniana Methanol Extract
- C. sorokiniana
-
Chlorella sorokiniana
- MRSA
-
Methicillin resistant S. aureus
- S. aureus
-
Staphylococcal aureus
- CFU
-
colony forming units
- SCCmec
-
(staphylococcal chromosome cassette mec)
- ClpP
-
(caseinolytic protease P)
- MIC
-
Minimum inhibitory concentration
- MBC
-
Minimum bactericidal concentration
- TLC-BA
-
(Thin-layer bioautography)
- CC
-
(Column chromatography)
- GCMS
-
(gas liquid chromatography
Abbreviations
1 Introduction
Staphylococcus aureus are Gram-positive cocci. They have pathogenic potential and may possess antibiotic resistance and virulence genes. Infections of the skin, soft-tissues, bone, blood and lower respiratory tract may exacerbate into severe illnesses such as osteomyelitis, endocarditis and toxic shock syndrome (Brinkac et al. 2017). Methicillin resistant S. aureus (MRSA) account for 25–50% of infections acquired in hospital settings and are a major cause of treatment failure. MRSA are resistant to both beta-lactam antibiotics and semisynthetic β-lactamase-insensitive drugs, such as methicillin, oxacillin and nafcillin. Drug resistance occurs by acquisition of the mecA gene, which encodes the PBP2A (Penicillin binding protein 2A), that possesses lower-affinity for methicillin. The mecA and related genes when present (mecB, mecC), are horizontally transferred on the mobile genetic element SCCmec (staphylococcal chromosome cassette mec). MRSA are known to be simultaneously resistant to aminoglycosides, macrolides and quinolones. Vancomycin, daptomycin, linezolid and newer drugs such as fifth-generation cephalosporins (ceftaroline, ceftobiprole), dalbavancin, oritavancin, iclaprim and delafloxacin are often the last resort to save critically ill patients infected with MRSA strains (Ohlsen et al. 1998; Kırmusaoğlu, 2017). Thus, there is a dire need for alternative sources of antimicrobials and efforts to focus on non-traditional drug targets. Among > 300 such candidates (Hossain et al., 2013), a major homeostatic protein, ATP-dependent ClpP (caseinolytic protease P), is a subject of interest. ClpP protease degrades and recycles misfolded proteins that are produced by immunological and treatment stress, thus prolonging survival time (Farrand et al. 2015; Ohlsen et al. 1998). In its absence, as in clpP- strains, regulons that control important stress-response functions such as stationary phase adaptation, nutrient starvation, cell motility and temperature challenges are inactivated. Additionally, hemolysin and biofilm formation are reduced. Owing to its indispensability in homeostasis, ClpP is thought to be a promising target for new anti-MRSA drugs. Currently beta-lactones and depsinopeptide are among the few already studied compounds with anti-ClpP function. They are known to eliminate biofilms and cure persistent infections (Farrand et al. 2015; Moreno-Cinos et al. 2019; Böttcher et al. 2008).
Scientists often look to nature for new sources of antimicrobials. Traditionally, research may involve solvent extraction of natural sources (example - plants, algae, fungi) and bioactivity guided fractionation. When coupled with bioinformatics, numerous drug candidates can be tested for their potential to engage with targets of interest. This helps narrow down candidates that can enter the pipeline for further in vitro and in vivo testing (Pinz & Rastelli, 2019) and reduce costs invested. Photo-autotrophic green microalgae are a rich natural resource capable of synthesizing a plethora of compounds of biomedical interest (Khan et al. 2018).
Our Microalgal laboratory at Ngee Ann Polytechnic screens locally isolated algae for compounds of pharmaceutical value. The focus of this paper is on one laboratory maintained local isolate of Chlorella sorokiniana [CS], which was studied in-depth due to antimicrobial activities detected in preliminary studies. CS microalgae are known fast-growers that possesses saturated fatty acid contents and are reported to have antioxidant, biofuel feedstock potential, biofertilizer, emulsification and wastewater treatment potential (Yun et al., 2020). Recent studies on CS have shown antimicrobial activity on Escherichia coli (Shaima et al., 2021), cell & cytokine modulatory action (Cilliberti et al., 2019); synergistic roles with probiotics (Cantu-Bernal et al., 2020), and chemo-protective action (Lin et al., 2020). This study focusses on traditional antimicrobial in vitro investigations of a laboratory-maintained freshwater isolate of CS, followed by in silico molecular docking analysis of peak compounds with staphylococcal ClpP protease (Fig. 1).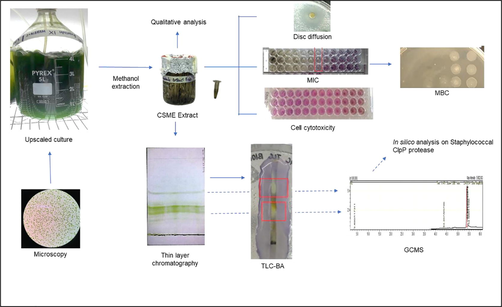
Legend: CSME – Chlorella sorokiniana extract; MIC – Minimum inhibitory concentration; MBC – Minimum bactericidal concentration; TLC-BA – Think layer chromatography-bioautography; GCMS – Gas chromatography-Mass spectrometry.
2 Materials and Methods
2.1 Microalgal processing
2.1.1 Identification and laboratory cultivation
Identification of the laboratory maintained microalgal isolate Chlorella sorokiniana [CS] was performed in a previous study (Lloyd et al. 2021), wherein pure cultures were identified based on morphology and sequencing of the D1-D2 LSU rDNA sequence. The strain CS, was maintained in Bold’s Basal Medium (BBM) (Sigma) supplemented with 2% Bacto agar (BD) on streak-plates in a plant tissue culture facility, at 23 °C with 12-hour cycles of light–dark alternation. For upscaling purposes, pure colonies were grown in 100 mL BBM flasks for 10-days in a shaker incubator. After testing for purity, the contents were inoculated into 5l in-house bioreactors with constant sterile air aeration, for 10–15 days. Late exponential phase cultures were pelleted in batches by centrifugation.
2.1.2 Extraction of microalgal compounds
Wet algal pellets were resuspended in 1x phosphate buffered saline (PBS) and washed thrice before being lyophilized. Dried pellets were resuspended in methanol at a 1:3 ratio and subjected to sonication in a Microson Ultrasonic Cell Disruptor to lyse algal cell walls (Araujo et al. 2013) at 8 W for 10 mins, on ice. The sonicated mixture was transferred into a sterile flask containing sterile glass beads (0.2–2.5 mm) and methanol (1:3 ratio) and was incubated in a shaker at 23 °C, at 400 rpm for 24 h. Successive methanol extraction was performed until the algal pellets were pale. The CS methanol extract [CSME] was dried in a rotary evaporator.
2.2 Testing of CSME
2.2.1 Disc diffusion
Dried CSME was weighed and resuspended in methanol. Whatman paper 1 discs were impregnated with CSME at 20 mg/disc and 10 mg/disc concentration and left to air-dry completely. Solvent control discs comprising methanol were also dried. Penicillin (10U) was used as a positive control. Sterile Mueller Hinton agar (MHA) plates were swabbed with S. aureus ATCC 25923 log phase culture (adjusted to McFarland 0.5 turbidity standard) and discs were placed on the seeded plate (Myers & Koshi, 1982; Lloyd et al. 2005). The plates were incubated at 37 °C for 18–24 h. The diameter (mm) of the zone of inhibition around each disc was measured.
2.2.2 Determination of Minimum inhibitory concentration (MIC) and Minimum bactericidal concentration (MBC)
Minimum inhibitory concentration (MIC) of CSME was determined using microbroth dilution followed by an MTT (3-(4,5-dimethylthiazol-2-yl-2,5-diphenyl tetrazolium bromide) (Sigma) assay (Benov, 2019) on S. aureus ATCC 25923 and three MRSA human clinical isolates (obtained from National University Hospital, Singapore). All MRSA isolates were blood-stream isolates and positive for mecA.
Dilutions of CSME were prepared in Mueller-Hinton Broth (MHB) (Difco) in a 96-well microtiter plate (20 to 0.07 mg/L). To the extract-broth dilutions (volume: 150 μl) in each well, 2 μl of broth culture of the test strain, adjusted to McFarland’s turbidity standard 0.5, was added. After incubation for 18–24 h at 37 °C, 40 μl of 0.5 mg/ml MTT (prepared in PBS) was added into each well, followed by incubation of the plate in the dark, at room temperature (RT), until a color change was observed in control wells containing S. aureus culture. Wells where microbial growth was inhibited, showed no colorimetric change and the highest dilution at which growth inhibition occurred, was taken as the Minimum inhibitory concentration (MIC). Five microliter volumes were sub-cultured from the wells onto Nutrient agar (before the MTT step for MIC) to determine the minimum bactericidal concentration (MBC), which is the highest dilution of extract at which there is no growth of the test bacteria upon subculture.
2.2.3 Cytotoxicity assay
The crude methanolic extracts were tested for cell cytotoxicity against an available healthy laboratory cell line Raw 264.7 using a MTT assay (Karakas et al. 2017). Briefly, in a 96-well microtiter plate, a cell suspension of Raw 264.7 in DMEM containing 5 × 104 cells/ml was added to all wells of the microplate to a final volume of 200 μl per well and incubated for 24 h at 37 °C in 5% CO2. The media was aspirated, and the cells were washed with 1x PBS. The CSME dilutions (20 to 0.07 mg/L) were prepared for this assay in Dulbecco’s Modified Eagle Medium (DMEM) in a second microtiter plate. Volumes of 100 μl were transferred from the second plate to the corresponding wells in the first plate containing cell growth.
A solution of 0.5 mg/ml MTT in DMEM was prepared and 100 μl was added to each well, followed by incubation at 37 °C for 1 h. The MTT solution was then removed, 100 μl of DMSO was added to each well, followed by incubation at RT for 10 mins before the wells were visualized under a light microscope.
2.2.4 Qualitative screening for phytochemicals
First, a series of phytochemical tests was performed on CSME to detect the presence of simple phenols, flavonoids, tannins, furanocoumarins, phenylpropenes, anthraquinones, tropolenes, iridoids, diterpenoids, triterpenes, phytosterols, cardiac glycosides, saponins, gibberellins, sesquiterpene lactones, fatty acids, alkaloids and tannins (Harborne, 1980). Second, the Folch method to screen for types of lipids was performed. A column was packed using silica gel mixed with dichloromethane in a CR30/50 glass column. 1 mL of extract in dichloromethane was layered on the column and was first eluted with 10 mL of dichloromethane for neutral lipids; next with 15 mL of acetone: methanol (9:1) to elute glycolipids; and last, 10 mL methanol for the elution of phospholipids (Heinzelmann et al. 2014). The three eluates were collected in glass tubes and were analyzed spectrophotometrically at 260 nm in a quartz cuvette. The fractions were dried and tested for antimicrobial activity.
2.2.5 Thin layer chromatography bioautography (TLC BA)
TLC sheets (Sigma) were used to fractionate compounds in the methanolic algal extract. Dried CSME was resuspended in methanol and spotted on TLC sheets in duplicate. A mobile phase of 7:3 hexane: acetone was used for extract fractionation in a sealed TLC tank. Sheets were left to air dry in a laminar air flow hood and then cut into strips. A bacterial agar suspension was prepared by mixing molten MHA cooled to 40 °C and the test S. aureus broth culture (3 × 108 CFU/ml) in a 5:1 ratio. This was poured over one of the strips and left to solidify in a closed sterile petri plate, followed by incubation 37 °C for 16 h. The overlay was then sprayed with 0.5 mg/ml MTT and developed at RT, in a dark chamber. Inhibition of microbial growth was indicated by the presence of clearings against a purple background and was used to identify the fractions A and B containing antimicrobial compounds. Rf values were calculated, where Rf = distance travelled by the active band divided by distance travelled by the solvent (Mcgaw et al. 2013).
Active bands from the duplicate TLC plates were cut and eluted into a tube with methanol. The mixture was spun at 10,000 rpm for 15 mins to separate the silica from the extract. The supernatant was examined for GCMS.
2.3 Characterization of active fractions A and B
2.3.1 GCMS analysis of the antimicrobial TLC polar fraction A
GCMS of the polar fraction A was facilitated using an Agilent GCMS 7890 gas chromatograph-mass spectrometer with a DB-5 MS column (30 m × 0.25 mm ID., 0.25 µm film thickness) through Setsco services, Singapore. Samples were reconstituted in 5 µl of HPLC grade methanol and injected under the in-house standardized conditions: Carrier gas: Helium; Flow rate: 1 mL/min. A splitless injection was performed in splitless mode. Ion source temperature and interface temperature was set at 200 °C. Detector voltage was maintained relative to the tuning result. The GC temperature program started at 40 °C, elevated to 280 °C after 2 min at a rate of 10 °C/min with a 20 min hold at 280 °C. Injector temperature was set at 250 °C, and injection volume was 1 µl.
2.3.2 GCMS analysis of the antimicrobial TLC non-polar fraction B
The non-polar fraction B sample was prepared as a 10% (w/v) solution in n-hexane. Saponification steps were as follows - equal volume of 3 M methanolic KOH added and incubated in a 37˚C shaker incubator for 30mins, followed by treatment with equal volume of saturated NaCl solution and additional 30 min incubation. The hexane layer was extracted and analyzed using an in-house Shimadzu single quadrupole GCMS-QP2010 Ultra gas chromatograph-mass spectrometer with a STABILWAX column (30.0 m × 0.5 µm thickness). Samples were injected under the in-house standardized conditions: Carrier gas: Helium; Flow rate: 2 mL/min. Normal injection mode was used, split injection 30.0. Ion source temperature and interface temperature was 200˚C, with a solvent cut-time of 2 min, and scanning range of 35–500 m/z. The detector voltage was maintained relative to the tuning result. The GC temperature program started at 40˚C, was then elevated to 60˚C, at a rate of 5˚C /min with a 20 min hold at 60˚C, then elevated to 200˚C, at a rate of 10˚C/min with a 20 min hold at 200˚C. The injector temperature was set to 240˚C, and injection volume was 1 µl. Compounds were identified via their individual retention times and mass fragmentation patterns by comparison with data from the NIST11 (National Institute of Standards and Technology) mass spectral database.
2.4 In-silico analysis of potential targets for select antimicrobial components
Nine compounds namely compound (1) 3-penten-2-one,4-methyl-, (2) 2-pentanone,4-hydroxy-4-methyl- (3) benzaldehyde,2,4-dimethyl-, (4) pentadecanoic acid,14-methyl-, methyl ester, (5) octadecanal, (6) 9,12-octadecadienoic acid (Z,Z)-, methyl ester, (7) 1,2-benzenedicarboxylic acid, mono(2-ethylhexyl) ester, (8) l-(+)-ascorbic acid 2,6-dihexadecanoate and (9) pentadecanoic acid on ClpP was attempted and were chosen for docking analysis based on their GCMS peak % concentration (Tables 1 and 2) and structures were generated and optimized using Chemsketch. The target structure of ClpP was retrieved from PDB database with the PDB ID as 5DL1 (https://www.rcsb.org/pdb, PDB ID: 5DL1, A Chain). For optimization, water molecules and complex moieties were removed. The drug likeliness properties of the 9 compounds were predicted using Molinspiration tool. Structures were saved in MDL-mol format and converted to pdb format using Open Babel molecular converter program. The structure of ClpP was uploaded in Autodock software version 4.2 with the addition of polar hydrogen and Kollman charges, with the final saved pdbq format. The structures of the compounds were also uploaded with the centre node and torsional bonds selection and the file was saved in pdbqt format. In grid preparation, the grid box was set as 126x126x126Å points with a grid spacing of 0.560 Å, whole protein was covered, and the initial search was carried out. The region of the most populated of the first ten clusters was selected as the probable binding region and the accurate search was executed in the probable binding region with a smaller grid map. Lamarckian genetic algorithm (GA) was applied in the drug-ligand interactions and 10 GA runs were performed with the following parameters: population size of 150, maximum number of 2.5 × 106 energy evaluations, and maximum number of 27,000 generations, and other parameters were default. The resulting conformations were clustered using a root-mean square deviation (RMSD) of 2.0 A. The least energy conformation among the ten conformations was chosen and saved in pdb format. The complex formed was visualized using Biovia discovery studio visualize software programme.
Retention time (min)
Compound (IUPAC Name)
Area%
6.62
3-Penten-2-one,4-methyl-
1.77
7.24
2-Pentanone,4-hydroxy-4-methyl-
65.61
9.01
Benzaldehyde
0.39
9.46
Decane,2,3,5,8-tetramethyl-
0.18
9.54
Dodecane,4,6-dimethyl
0.18
9.67
Dodecane,2,6,11-trimethyl-
0.43
12.95
Benzaldehyde,2,4-dimethyl-
1.43
13.15
Dodecane,2,7,10-trimethyl-
0.16
13.24
Dodecane,2,6,11-trimethyl-
0.15
14.01
Cyclohexasiloxane, dodecamethyl-
0.55
14.30
Hexadecane
0.16
15.27
3-Octadecene, (E)-
0.09
15.51
Pentadecane
0.09
16.10
Heptadecane
0.16
16.20
Pentadecane, 2,6,10-trimethyl-
0.26
16.25
Cycloheptasiloxane,tetradecamethyl-
0.17
16.67
Heptadecane
0.24
17.08
Hexadecane,2,6,10,14-tetradecamethyl-
0.24
19.05
Hexadecane,2,6,10,14-tetramethyl-
0.6
19.55
Heneicosane
0.16
20.37
Hexadecanal
0.22
21.15
Heptadecane,2,6,10,15-tetramethyl-
0.11
21.33
Eicosane
0.25
21.46
Pentadecanoic acid,14-methyl-,methyl ester
0.53
21.76
Tetratriacontane
0.23
22.45
Octadecanal
0.51
23.13
9,12-Octadecadienoic acid (Z,Z)-,methyl ester
0.52
23.26
Heptadecanenitrile
0.4
23.39
Pentadecane, 8-hexyl-
0.27
24.17
Cyclononasiloxane,octadecamethyl-
0.72
27.00
1,2-Benzenedicarboxylic acid, mono(2-ethylhexyl) ester
0.5
Retention time (min)
Compound (IUPAC Name)
Area%
3.480
3-Methylbenzyl alcohol, tert-butyldimethylsilyl ether
0.09
43.357
Hexadecanoic acid, methyl ester
<1
53.876
Pentadecanoic acid
45.57
53.902
Tridecanoic acid
0.16
53.880
l-(+)-Ascorbic acid 2,6-dihexadecanoate
45.09
54.500
n-Octanoic acid, methyl (tetramethylene) silyl ester
0.01
3 Results
3.1 Analysis of anti-staphylococcal activity
3.1.1 Kirby-Bauer disc diffusion
Mean inhibition diameter produced by a 20 mg/disc was 21 ± 0.5 mm, whereas the 10 mg/disc gave 8 ± 0.8 mm This was less than the positive control of Penicillin 10U which gave 32 mm. Methanol disc controls did not produce any zones.
3.1.2 Determination of MIC and MBC
For broth microdilution tests for MIC and MBC on standard and clinical MRSA strains, visual read-outs of inhibition (without MTT dye) were unreliable due to the greenish turbidity caused by colored extracts. Upon addition of MTT, growth was indicated by a violet colour (Fig. 2) and was 0.313 mg/L for Staphylococcus aureus ATCC 25923 and MRSA strains and was taken as the MIC. Upon subculture, the MBC was 2.5 mg/L for all strains. Due to the low concentrations of extract, these tests were performed only in duplicate and were reproducible.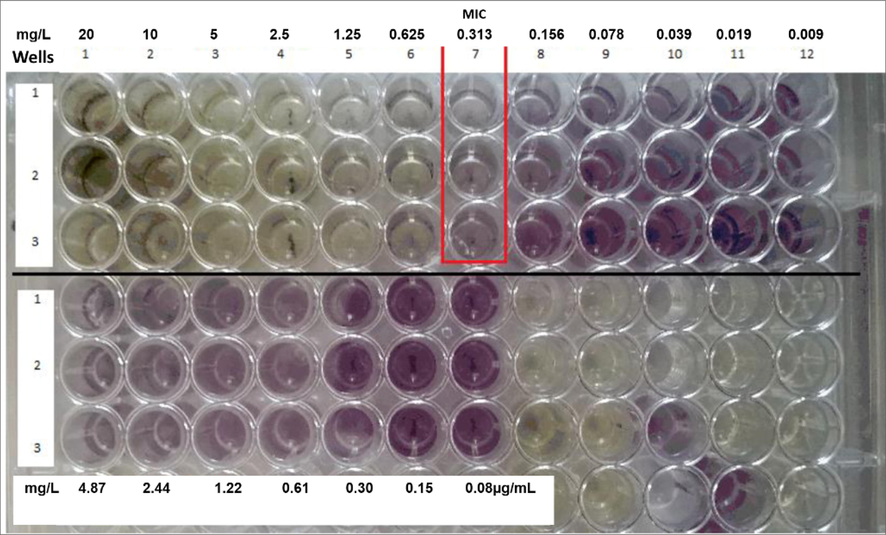
Minimum Inhibitory Concentration (MIC) of MRSA isolates using microbroth dilution method and MTT assay. Legend: Purple coloured wells are indicative of bacterial growth.
3.2 Cytotoxicity
Microscopy was used to assess the viability of cells in each dilution, instead of optical density, as the MTT assay in the presence of greenish algal extract produced false cell growth positives as also described by Karakas et al. (2017) with plant extracts. The cytotoxicity tests of CSME on the cell lines revealed that lower concentrations of CSME active on S. aureus seemed to be cytotoxic to the Raw 264.7 cell lines at 0.313 ± 0.3 mg/L.
3.3 Qualitative analysis of CSME
3.3.1 Phytochemical analysis
Qualitative analysis of the CSME indicated only the presence of sesquiterpene lactones, fatty acids and alkaloids. The extracts were negative for the other compounds tested.
3.3.2 Lipid analysis
Analysis of the types of lipids gave highest absorbance for the third eluate > first eluate > second eluate (2.6 > 0.162 > 0.0016) interpreted as the concentration of phospholipids > neutral lipids > glycolipids.
3.4 Thin layer chromatography-bioautography
Two clear antimicrobial bands were obtained in TLC bioautography with all test strains. The band closer to the start-line was labelled TLC fraction A band and the non-polar band further away, as TLC fraction B band. Rf values of locations that flanked the inhibitory fractions could be found in the range 0.46–0.61 for fraction A and 0.7–0.9 for fraction B. The bands A and B were extracted and analyzed using GCMS.
3.5 GCMS analysis
The polar fraction A when analyzed by GCMS gave 31 products of which 2-pentanone,4-hydroxy-4-methyl- was in the highest concentration (65.61%) among 30 other peaks (Fig. 3, Table 1). This was speculated to be a key antimicrobial in the fraction. The non-polar fraction gave 4 peaks of which pentadecanoic acid (45.57%) and l-(+)-Ascorbic acid 2,6-dihexadecanoate (45.09%) were in highest concentration (Fig. 4, Table 2). There were 25 peaks that could not be identified, of which only one was > 2% (2.1%).
GCMS characterization of the CSME TLC-BA fraction A.
GCMS characterization of the CSME TLCBA fraction B.
3.6 In-silico analysis
Based on Lipinski’s rule of five (Lipinski et al., 1997), hydrogen-bond donors are to be below 5, hydrogen-bond acceptors below 10, molecular mass below 500 Da, XlogP3 (lipophilicity) to be less than 5, and the total polar surface area (TPSA) not>140 Å. Four candidates (1), (2), (3), (7) possessed zero violations of the Lipinski’s rule of five (Table 3), suggesting the efficiency of these molecules to act as drug candidates with good pharmacokinetics, whereas (4), (5), (6), (9) had slight deviation in Log P and (8) had > 5 hydrogen bond donors. Fig. 5 represents the schematic representation of the docking interactions of the compounds (1, 2, 3, 7) with ClpP that were visualized and generated using the software (Biovia Discovery studio visualizer). Key residues in ClpP forming hydrophobic, hydrogen and Van der Waal interactions with atoms of bioactive compounds are provided in the figure. The ClpP bound with compound (2) shows least binding energy with −6.93 kcal/mol with two hydrogen bonds with THR169 and GLN124; compound (4) forms two hydrogen bonds with GLN132, THR146 (OG1…O); compound (6) forms two hydrogen bonds with THR146, HIS142 with binding energy of −6.52 and −6.37 kcal/mol respectively; compound (7) forms four hydrogen bonds with TYR18, LYS26 amino acids of ClpP with the binding energy of −5.74 kcal/mol. Key residues in ClpP forming hydrophobic, hydrogen and Van der Waal interactions with atoms of bioactive compounds are provided in the figure. The ClpP bound with compound (2) shows least binding energy with −6.93 kcal/mol with two hydrogen bonds with THR169 and GLN124; compound (4) forms two hydrogen bonds with GLN132, THR146 (OG1…O); compound (6) forms two hydrogen bonds with THR146, HIS142 with binding energy of −6.52 and −6.37 kcal/mol respectively; compound (7) forms four hydrogen bonds with TYR18, LYS26 amino acids of ClpP with the binding energy of −5.74 kcal/mol (Table 4). Compounds (1, 3, 5 and 9) form one hydrogen bond with binding energy of −4.69, −4.99, −5.54 and −5.6 respectively. Compound 8 had the least binding energy at 0.82 kcal/mol. Compounds that did not satisfy Lipinski’s rule of 5 (4,5,6,8,9) and had low binding energies with ClpP (1,3) were not be studied further. Compounds (2 and 7) satisfy both criteria and therefore are chosen to be studied in future. Additionally they have below 10 rotatable bonds, which makes them stable (Anza et al, 2021).
Compound number
Compound name
Molecular weight (g/mol)
Hydrogen Bond Donor
Hydrogen Bond Acceptor
XLogP3
Rotatable bonds
TPSA Å2
1
3-Penten-2-one,4-methyl-
98.14
0
1
1.53
1
17.07
2
2-Pentanone,4-hydroxy-4-methyl-
344.4
1
2
4.79
6
37.30
3
Benzaldehyde,2,4-dimethyl-
148.20
0
1
2.93
1
17.07
4
Pentadecanoic acid,14-methyl-, methyl ester
270.5
0
2
6.84
14
26.30
5
Octadecanal
268.5
0
1
8.50
16
17.07
6
9,12-Octadecadienoic acid (Z,Z)-, methyl ester
294.5
0
2
7.17
15
26.30
7
1,2-Benzenedicarboxylic acid, mono(2-ethylhexyl) ester
278.34
1
4
4.49
9
63.60
8
Ascorbic acid
176.12
4
6
−1.6
2
107
9
Pentadecanoic acid
242.4
1
2
5.8
13
37.3
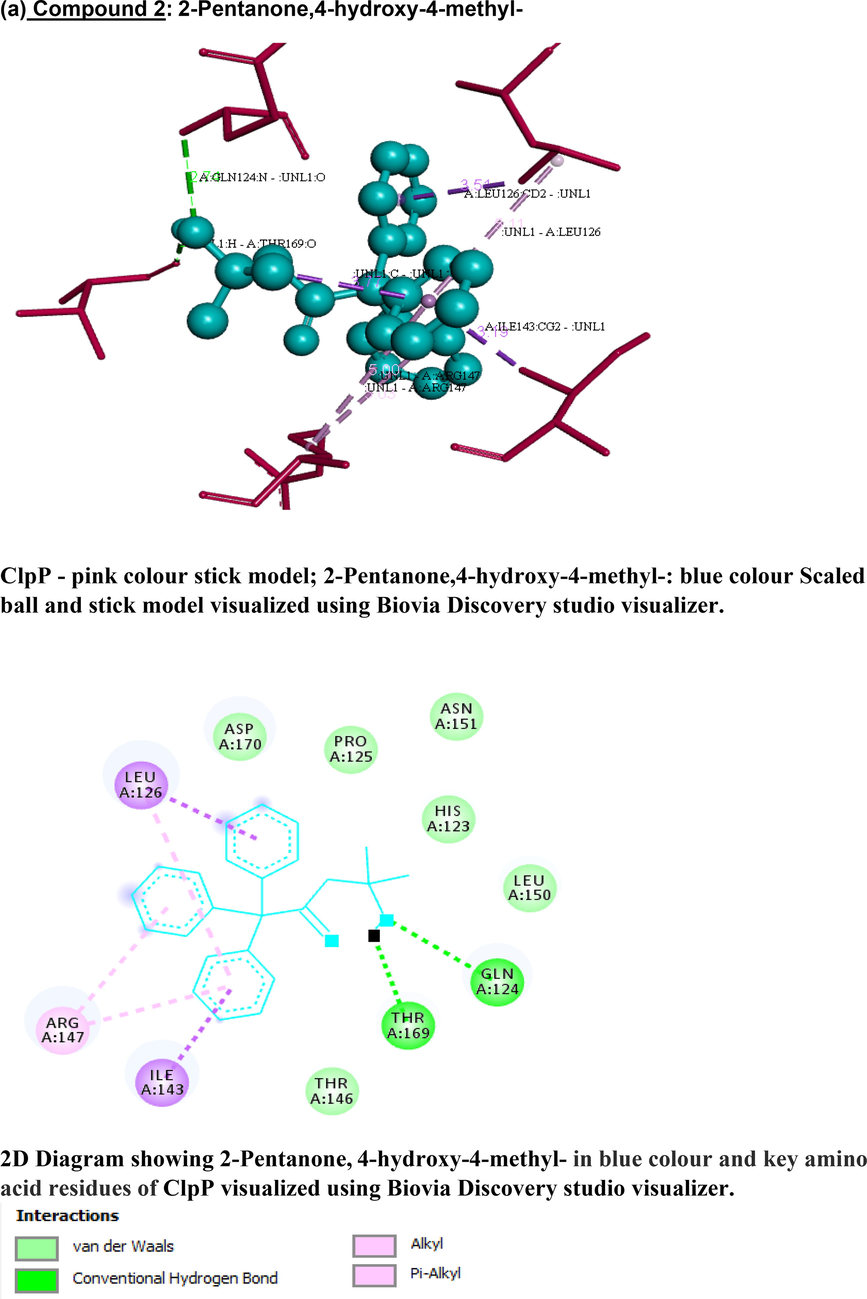
3-D and 2-D images of (a) compound 2 and (b) compound 7 interacting with staphylococcal ClpP protease using Biovia Discovery studio visualizer. (a) Compound 2: 2-Pentanone,4-hydroxy-4-methyl- ClpP - pink colour stick model; 2-Pentanone,4-hydroxy-4-methyl-: blue colour Scaled ball and stick model visualized using Biovia Discovery studio visualizer. 2D Diagram showing 2-Pentanone, 4-hydroxy-4-methyl- in blue colour and key amino acid residues of ClpP visualized using Biovia Discovery studio visualizer. (b) Compound 7: 1,2-Benzenedicarboxylic acid, mono(2-ethylhexyl) ester ClpP - pink colour stick model; 1,2-Benzenedicarboxylic acid, mono(2-ethylhexyl) ester: blue colour scaled ball and stick model visualized using Biovia Discovery studio visualizer.
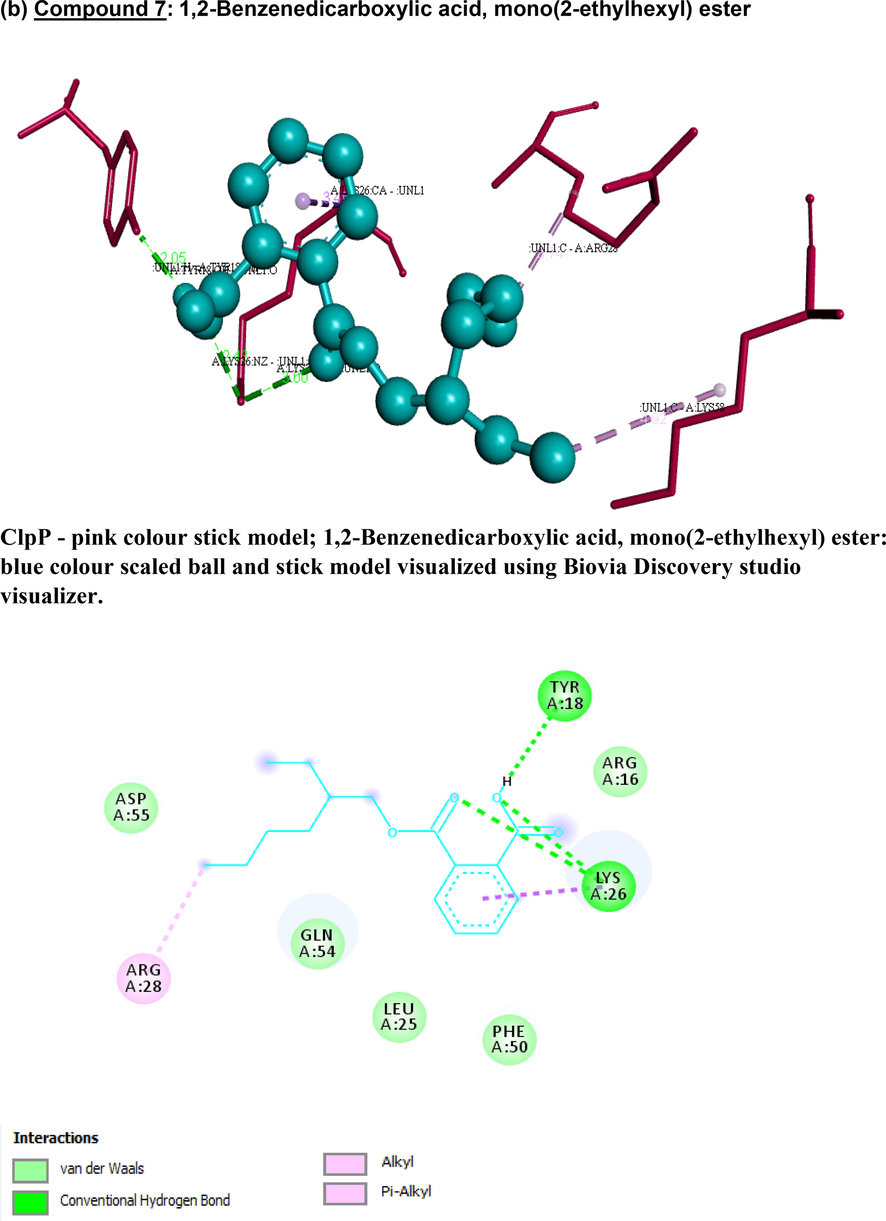
3-D and 2-D images of (a) compound 2 and (b) compound 7 interacting with staphylococcal ClpP protease using Biovia Discovery studio visualizer. (a) Compound 2: 2-Pentanone,4-hydroxy-4-methyl- ClpP - pink colour stick model; 2-Pentanone,4-hydroxy-4-methyl-: blue colour Scaled ball and stick model visualized using Biovia Discovery studio visualizer. 2D Diagram showing 2-Pentanone, 4-hydroxy-4-methyl- in blue colour and key amino acid residues of ClpP visualized using Biovia Discovery studio visualizer. (b) Compound 7: 1,2-Benzenedicarboxylic acid, mono(2-ethylhexyl) ester ClpP - pink colour stick model; 1,2-Benzenedicarboxylic acid, mono(2-ethylhexyl) ester: blue colour scaled ball and stick model visualized using Biovia Discovery studio visualizer.
Compound
Compound name
Binding energy (kcal/mol)
Interactions between compounds and staphylococcal ClpP protease
Hydrogen bond
Hydrophobic interactions
Van der Waal’s reactions
Amino acid (Donor⋅⋅⋅Acceptor)
Distance (A˚)
1
3-Penten-2-one,4-methyl-
−4.69
GLN124 (N⋅⋅⋅O)
2.81
ILE122, LEU150,
HIS123, THR169, SER98, SER101, ASN151
2
2-Pentanone,4-hydroxy-4-methyl-
−6.93
THR169 (O⋅⋅⋅H)GLN124
(N⋅⋅⋅O)2.13
2.74ILE143, LEU126
ARG147, ARG147
LEU126ASP70, PRO125, ASN151, HIS123, LEU150, THR146
3
Benzaldehyde,2,4-dimethyl-
−4.99
GLN132 (NE2⋅⋅⋅O)
3.06
ILE143, LEU126
VAL71, LEU150
HIS142, ARG147LEU150, THR146, PRO125, GLY128, GLY127
4
Pentadecanoic acid,14-methyl-, methyl ester
−6.52
GLN132 (NE2⋅⋅⋅O)THR146
(OG1⋅⋅⋅O)
2.99
2.90HIS142, LEU150
ILE122, VAL71
LEU154, PHE102HIS123, ARG147, ILE143, LEU126, GLN124, ASN141, THR169
5
Octadecanal
−5.54
GLN132 (NE2⋅⋅⋅O)
3.01
MET99
HIS123, THR169, SER98, ASN151, LEU150, THR146, VAL71, HIS142, ILE143, GLN124, LEU126, PRO125
6
9,12-Octadecadienoic acid (Z,Z)-, methyl ester
−6.37
THR146 (OG1⋅⋅⋅O)HIS142
(CD2⋅⋅⋅O)2.97
3.07VAL71, HIS142
LEU126, GLN132, THR169, SER98, ASN151, LEU150, HIS123, ILE143, GLN124, SER101, ARG147, PHE102, ILE122
7
1,2-Benzenedicarboxylic acid, mono(2-ethylhexyl) ester
−5.74
TYR18 (OH⋅⋅⋅H)TYR18
(OH⋅⋅⋅H)LYS26
(NZ⋅⋅⋅O)LYS26
(NZ⋅⋅⋅O)2.92
2.05
2.43
3.00LYS26, ARG28
LYS58
ARG16, ASP55, GLN54, LEU25, PHE50, ARG16
8
l-(+)-Ascorbic acid 2,6-dihexadecanoate
0.82
SER70(OG ⋅⋅⋅O)GLY127
(N⋅⋅⋅O)GLN132
(NE2 ⋅⋅⋅O)PRO125
(O⋅⋅⋅H)3.05
2.81
2.78
2.06NIL
ASN39, ASP38, THR72, ASP37, LEU126, GLN124, GLN35, GLY69, VAL71, HIS142, GLY128
9
Pentadecanoic acid
−5.6
GLN132(HE22…O)
2.11
LEU150
MET99
PHE102GLY128, HIS142, ASN151, ILE122, THR169, SER98, GLN124, HIS123, THR146, ILE143, LEU126
4 Discussion
MRSA infections, are still a major concern in hospital and community settings. Preliminary screening of microalga C. sorokiniana crude methanol extract CSME by disc diffusion against S. aureus ATCC 25923 showed antimicrobial activity. Preliminary qualitative phytochemical analyses of the crude extract CSME, revealed fatty acids, alkaloids, sesquiterpene lactones and phospholipids.
Prior to investing time and funds into testing the individual GC components, computational analysis to filter compounds with good pharmacokinetic properties out of the 35 GCMS peaks obtained was attempted. Among the major 9 compounds studied, (1 to 7) were identified in TLC fraction A and (8) and (9) from TLC fraction B. Computational analysis of docking properties eliminated compounds that did not satisfy Lipinski’s rule of 5 (4,5,6,8,9) and had low binding energies with ClpP (1,3) leaving compounds (2 and 7) as prospective candidates.
We can speculate that compound (2) and (1) in TLC fraction A might have contributed to the antimicrobial activity of CSME wherein the MIC/MBC was 0.313 mg/L/ 2.5 mg/L. Our in silico studies show that it has ideal pharmacokinetics and binding energy values. Literature shows that compound (2) which was retrieved at 65.61% (area %) in TLC fraction A, is commonly referred to as diacetone alcohol or 2-pentanone, 4-hydroxy-4-methyl- (HMP), and compound (1) 1.77% (area %) is a breakdown product of (2). It is interesting to note that these compounds were previously reported from extracts of marine species, as antagonists to fish pathogens (Wefky & Ghobrial, 2008), also with anti-biofilm potential (El Zawawy et al., 2020). Further support to include this compound in future animal studies is that it was reported to be non-toxic up to 50 mg/kg in rats (Johnson, 2004) in an unrelated study.
TLC fraction B could have contributed to the cell cytotoxicity in our crude extract CSME, and could have been due to fatty acids (compound 9, area % = 45.57), as fatty acids with > 10C atoms are capable of lysing cell membranes (De Morais et al. 2015). Fatty acids are also part of alkaloids (as seen in the phytochemical tests) and are reported in algae e.g. Spirulina spp. (Kata et al. 2018; Güven et al. 2010). Compound (7), which is our next promising compound, is commonly referred to as a phthalate, and is also found in weeds with antimicrobial potential (Shanab et al., 2010). In our study, it showed good docking and binding energy properties as well.
Compounds that did not qualify through bioinformatics analysis included compound (4) also previously reported to have antibacterial and antifungal activity (Ghazala et al. 2004); compounds (5, 6) along with other minor compounds have been reported in red algae (Qhairul et al., 2011; Mohy El-Din and El-Ahwany, 2016), and compound (9)-coupled with fatty acid moieties are reported to have natural antioxidant potential (Bose et al. 2018).
Apart from the 9 major compounds studied in detail, some of the minor compounds have also found mention in past reports, for example - benzaldehyde in seaweeds (Yamamoto et al. 2014); decane,2,3,5,8-tetramethyl-, dodecane,2,6,11-trimethyl-, octadecanal and cyclononasiloxane, octadecamethyl- in red algae (Izreen et al. 2011; Mohy El-Din et al. 2016); decane, 2,3,5,8-tetramethyl-, dodecane, 2,6,11-trimethyl-; hexadecane, eicosane, hexadecane 2,6,10,14-tetradecamethyl-, hexadecane, 2,6,10,14-tetramethyl-, in sea buckthorn oils (Yue et al. 2017); 3-Octadecene, (E)- in leaf extracts of Ricinus spp, (Altameme et al. 2015); heptadecane, 2,6,10,15-tetramethyl- in Gymnema spp. (Subramanian et al. 2020); heneicosane, penta- and heptadecanes, hexadecanal in brown algae (Ávila et al. 2019); eicosane an antifungal from Streptomyces spp. (Ahsan et al. 2017); tetratriacontane from traditional Ashtawarga plants (Singh and Patra, 2019); heptadecanenitrile from Spirulina sp. (Chen et al. 2019); and pentadecane, 8-hexyl- from chrysanthemum flowers (Pubchem, n.d.). Dodecane, 2,6,11-trimethyl- or Farnesan and pentadecane, 2,6,10-trimethyl- was previously documented in walnuts (Centre, n.d.), microalgae (Gutierrez et al. 2012), jasmines and diatoms (CSIR, n.d.; Peters et al. 2008) and are examples of sesquiterpene lactones, that were identified in our qualitative analysis studies. Phospholipids, are also associated with antimicrobial activity in red algae (Alves et al. 2020) and were also detected in the qualitative analysis.
Computational analysis showed that among the candidates that satisfy Lipinski’s rule of 5, two compounds (2) and (7), from the non-lipid polar fraction A, displayed better binding energy properties with ClpP and can be considered candidates for further in vitro and in vivo analysis. We acknowledge that we have used the wild-type staphylococcal ClpP protease for analysis in this study, and the outcomes may not be applicable to possible mutants that lack ClpP or possess a defective ClpP.
5 Conclusions
CSME extract inhibited MRSA strains at MBC/MIC of 0.313 mg/L/ 2.5 mg/L and was cytotoxic at the same concentrations. Fraction B which possesses a high percentage of fatty acids could have contributed to cytotoxicity in vitro and need not be considered further from fractionated extracts. Fraction A, particularly compounds (2): 2-pentanone,4-hydroxy-4-methyl- and (7) 1,2 - benzenedicarboxylic acid, mono (2- ethylhexyl) ester, showed good docking and binding energy properties with the protein target ClpP protease, and may be upscaled for further studies in animal models. We do not attest any of these compounds for clinical use, as they have been tested in vitro only. Overall, our antimicrobial studies, coupled with bioinformatics analysis shows that CS is a source of valuable anti-staphylococcal compounds.
Funding
This research was funded by the Internal Ngee Ann Polytechnic Technology Development and Innovation Fund (NPTDIF).
Institutional Review Board Statement: Not applicable.
Informed Consent Statement: Not applicable.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Extraction and identification of bioactive compounds (eicosane and dibutyl phthalate) produced by Streptomyces strain KX852460 for the biological control of Rhizoctonia solani AG-3 strain KX852461 to control target spot disease in tobacco leaf. AMB Express. 2017;7(1)
- [CrossRef] [Google Scholar]
- Determination of alkaloid compounds of Ricinus communis by using Gas Chromatography- Mass Spectroscopy (GC-MS) J. Med. Plants Res.. 2015;9(10):349-359.
- [CrossRef] [Google Scholar]
- Alves, E., Dias, M., Lopes, D., Almeida, A., Domingues, M. D. R., Rey, F., 2020. Antimicrobial lipids from plants and marine organisms: An overview of the current state-of-the-art and future prospects. Antibiot. 2020, Vol. 9, p. 441, 9 (8), 441.
- Anza, M., Endale, M., Cardona, L., Cortes, D. , Eswaramoorthy, R., Zueco, J., Rico, H., Trelis, M., & Abarca, B., 2021. Antimicrobial Activity, in silico molecular docking, ADMET and DFT Analysis of secondary metabolites from roots of three Ethiopian medicinal plants. Adv. Appl. Bioinform. Chem., Volume 14, 117–132. https://doi.org/10.2147/aabc.s323657.
- Extraction of lipids from microalgae by ultrasound application: Prospection of the optimal extraction method. Ultrasonics Sonochemistry. 2013;20:95-98.
- [CrossRef] [Google Scholar]
- New antiproliferative polyunsaturated epoxy-heneicosane derivatives isolated from the brown alga Lobophora variegata. J. Braz. Chem. Soc.. 2019;30(2):406-412.
- [Google Scholar]
- Effect of growth media on the MTT colorimetric assay in bacteria. PLoS One. 2019;14(8):e0219713.
- [CrossRef] [Google Scholar]
- Chemical constituents of Sauropus androgynus and evaluation of its antioxidant activity. Res. J. Phytochem.. 2018;12(1):7-13.
- [CrossRef] [Google Scholar]
- Böttcher, T., Sieber, S. A, 2008. β-Lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J. Am. Chem. Soc. 130 (44), 14400–14401. https://doi.org/10.1021/JA8051365/SUPPL_FILE/JA8051365_SI_001.PDF.
- The threat of antimicrobial resistance on the human microbiome. Microb. Ecol.. 2017;74(4):1001-1008.
- [CrossRef] [Google Scholar]
- Cantu-Bernal, S., Dominiguez-Gamez, M., Medina-Peraza, I., Aros-Uzarraga, E., Ontiveros, N., Flores-Mendoza, L. et al, 2020. Enhanced viability and anti-rotavirus effect of Bifidobacterium longum and Lactobacilus plantarum in combination with Chlorella sorokiniana in a dairy product.Front. Microbiol. doi:10.3389/fmicb.2020.00875.
- Centre, T. M. I. (n.d.) showing compound 2,6,10-trimethyldodecane (FDB005822) - FoodDB https://foodb.ca/compounds/FDB005822 (accessed 2022 -03 -29).
- Chen, W.T. G., Wu, Z., Si, B., Zhang, Y., 2019. Renewable diesel blendstocks and bioprivileged chemicals distilled from algal biocrude oil converted via hydrothermal liquefaction. https://doi.org/10.26434/CHEMRXIV.9630695.V1.
- Extracts from microalga Chlorella sorokiniana exert an anti-proliferative effect and modulate cytokines in sheep peripheral blood mononuclear cells. Animals.. 2019;9:4.
- [CrossRef] [Google Scholar]
- CSIR, CIMAP AROMA (n.d.)| Compound Pentadecane, 2,6,10-trimethyl-http://bioinfo.cimap.res.in/aromadb/web_compound_detail.php?id=CRMOL125 (accessed 2022 -03 -29).
- El Zawawy, N. A., El-Shenody, R. A., Ali, S. S., El-Shetehy, M. A., 2020. Novel study on the inhibitory effect of marine macroalgal extracts on hyphal growth and biofilm formation of candidemia isolates. Sci. Reports 101, 10 (1), 1–10. https://doi.org/10.1038/s41598-020-66000-1.
- Farrand, A. J., Friedman, D. B., Reniere, M. L., Ingmer, H., Frees, D., & Skaar, E. P., 2015. Proteomic analyses of iron-responsive, Clp-dependent changes in Staphylococcus aureus. Pathogens and Disease. 73(3), ftv004–ftv004. https://doi.org/10.1093/femspd/ftv004.
- Phycochemistry and bioactivity of Tetraspora (Volvocophyta) from Sindh. Pak. J. Bot. 2004;36(3):531-547.
- [Google Scholar]
- Gutierrez, R. M. P., Anaya Sosa, I., Cruz Y Victoria, T., Flores, J. M. M., 2012. Sesquiterpene lactones: Antispasmodic principles of the freshwater algae Hydrodictyon reticulatum. Med. Chem. Res. 21 (7), 1023–1029. https://doi.org/10.1007/S00044-011-9601-9.
- Phytochemical methods: a guide to modern techniques of plant analysis. London, New York: Chapman and Hall; 1980.
- Critical assessment of glyco- and phospholipid separation by using silica chromatography. Appl. Environ. Microbiol.. 2014;80(1):360.
- [CrossRef] [Google Scholar]
- Identification of potential targets in Staphylococcus aureus N315 using computer aided protein data analysis. Bioinformation. 2013;9(4):187-192.
- [CrossRef] [Google Scholar]
- Johnson, W., 2004. Safety assessment of MIBK (Methyl Isobutyl Ketone). Int. J. Toxicol. 23 Suppl 1 (SUPPL. 1), 29–57. https://doi.org/10.1080/10915810490274298.
- Karakaş, D., Ari, F., Ulukaya, E., 2017. The MTT viability assay yields strikingly false-positive viabilities although the cells are killed by some plant extracts. Turk J Biol. 18,41(6):919-925. doi: 10.3906/biy-1703-104. PMID: 30814856, PMCID: PMC6353273.
- Therapeutic effect of the alkaloid extract of the cyanobacterium Spirulina platensis on the lipid profile of hypercholesterolemic male rabbits. Environ. Sci. Pollut. Res. Int.. 2018;25(20):19635-19642.
- [CrossRef] [Google Scholar]
- The promising future of microalgae: Current status, challenges, and optimization of a sustainable and renewable industry for biofuels, feed, and other products. Microb. Cell Factories. 2018;171, 17 (1):1-21.
- [CrossRef] [Google Scholar]
- MRSA and MSSA: The mechanism of methicillin resistance and the influence of methicillin resistance on biofilm phenotype of Staphylococcus aureus. In: Enany S., Alexander L.E.C., eds. The Rise of Virulence and Antibiotic Resistance in Staphylococcus aureus. London: IntechOpen; 2017.
- [CrossRef] [Google Scholar]
- Lin, S,H., Li, M,H., Chuang, K,A., Lin, N,H., Chang, C,H., Wu, H,C., Chao, Y,H., Lin, C,C., Pan, I,H., Perng, M,D., Wen, S,H., 2020. Chlorella sorokiniana extract prevents Cisplatin-induced myelotoxicity in vitro and in vivo. Oxidative medicine and cellular longevity. Oxidative Medicine and Cellular Longevity. 1–4. https://doi.org/10.1155/2020/7353618.
- Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev.. 1997;23:3-25.
- [CrossRef] [Google Scholar]
- Lloyd, C., Tan, K. H., Lim, K. L., Valu, V. G., Fun, S. M. Y., Chye, T. R., Mak, H. M., Sim, W. X., Musa, S. L., Ng, J. J. Q., Bte Nordin, N. S., Bte Md Aidzil, N., Eng, Z. Y. W., Manickavasagam, P., New, J. Y., 2021. Identification of microalgae cultured in Bold’s Basal Medium from freshwater samples, from a high-rise city. Sci. Rep. 11 (1). https://doi.org/10.1038/S41598-021-84112-0.
- Anticandidal activity of Azadirachta indica. Indian J. Pharmacol.. 2005;37(6):386.
- [CrossRef] [Google Scholar]
- Antifungal and antibacterial activity and chemical composition of polar and non-polar extracts of Athrixia phylicoides determined using bioautography and HPLC. BMC Complementary and Alternative Medicine. 2013;13:356.
- [CrossRef] [Google Scholar]
- Bioactivity and Phytochemical Constituents of Marine Red Seaweeds (Jania rubens, Corallina mediterranea and Pterocladia capillacea) J. Taibah Univ. Sci.. 2016;10(4):471-484.
- [CrossRef] [Google Scholar]
- ClpP Protease, a promising antimicrobial target. Int. J. Mol. Sci.. 2019;20(9)
- [CrossRef] [Google Scholar]
- Manual of diagnostic procedures in medical microbiology and immunology/serology. Pondicherry: All India Press; 1982. p. :71.
- Effects of subinhibitory concentrations of antibiotics on alpha-toxin (hla) gene expression of methicillin-sensitive and methicillin-resistant Staphylococcus aureus isolates. Antimicrob. Agents Chemother.. 1998;42(11):2817-2823.
- [Google Scholar]
- Peters, K.E., Walters, C.C., Moldowan, J.M., 2008. The Biomarker Guide: Volume 2, Biomarkers and isotopes in petroleum systems and earth history : K. E. Peters : 9780521039987.
- Molecular docking: Shifting paradigms in drug discovery. International Journal of Molecular Sciences. 2019;20(18):4331.
- [CrossRef] [Google Scholar]
- Pubchem, n.d., Pentadecane, 8-hexyl- | C21H44 - PubChem https://pubchem.ncbi.nlm.nih.gov/compound/Pentadecane_-8-hexyl (accessed 29-03-2022).
- Volatile compound extraction using Solid Phase Micro Extraction coupled with Gas Chromatography Mass Spectrometry (SPME-GCMS) in local seaweeds of Kappaphycus alvarezii, Caulerpa lentillifera and Sargassum polycystem. Int. Food Res. J.. 2011;18(4):1449-1456.
- [Google Scholar]
- Unveiling antimicrobial activity of microalgae Chlorella sorokiniana (UKM2), Chlorella sp. (UKM8) and Scenedesmus sp. (UKM9) Saudi Journal of Biological Sciences.. 2021;29:1043-1052.
- [Google Scholar]
- Allelopathic effects of water hyacinth [Eichhornia crassipes] PLoS One. 2010;5(10):e13200.
- [Google Scholar]
- Evaluation of adaptogenic potential of Polygonatum cirrhifolium (Wall.) Royle: In vitro, in vivo and in silico studies. South African J of Bot.. 2019;121:159-177.
- [Google Scholar]
- Effect of solvent on the phytochemical extraction and GC-MS Analysis of Gymnema sylvestre. Pharmacogn. J.. 2020;12(4):749-761.
- [CrossRef] [Google Scholar]
- Studies on the bioactivity of different solvents extracts of selected marine macroalgae against fish pathogens. Res. J. Microbiol.. 2008;3(12):673-682.
- [CrossRef] [Google Scholar]
- Determination of volatile compounds in four commercial samples of Japanese green algae using solid phase microextraction gas chromatography mass spectrometry. Sci. World J. 2014
- [CrossRef] [Google Scholar]
- Phytochemical composition and antibacterial activity of the essential oils from different parts of sea buckthorn (Hippophae rhamnoides L.) J of Food and Drug Analysis. 2017;25:327-332.
- [CrossRef] [Google Scholar]
- Characterization of Chlorella sorokiniana and Chlorella vulgaris fatty acid components under a wide range of light intensity and growth temperature for their use as biological resources. Heliyon. 2020;6(7)
- [CrossRef] [Google Scholar]







