Translate this page into:
Antimicrobial activities of organic solvent extracts of four seaweeds from Oman
⁎Corresponding author. aladwani@squ.edu.om (Salma Al-Adwani)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Seaweeds are valuable sources of bioactive compounds in biomedicine, cosmetics, food, and pharmacology. The purpose of this investigation was to study the antimicrobial properties of organic solvent extracts from two red seaweed species (Melanothamnus somalensis & Gelidium omanense), and two brown seaweed species (Jolyna furcata & Nizamuddinia zanardinii) compiled from the southern coastline of Oman against several bacterial strains of global health concern (Staphylococcus aureus, Klebsiella pneumonia, Escherichia coli, and Pseudomonas aeruginosa) and one fungal strain (Candida albicans). Five organic solvents were used sequentially to achieve extraction. The solvents were applied in the following order: hexane, dichloromethane, ethyl acetate, acetone, and methanol. Only the methanol extract of Nizamuddinia zanardinii (MeNZ) showed interesting antimicrobial activity; the inhibition zone was 13 ± 1 mm. Furthermore, MeNZ was fractionated, and fraction 1 (MeNZ-F1) was recognized to have antimicrobial activity; the inhibition zone was 14.66 ± 0.57 mm. The stock concentration exhibited higher antimicrobial activity compared to the diluted concentrations after 3 h of incubation. The TEM and SEM results indicated that E. coli treated with the active fraction exhibited irregular shape, rough surface, and leakage of cellular content. Additionally, ribosomes were clustered and directed toward the inner membrane of the bacteria, while the DNA clustered in the center of the cell. In conclusion, the methanol extract of Nizamuddinia zanardinii has shown high efficacy against pathogenic bacteria and fungi. Therefore, it can be a valuable candidate for improving/developing antimicrobial drugs in the pharmaceutical and food industries.
Keywords
Antibacterial
Organic extract
Oman
Seaweed
Nizamuddinia zanardinii
Antimicrobial
- Ac
-
Acetone
- C. albicans
-
Candida albicans
- CFU
-
Colony Forming Units
- DCM
-
Dichloromethane
- E. coli
-
Escherichia coli
- EA
-
Ethyl Acetate
- FTIR
-
Fourier-transform infrared
- H
-
Hexane
- HPLC
-
High-Performance Liquid Chromatography
- IZA
-
Inhibition Zone Assay
- K. pneumonia
-
Klebsiella pneumonia
- LB
-
Luria Broth
- Me
-
Methanol
- MeNZ
-
Methanol extract of Nizamuddinia zanardinii
- MeNZ-F1
-
Methanol extract of Nizamuddinia zanardinii Fraction-1
- OD
-
Optical Density
- P. aeruginosa
-
Pseudomonas aeruginosa
- PBS
-
Phosphate-Buffered Saline
- S. aureus
-
Staphylococcus aureus
- SEM
-
Scanning Electron Microscopy
- TEM
-
Transmission Electron Microscopy
Abbreviations
1 Introduction
Seaweeds or macroalgae are non-flowering, photosynthetic, and non-vascular plants with around 12,000 species identified worldwide (Kulshreshtha et al., 2020). They are distributed among three main clusters depending on natural colours or pigments: the red, brown and green algae (Rhodophyta, Phaeophyta and Chlorophyta, respectively) (Arias et al., 2023). Seaweeds are usually found in salty water and are frequently called benthic marine algae, which means attached algae (Fiori and Pratolongo, 2021). Seaweeds demonstrate a remarkable ability to thrive in diverse and challenging environments, including complex communities and hostile habitats. One survival strategy that marine organisms employ involves producing a wide range of bioactive compounds in response to various external factors. These factors include changes in tides, defense from predators, and ecological pressures such as competition for resources (Pérez et al., 2016). Many valuable active molecules have been isolated from seaweeds and have found applications in various industries such as biomedicine, nutraceuticals, cosmeceuticals, pharmaceuticals, and food. Those molecules are diverse and include phenolic compounds, phlorotannins, polyunsaturated fatty acids, carotenoids, and polysaccharides (Suleria et al., 2015).
Examples of pharmaceutical applications include antimicrobial activities, which involve inhibiting or ceasing the growth and development of microorganisms (Pérez et al., 2016). Several seaweed crude extracts showed interesting antibacterial activities against many pathogenic bacteria, such as Enterobacter aerogens, Klebsiella pnemoniae, Bacillus subtilis, Staphylococcus aureus, Streptococcus faecalis, Enterobacter faecalis, Micrococcus luteus, Pseudomonas aeruginosa and Escherichia coli (Lomartire and Gonçalves, 2023). Several compounds found in seaweeds, including alkaloids, terpenes, halogenated compounds, and lectins, have demonstrated antimicrobial activities (Jin et al., 2014).
Approximately 5 million people died in 2019, due to bacterial antimicrobial resistance (Murray et al., 2022). Six bacterial strains (Acinetobacter baumannii, Pseudomonas aeruginosa, Staphylococcus aureus, Escherichia coli, Klebsiella pneumonia, and Streptococcus pneumonia) were responsible for 72 % (3.57 million) of the deaths (Murray et al., 2022). Multidrug-resistant pathogens are considered a worldwide clinical issue, and by 2050, global deaths are estimated to reach 10 million annually, which will cost the global economy around US$100 trillion (de Kraker et al., 2016). Combating antibiotic resistance is a complex problem that requires a comprehensive approach. Several strategies have been proposed to tackle antibacterial resistance, including prudent antibiotic use, developing new antibiotics, using a combination of therapies, implementing vaccination programs, promoting diagnostic tools, monitoring and surveillance, global cooperation and policy measures, and adopting a one-health approach (Shankar, 2016). Therefore, the development of new antimicrobial drugs or alternatives to conventional antibiotics is necessary to combat the problems of antimicrobial resistance.
Oman’s seaweeds have been demonstrated to be remarkably diverse, especially in the southern part of Oman (Dhofar Province) due to monsoon effects. Wynne, In 2018, published a comprehensive list of seaweeds in Oman, which showed 238 taxa of Rhodophyta, 89 taxa of Chlorophyta, and 75 taxa of Phaeophyceae (red, green, and brown algae, respectively). The list included several species reported worldwide as potential sources of valuable ingredients, such as carrageenan, agar, alginate, fucoidan, pigments, etc. (Wu, 2016). In addition, high sunlight intensity (typical to Oman) was found to be a stimulating factor for faster growth rate and more pigment production (Wu, 2016). Although the worldwide production of seaweed has increased in response to the rise in demand (Labban et al., 2019), the full potential of Omani seaweeds has not yet been studied. The current study aims to investigate the antimicrobial activity of organic solvent seaweed extracts from four species found in the southern part of Oman (Dhofar Province). The findings of this study will open the door for more intensive research in this field.
2 Materials and methods
2.1 Reagents and chemicals
All reagents were of analytical grade and purchased from Sigma-Aldrich (Germany). Methanol LC-MS grade from Honeywell (France), acetonitrile LC-MS grade from HiPerSolv CHROMMANORM (USA), ammonium acetate LC-MS grade from Sigma-Aldrich (Germany), and formic acid LC-MS grade from CARLO ERBA (France).
2.2 Collection and identification of raw materials
Two red seaweed species (Melanothamnus somalensis & Gelidium omanense) and two brown seaweed species (Jolyna furcata & Nizamuddinia zanardinii) were collected in September 2021 from Sadh (Dhofar governorate, Oman), at coordinates: 16°96′07.90″N 54°75′60.32″E, 17°04′82.96″N 55°07′65.3″E, 16°95′15.19″N 54°81′85.18″E, and 16°94′52.18″N 54°80′47.89″E) by expert divers (Salalah Diving Services Company, Sultanate of Oman, Salalah). Seaweed plants were identified based on their anatomical and morphological characteristics. At the collection site, the samples were washed by hand with seawater to get rid of dirt. The samples were delivered to a drying site in cool boxes and cleaned with fresh water to remove seawater. The samples were dried under the sun for three days and transferred to the Food Chemistry Laboratory at Sultan Qaboos University in cool boxes. The samples were kept at 4 °C until further handling.
2.3 Treatment of seaweeds
The sun-dried samples were milled using a coffee grinder (Ikon, Model No: IK-ZCG150, China) and sieved to pass through a 250-µm mesh. The milled samples were placed in plastic containers and kept at 4 °C till further analysis.
2.4 Algal extract
The extract was obtained using several solvents having different polarity, namely: hexane (H), dichloromethane (DCM), ethyl acetate (EA), acetone (Ac), and methanol (Me) in sequential bases as previously described by Chia et al. (2015) with some variations. Twenty grams (20 g) of the seaweed powder were placed in a 500 ml amber bottle. Four hundred milliliters (400 ml) of the first solvent (hexane) were added to the powder and the mixture was shaken at 120 rpm (HS 501 digital, IKA-Werke GmbH & Co. KG, Germany) for 24 h and then passed through a filter paper (Whatman No. 1). The filtrate was retrieved and the solid part was re-extracted twice following the same procedure used in the first extract before shifting to the second solvent with higher polarity. The filtrates were combined and evaporated using a SpeedVac vacuum concentrator (Concentrator Plus, Eppendorf AG, Germany). The dried extracts were reconstituted in 10 ml methanol and kept in a refrigerator at 4 °C until further processing.
2.5 Fractionation using preparative HPLC
The organic extract was fractionated using a preparative HPLC system (Shimadzu Corporation, Japan) composed of an auto-sampler (SIL-10AP), a pump (LC-20AP), a detector (SPD-20 M), a fraction collector (FRC-10A) and a recycling valve (valve unit, FCV-12AH). The LC column Shim-pack PREP-ODS (250 × 20 mm, 15 µm) was used to achieve separation. The mobile phase consisted of 0.1 % formic acid (mobile phase A) and methanol with 0.1 % formic acid (mobile phase B). The gradient for mobile phase B was as follows, 10 %-50 % (0–208 min), 50 % (208–291 min), 50 %-100 % (291–316 min), 100 % (316–458 min), 100 % − 10 % (458–475 min) and 10 % (475–500 min). The run was fractionated based on time; hence, 12 fractions were collected (Bauer et al., 2019).
2.6 Bacteria & fungus species
One fungal strain (Candida albicans, C. albicans) and four bacterial strains (Escherichia coli ATCC 25922 (E. coli), Staphylococcus aureus ATCC 25923 (S. aureus), Klebsiella pneumonia ATCC 1706 (K. pneumonia) and Pseudomonas aeruginosa ATCC 27853 (P. aeruginosa)) were used in this study. The pathogens were provided by Dr. Zaaima Al Jabri, College of Medicine and Health Sciences, Sultan Qaboos University. Microbes were taken from glycerol stock stored at −20 °C, streaked out on Luria Broth (LB) agar plates, and incubated (Gallenkamp Model INC.200.210C Plus Series Incubator, England) overnight at 37 °C.
2.7 Inhibition zone assay (IZA)
Bacteria and fungi colonies were added to 10 ml of Luria Broth (LB) media and incubated at 37 °C for 2–3 h while shaking. Growth of the microbes was monitored using a spectrophotometer (Thermo Electron Corporation Helios Beta Spectrophotometer Model 9423 UVB 133214, England). Once the optical density (OD) reached 0.6, the microbes were diluted with LB media at a ratio 1:100. Then, 40 µl was added to 6 ml LB media with 1 % agarose, poured onto a 90 mm petri dish (1 mm thick) and left for 30 min to solidify. Small wells (3 mm diameter wide) were made and then loaded with 3 µl of the reconstituted seaweed extracts. Hydrogen peroxide (H2O2) was used as a positive control and phosphate-buffered saline (PBS) was used as a negative control. In addition, pure methanol was tested to examine its lethality against the tested microbes. All plates were incubated (Gallenkamp Model INC.200.210C Plus Series Incubator, England) overnight at 37 °C and the diameter of the microbes’ free zone was measured (Al Adwani et al., 2021).
2.8 Colony forming units (CFU) assay
Microbes’ colonies were placed in 10 ml of LB media and incubated at 37 °C while shaking for 2–3 h. The OD was adjusted to 0.1, then 180 µl of the culture and 20 µl of the reconstituted extract were placed in 96-well plates and incubated at 37 °C for 3 h. Then, a series of dilutions of the incubated culture was made with PBS to produce 1:1–10 dilutions. Next, 10 µl was placed on LB agar plates and incubated overnight at 37 °C. Finally, the CFU of microbes was calculated (Al Adwani et al., 2021).
2.9 Time-dependent killing
E. coli colonies were placed in 10 ml of LB media and incubated at 37 °C while shaking for 2–3 h. Then, the broth was diluted with LB media to OD 0.1. Then, 180 µl of the diluted broth and 20 µl of the reconstituted extract were placed in 96-well plates and incubated at 37 °C for 15, 30, 60, 120, and 180 min. Then, a series of dilutions of the incubated culture was made with PBS to produce 1:1–10 dilutions. Next, 10 µl was placed on LB agar plates and incubated overnight at 37 °C. Finally, the CFU of E. coli was calculated (Al Adwani et al., 2021).
2.10 Dose-dependent killing
E. coli colonies were placed in 10 ml of LB media and incubated at 37 °C while shaking for 2–3 h. Then, the broth was diluted with LB media to OD 0.1. A serial dilution of the reconstituted extract was made by methanol to produce 1:1–4 dilutions. Then, 20 µl of the following: the non-diluted reconstituted extract, the diluted reconstituted extracts and H2O2 were placed in separate wells on 96-well plates. 180 µl of the diluted broth were added to each well, and incubated at 37 °C for 3 h. Then, serial dilutions of the incubated culture were made with PBS to produce 1:1–10 dilutions. Next, 10 µl was placed on LB agar plates and incubated overnight at 37 °C. Finally, the CFU of E. coli was calculated (Al Adwani et al., 2021).
2.11 Scanning electron microscope (SEM)
SEM was performed based on Rahman et al. (2019) with minor changes. In brief, 2 ml of E. coli broth treated or non-treated with 1 ml of EM 2.5 % Karnovsky's fixative were placed in an Eppendorf tube, thoroughly mixed in a rotary mixer (roller mixer, SRT9, Germany) for 2 h and then centrifuged at 5,000 rpm for 5 min. The supernatant was discarded and the pellets were re-suspended in 1 ml sodium cacodylate washing buffer for 10 min and then centrifuged at 5,000 rpm for 5 min. The supernatant was discarded and the pellets were fixed using 1 ml 2 % osmium tetroxide and dehydrated using a series of alcohol concentrations starting from two washes of distilled water then 25 % ethanol, 75 % ethanol, 95 % ethanol, and finally two washes of 99.9 % ethanol. Later, 1 ml of hexamethyldisilazane (HMDS)/ethanol mixture (1:1, v/v) was added and left to stand for 30 min, and then 1 ml of HMDS was added and left for 20 min. Then, the samples were treated with pure HMDS and left at room temperature for 3 h to dry. Dried bacterial samples were then transferred to 10 mm aluminium stubs and adhered to the top of the stubs using double-side carbon adhesive. They were then coated with gold particles using a BioRad coating system (BIO-RAD, Microscience Division Serial No 88091, England) and examined by Jeol JSM-5600LV scanning electron microscope (Japan). Topographic micrographs of bacteria samples were retrieved and saved as images.
2.12 Transmission electron Microscopy (TEM)
Bacteria samples were processed for transmission electron microscopy (TEM) based on the protocol described by Al Adwani et al. (2021) with minor modifications. In brief, 1 ml of each broth was collected and placed in an Eppendorf tube, and immediately, 1 ml of 2.5 % glutaraldehyde fixative was added into every sample tube and then mixed for 2 h using a rotary mixer (roller mixer, SRT9, Germany). After that, the tubes were centrifuged (Centrifuge 5702, Eppendorf, England) at 5,000 rpm for 5 min and the supernatants were discarded. The formed pellets were mixed with 1 ml of cacodylate washing buffer twice for 10 min. Later, the samples were centrifuged and the pellets were retained in the same way as explained in the previous step. Then, the pellets were post-fixed using 1 ml of osmium tetroxide and left in the mixer for 1 h. Dehydration steps were carried out using a series of acetone concentrations, starting from two washes of distilled water, 25 %, 75 %, 95 %, and finally, two washes of 99.9 % acetone. The samples were then infiltrated by a mixture of 1:1 acetone to epoxy resin for 1 h, then 1:3 acetone to epoxy resin for 30 min, and finally mixed with pure resin for 1 h. The samples were then polymerized in an oven incubator (Gallenkamp Model INC.200.210C Plus Series Incubator, England) overnight. The next day, the resin blocks were retrieved, and ultrathin sections were produced using an ultramicrotome (Leica UCT Ultracut Ultramicrotome, Austria) to produce 70 nm thick sections. Sections of samples were picked on 300 mesh copper grids and then stained with uranyl acetate and lead citrate. The samples’ sections were screened using Jeol JEM-1230 transmission electron microscope (Japan). Micrographs were recorded and saved.
2.13 Fourier-transform infrared spectroscopy (FTIR)
Methanol extract fraction (1) from Nizamuddinia zanardinii (MeNZ-F1) was analyzed using a Cary 670 FTIR spectrometer (Agilent, Cary 670 FTIR spectrometer) attached with a diamond single bounce ATR cell (GladiATR, PIKE technologies, USA) was used. One drop of the extract fraction was placed on the ATR crystal and left to evaporate. Infrared spectrum was measured by averaging 32 scans at a resolution of 4 (Al-Alawi et al., 2011).
2.14 Statistical-analysis
The organic solvent extraction, colony forming assay, dose-depending assay, and time-depending assay were performed in triplicate and reported as mean values ± standard deviation on a sun-dried basis. The GraphPad Prism software, version 10.2.3, was used to analyze the data. Testing the data for normality was done, and then one-way ANOVA, unpaired t-test, and multi-t-test were applied to analyze the results. P-value of <0.05 was considered statistically significant.
3 Results
3.1 Fractionation of seaweed extracts and screening for antibacterial activity
Seaweeds were subjected to solvents with different polarities to get fractions with different compounds and properties. As a result, five fractions of each plant were collected and screened for antibacterial activities against E. coli. Based on the inhibition zone assay (IZA), antibacterial activity was detected in the methanol fraction from Nizamuddinia zanardinii (MeNZ); the inhibition zone was 13 ± 1 mm. On the other hand, there was no detectable inhibition of bacterial growth in any of the other fractions. The other plants also didn’t show any antibacterial activity (Table 1). –, no inhibition zone detected, data presented as Mean ± SD (n = 3).
Seaweeds species
Fractions (Solvents)
Inhibition zone (mm) in E. coli
Melanothamnus somalensis
Hexane (H)
−
Dichloromethane (DCM)
−
Ethyl Acetate (EA)
−
Acetone (Ac)
−
Methanol (Me)
−
Gelidium omanense
Hexane (H)
−
Dichloromethane (DCM)
−
Ethyl Acetate (EA)
−
Acetone (Ac)
−
Methanol (Me)
−
Jolyna furcata
Hexane (H)
−
Dichloromethane (DCM)
−
Ethyl Acetate (EA)
−
Acetone (Ac)
−
Methanol (Me)
−
Nizamuddinia zanardinii
Hexane (H)
−
Dichloromethane (DCM)
−
Ethyl Acetate (EA)
−
Acetone (Ac)
−
Methanol (Me)
13 ± 1 mm
MeNZ was further fractionated using a preparative reversed-phase HPLC system which resulted in 12 fractions. The fractions were screened for antibacterial activity using E. coli to identify the fraction containing the compounds responsible for the inhibition. IZA revealed that Fraction 1 from MeNZ (MeNZ-F1), which is expected to have high hydrophilic and polar compounds, had the antibacterial activity (the inhibition zone was 14.66 ± 0.57 mm). By contrast, there was no detectable inhibition of bacterial growth in any of the other fractions (Table 2). As a result, further research focused solely on MeNZ-F1. –, no significant activity detected, data is presented as Mean ± SD (n = 3).
Seaweed/solvent
Fractions
Inhibition zone (mm)
E. coli
Nizamuddinia zanardinii
(methanol fraction)1
14.66 ± 0.57 mm
2
−
3
−
4
−
5
−
6
−
7
−
8
−
9
−
10
−
11
−
12
−
3.2 Time-dependent and dose-dependent antimicrobial activity of MeNZ-F1 against E. coli
MeNZ-F1was evaluated for its potential antibacterial activity against E. coli using colony count assay and it showed a significant difference (F (df) = 2, P = 0.0001) as 1.8 log10 reduction in the growth of E. coli observed after 3 h of incubation compared to the negative control (Fig. 1).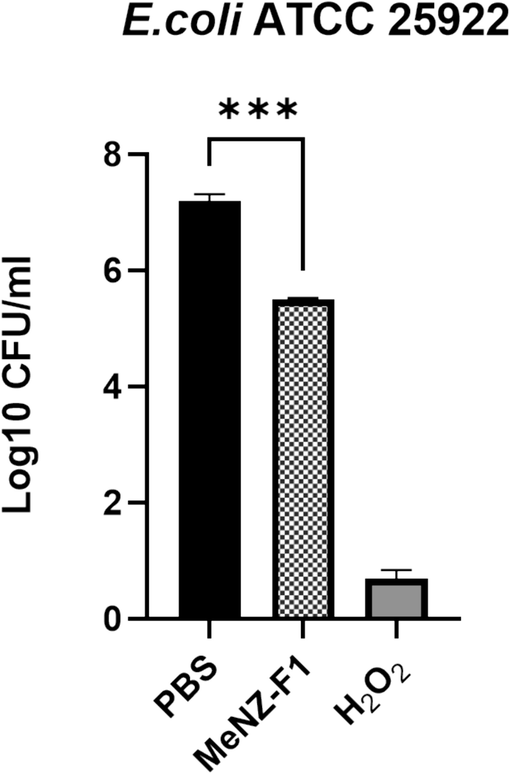
Colony forming units (CFU) of E. coli submitted to MeNZ-F1, C = negative control (PBS), T = treatment, C+ = positive control (H2O2), data is presented as Mean Log10 ± SD (n = 3), (P value: 0.05 > *, 0.01 > **, 0.001 > ***, 0.0001 > ****).
The time-dependent killing assay of MeNZ-F1 was also performed against E. coli. There was a rapid bacterial killing kinetics starting from 30 min of incubation until the first hour. There were no significant differences in the killing power between 60, 120, and 180 min. However, a log10 reduction of 1.7, 1.8 and 2.2 in bacterial growth was attained in the periods 60, 120, and 180 min, respectively (Fig. 2).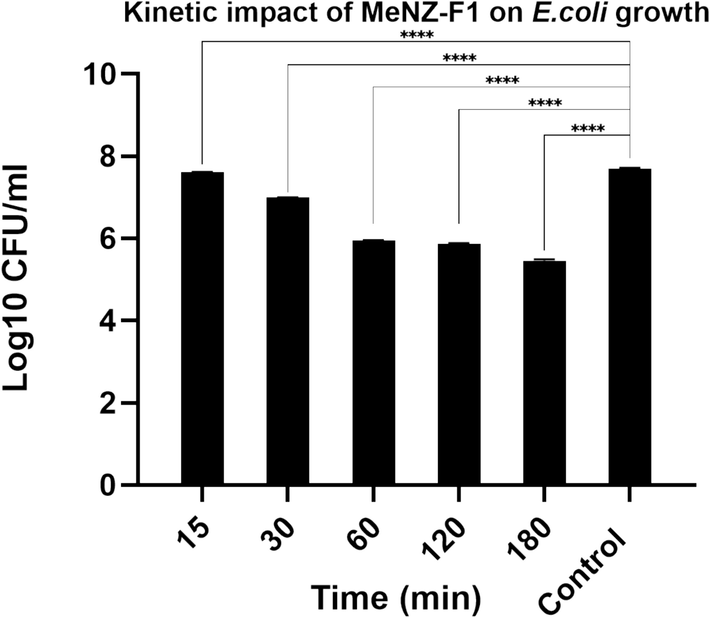
Kinetic impact of MeNZ-F1 on E.coli growth (time-dependent killing assay), data is presented as Mean Log10 ± SD (n = 3), (P value: 0.05 > *, 0.01 > **, 0.001 > ***, 0.0001 > ****).
The killing effect of dilutions of MeNZ-F1 on E. coli was tested, with the bacteria being incubated for 3 h. With increasing dilution, the antibacterial activity showed a decline. There were significant differences between the stock, 1:1, 1:2, and 1:3 dilutions when compared to a negative control. However, no significant differences existed between the 1:4 dilutions and the negative control (Fig. 3) meaning that the active compound is very diluted.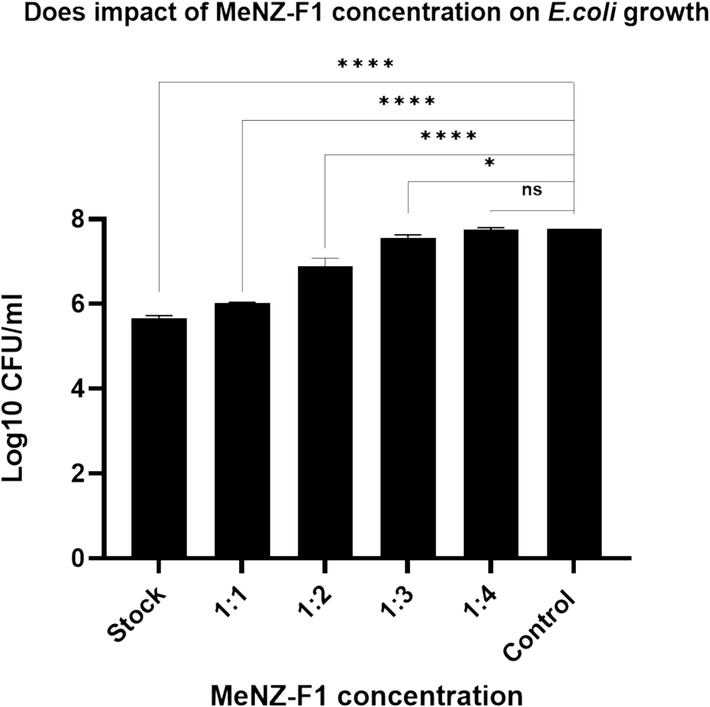
Does impact of MeNZ-F1 concentration on E.coli growth (dose-dependent killing assay), data is presented as Mean Log10 ± SD (n = 3), (P value: 0.05 > *, 0.01 > **, 0.001 > ***, 0.0001 > ****).
3.3 Extracellular and intracellular changes on E. coli caused by MeNZ-F1
Morphological changes of E. coli cells after 3 h incubation with the reconstituted crude extract of MeNZ-F1 were determined using electron microscopy techniques. Extracellular changes in bacteria were observed using Scanning Electron Microscopy (SEM). The untreated E. coli had a smooth and regular shape. However, E. coli treated with MeNZ-F1 had an irregular shape and rough surface compared to the control. Moreover, leakage of cellular content (DNA) was observed compared to the control E. coli (Fig. 4).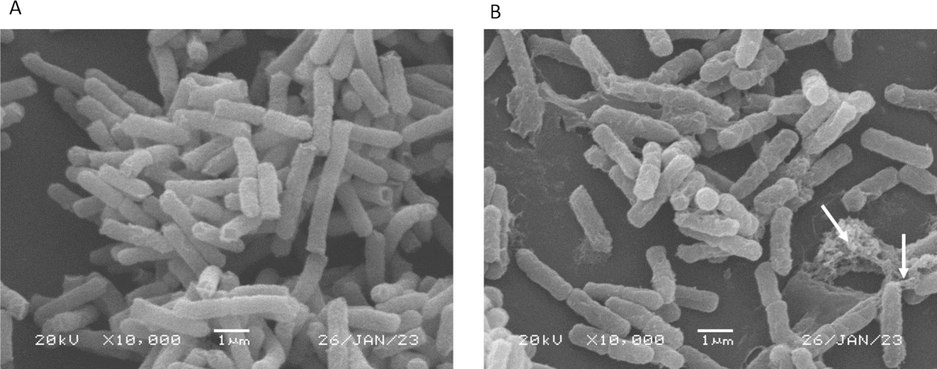
Scanning electron micrograph of E.coli. (A) Control (B) Treated E.coli with MeNZ-F1. Arrows = Cell content leakage. The scale bar represents 1.0 μm.
Intracellular changes were observed using Transmission Electron Microscopy (TEM) on both MeNZ-F1 treated and non-treated E. coli. Untreated E. coli had intact-undamaged membranes, and their ribosomes and DNA were evenly distributed in the cytoplasm (Fig. 5; dark and light area, respectively). Moreover, exposure to MeNZ-F1 resulted in wrinkling of E. coli membranes. Additionally, there was some degree of dissociation of membrane fragments. Notably, small vesicles or blebs were observed to be released from the membrane of the MeNZ-F1-treated E. coli. Furthermore, ribosomes were clustered and directed toward the inner membrane of the bacteria, and the DNA clustered in the center of the cell, whereas the double membrane was not destroyed (Fig. 5).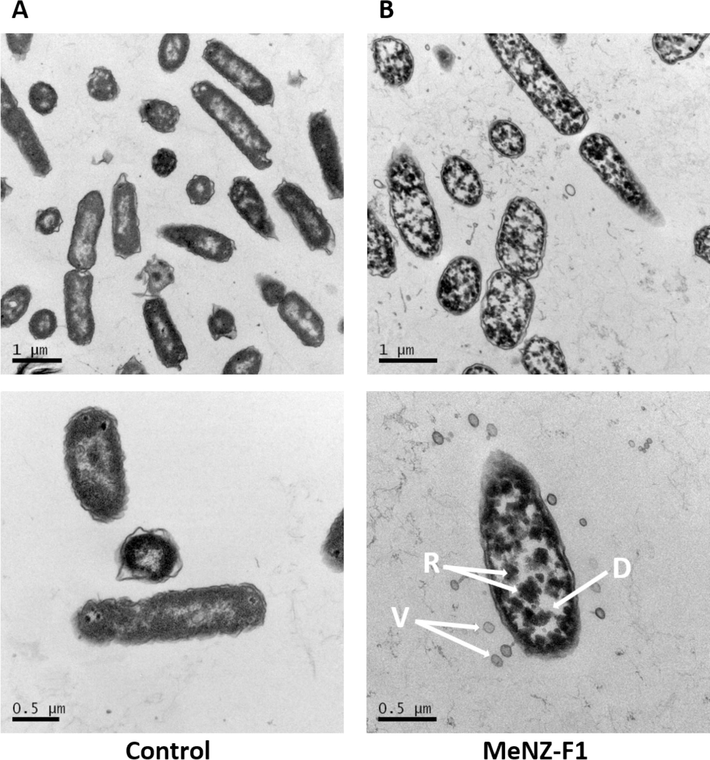
Transmission electron micrograph of E.coli. Upper and lower panel (A) Control (B) Treated E.coli with MeNZ-F1. Upper panel magnification at 1.0 μm and lower panel magnification at 0.5 μm. D: clustered DNA; R: clustered ribosomes; V: vesicles.
3.4 Antimicrobial activity of MeNZ-F1 against several bacterial and fungal pathogens.
The antimicrobial activities of MeNZ-F1 were tested against the bacterial strains S. aureus, P. aeruginosa, and K. pneumonia and the fungal strain C. albicans. Based on IZA, activity was detected on all tested pathogens, and the inhibition zones were as follows: 12.33 ± 0.5 mm, 14.33 ± 0.5 mm, 12.33 ± 0.52 mm, and 9.33 ± 1.5 mm, respectively (Fig. 6). Furthermore, the CFU results showed a growth reduction in S. aureus, P. aeruginosa and K. pneumonia, and C. albicans (1.2 log, 1.1 log, 1 log, and 2.7 log, respectively) after 3 h of incubation (Fig. 7). There were significant differences between the control and treatments in all microbe species which indicated broad-spectrum antimicrobial activity.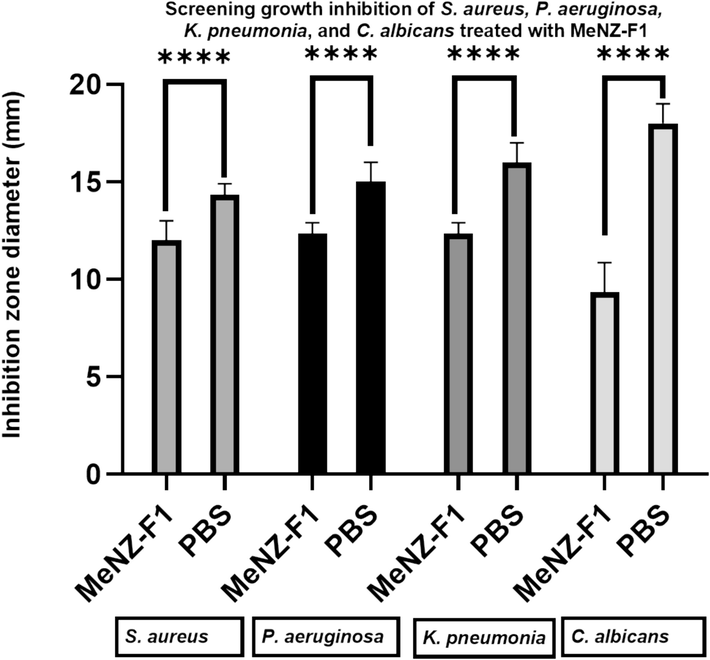
Screening growth inhibition of S. aureus, P. aeruginosa and K. pneumonia, and C. albicans treated with MeNZ-F1 was expressed as diameter of bacteria-free zone (mm). Data is presented as Mean ± SD (n = 3).
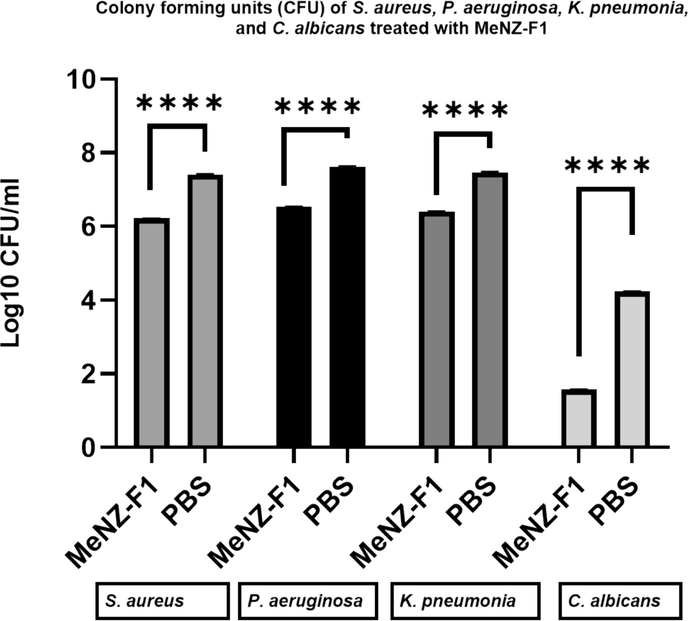
Colony forming units (CFU) of S. aureus, P. aeruginosa and K. pneumonia, and C. albicans treated with MeNZ-F1, T = Treatment, C=negative control (PBS), data is presented as Mean Log10 ± SD (n = 3), (P value between treatment and negative control: 0.05 > *, 0.01 > **, 0.001 > ***, 0.0001 > ****).
3.5 FTIR analysis of MeNZ-F1
The FTIR-ATR spectroscopy of the MeNZ-F1was recorded to identify the major chemical compounds in the fraction. Presence of various peaks in the FTIR spectrum is an indication of different functional groups. The large and broad peak with maximum absorption at ∼3400 cm−1 is assigned to the O–H stretch of H-bonded alcohols and phenols. The broad and weak band in the region 2000–2400 cm−1 is a common feature in our FTIR machine and it is not correlated to any specific group. The peak at ∼1600 cm−1 represents C⚌C stretching vibrations and is indicative of the presence of alkenes. This band also overlaps with the OH bending vibration that comes from water. The broadband in the region 400–900 cm−1 is related to the vibrations of C–H from aromatic rings in the phenolic compounds (Fig. 8).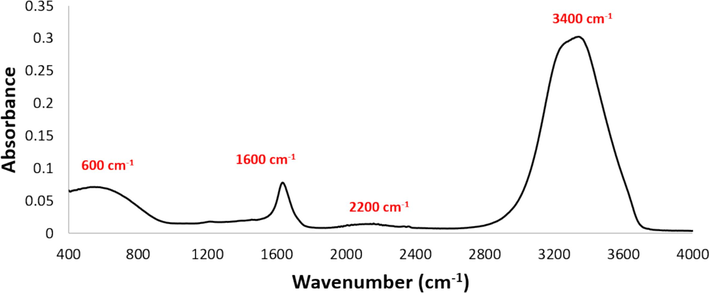
FTIR-ATR spectra of MeNZ-F1 showing the positions of specific functional groups.
4 Discussion
In general, seaweeds contain numerous biologically active and unique metabolites that may have applications in different fields, such as pharmacology and food production. For example, several ingredients of various seaweeds have shown antimicrobial activities in vitro against fungi, gram-positive and gram-negative bacteria (Gomes et al., 2022; Madkour et al. 2019; Manivannan et al., 2011; Moubayed et al., 2017; Pérez et al., 2016). Nevertheless, there is a lack of studies on the antimicrobial properties of seaweed extracts collected from Oman’s coastline especially those unique to Oman, such as Gelidium omanense (Wynne, 2018). Hence, the present study investigated the antimicrobial activities of two red seaweed species (Melanothamnus somalensis and Gelidium omanense) and two brown seaweed species (Jolyna furcata and Nizamuddinia zanardinii). Our study revealed that Nizamuddinia zanardinii (MeNZ) methanol fraction had broad antimicrobial action against pathogenic bacteria and fungus.
In this study, MeNZ showed a high antibacterial action against E. coli (Table 1), and the fraction MeNZ-F1 contained the active compounds responsible for the antibacterial activity (Table 2 & Fig. 1). MeNZ-F1 showed rapid bacterial killing kinetics against E. coli starting from 30 min of incubation until the first hour. Moreover, the reconstituted extract of MeNZ-F1 showed high antibacterial activity against E. coli, and with increasing dilution of MeNZ-F1, the antibacterial activity gradually decreased (Fig. 2 & Fig. 3). The work on Nizamuddinia zanardinii is very limited worldwide; however, Alboofetileh et al. (2019) stated that fucoidan (a polysaccharide) extracted from Nizamuddinia zanardinii showed interesting antimicrobial, antiviral, anticancer and immunomodulatory activities. With regard to the work done on other Omani seaweeds, Anjali et al. (2019) studied the antimicrobial properties of two seaweeds, Stoechospermum marginatum (S. marginatum, brown seaweed) and Ulva lactuca (U. lactuca, green seaweed) against five multidrug-resistant bacteria (K. pneumonia, S. aureus, Salmonella typhi, Proteus vulgaris and E. coli). They found that the aqueous extracts showed enhanced antibacterial activities paralleled to methanol extracts and the maximum activity against E. coli was given by U. lactuca extract. Concerning the work done in other parts of the world, there is very limited antibacterial work done on the seaweed plants studied in the present work. Therefore, it was not possible to make much comparison.
Nevertheless, the literature is rich with work on other plants using various extraction solvents. For example, Madkour et al. (2019) stated that acetone, isopropyl alcohol and ethanol extracts of three brown algae (Cystosiera myrica, Padina pavonica, and Turbinaria ornate) exhibited antibacterial actions against three pathogenic bacteria (E. coli, P. aeruginosa and S. aureus). Bhuyar et al. (2020) reported that hot water and ethanolic extracts of Kappaphycus alvarezii displayed antibacterial action against B. cereus. In another investigation, the ethanolic extracts of Ascophyllum nodosum, Saccharina longicruris and Ulva lactuca showed antibacterial activity against E. coli (Boisvert et al., 2015). Methanolic extracts of Jania rubens, Padina pavonica, Ulva lactuca and Enteromorpha compressa showed antibacterial activity against S. aureus, Proteus vulgaris, K. pneumoniae, E. coli, P. aeruginosa, and B. subtilis (Moghadam et al., 2013). The variation in antibacterial activity is attributed to the variance in the active compounds present. This variance is influenced by factors such as species type, extraction conditions, post-harvest treatments, and the type of bacteria being tested (Cox et al., 2010; Kandhasamy and Arunachalam, 2008; Moubayed et al., 2017). For example, dried seaweed extracts (due to the lyophilization of seaweed samples) showed a higher inhibitory effect, whereas fresh seaweed samples, which had high water content, showed negligible inhibitory activity (Moubayed et al., 2017).
In our findings, the treated E. coli with MENZ-F1 had shown structural changes such as irregular shape, rough surface, leakage of cellular content, wrinkling of membranes and ribosomes clustering, (Fig. 4 & Fig. 5). Phenolic compounds are one of the main recognised active antibacterial compounds in seaweeds (Vatsos and Rebours, 2015). Overall, these active compounds change extracellular pH and cause leakage of cell constituents, including inorganic ions such as phosphate and potassium, proteins, and nucleic acids (Pernin et al., 2019). Moreover, they could alter enzymes’ activities, DNA structure and ribosome translation (Zhang et al., 2019).
Comparison of the results presented in this study with the other studies shown above indicates a wide spectrum of antibacterial activity of the MeNZ extract (Fig. 6 & Fig. 7). In addition, our findings indicated that MeNZ had an added potent inhibitory effect on S. aureus than on E. coli (Fig. 7). The differences in capability among tested bacterial strains could be attributed to differences in bacterial membrane composition (Taskin et al., 2007). High antibacterial activity in seaweeds shows potential for developing drugs to treat human pathogens.
The FTIR-ATR spectra of the MeNZ-F1 are shown in Fig. 8. The spectra demonstrated that MeNZ-F1 is rich in phenols; however, it was impossible to determine the identity of the phenol compounds from the FTIR spectrum. This was apparent from the presence of characteristic peaks of phenols in the spectrum, such as the large and broad peak with a maximum absorption at around 3400 cm−1 (represents OH group in the phenols), peak at ∼1600 cm−1 (representing alkenes in an aromatic ring) and broadband in the region 400–900 cm−1 (vibrations of C–H from aromatic rings in the phenolic compounds) (da Silva et al., 2018; Mbonyiryivuze et al., 2015; Wongsa et al., 2022). Phenolic compounds are proposed to be the main constituents of organic solvent extracts, and the inhibition effects on microbial growth depends on concentration and constitution (Moubayed et al., 2017). The concentration and constitution of the antimicrobial agents depend on the type of seaweed as well as the solvents used, since some biologically active compounds may be soluble in one solvent but not in another (Moubayed et al., 2017; Salem, 2011). Cox et al. (2010) demonstrated that the extraction of compounds with high antimicrobial activities was dependent on the solvent used. Methanol was suggested to be the best solvent to extract compounds with high antimicrobial activities from brown seaweeds; whereas acetone was the best to use with green seaweeds. Manilal et al. (2009) suggested that methanolic extraction leads to higher antimicrobial activity. Moreover, Vlachos et al. (2002) demonstrated that brown seaweed exhibits the highest level of antibacterial activity, followed by red seaweed and then green seaweed. In contrast, Kandhasamy and Arunachalam (2008) showed that green and brown seaweeds have more active compounds compared to the other seaweed groups. In the present study, higher antimicrobial activity was obtained with methanol extraction of the brown seaweed Nizamuddinia zanardinii compared to the other solvents and other species, which is in line with the previous studies.
Recently, global trends have shown an increase in the use of biological agents, such as microorganisms and natural extracts, for synthesizing nanoparticles. This approach offers an eco-friendly alternative to traditional chemical methods (Mughal et al., 2021). This approach involves the use of microorganisms such as bacteria and algae, as well as plant extracts, which serve as both reducing and stabilizing agents during the synthesis process. The natural sources contain biomolecules and phytochemicals, such as enzymes, flavonoids, alkaloids, phenols, and terpenoids, which effectively aid in the incorporation of metal ions into nanoparticles (Kaningini et al., 2022). This method of biosynthesis offers numerous benefits, including reduced toxicity, lower energy requirements, and cost-effectiveness compared to traditional methods. The nanoparticles that are produced have wide range of applications in fields such as medicine, biosensors, and pharmaceuticals. Additionally, researchers have explored the use of agricultural waste as a potential bioresource for synthesizing nanoparticles (Maaza, 2014). For example, a study conducted by Madubuonu et al. (2020) revealed antibacterial activity of iron oxide nanoparticles (FeONPs), which were made from aqueous extract of Psidium guajava (P. guajava), against six human pathogenic strains (Gram-negative (P. aeruginosa, S. typhi, Shigella, Pasteurella and E. coli) and Gram-positive (S. aureus). FeONPs showed high antibacterial activity when compared with standard antibiotic drugs using IZA. These results are in agreement with our findings, as we found that the use of MeNZ-F1 was effective in inhibiting the growth of not only bacteria but also fungus as shown by both the IZA and the CFU killing assay. Therefore, natural extracts, such as MeNZ-F1, have the potential to be used as alternative antimicrobial drugs.
5 Conclusion
The current investigation showed an appreciable antimicrobial activity by MeNZ-F1 against specific human pathogens. MenZ-F1 exhibited broad antimicrobial activity, specifically against bacterial strains S. aureus, E. coli, P. aeruginosa and K. pneumoniae as well as the fungal strain C. albicans. TEM and SEM results revealed the effect of MeNZ-F1 on the tested pathogens at the cell level. The results showed an irregular shape, rough surface, and leakage of the cellular content. Furthermore, ribosomes were clustered and directed toward the inner membrane of the bacteria, and the DNA clustered in the center of the cell. Although MeNZ-F1 was demonstrated to be applicable to a wide range of pathogenic microbes, more bacteria and fungi need to be tested in the future to establish solid conclusions. In addition, future studies are required to examine the antibacterial activity of MeNZ-F1 over a longer period to determine its efficacy and activity. Furthermore, MeNZ-F1 requires a detailed study to isolate and characterize the active antimicrobial compounds.
CRediT authorship contribution statement
Abdullah Al-Nassri: Writing – original draft, Software, Methodology, Investigation, Formal analysis, Data curation. Ahmed Al-Alawi: Writing – review & editing, Validation, Supervision, Resources, Project administration, Investigation, Funding acquisition, Conceptualization. Salma Al-Adwani: Writing – review & editing, Validation, Supervision, Resources, Project administration, Investigation, Funding acquisition, Formal analysis, Conceptualization.
Acknowledgment
The authors extend their appreciation to Sultan Qaboos University (Oman) for funding this project: SR/AGR/FOOD/21/01.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Citrullination alters the antibacterial and anti-inflammatory functions of the host defense peptide canine cathelicidin K9CATH in vitro. J. Immunol.. 2021;207:974-984.
- [CrossRef] [Google Scholar]
- Characterization of Carrageenan Extracted from Hypnea bryoides in Oman. Mar. Biotechnol.. 2011;13:893-899.
- [CrossRef] [Google Scholar]
- Bioactivities of Nizamuddinia zanardinii sulfated polysaccharides extracted by enzyme, ultrasound and enzyme-ultrasound methods. J. Food Sci. Technol.. 2019;56:1212-1220.
- [CrossRef] [Google Scholar]
- Bioprospecting of seaweeds (Ulva lactuca and Stoechospermum marginatum): the compound characterization and functional applications in medicine-a comparative study. J. Photochem. Photobiol. B Biol.. 2019;200:111622
- [CrossRef] [Google Scholar]
- Macroalgae biorefineries as a sustainable resource in the extraction of value-added compounds. Algal Res.. 2023;69:102954
- [CrossRef] [Google Scholar]
- A preparative method for the isolation and fractionation of dissolved organic acids from bitumen-influenced waters. Sci. Total Environ.. 2019;671:587-597.
- [CrossRef] [Google Scholar]
- Antioxidant and antibacterial activity of red seaweed; Kappaphycus alvarezii against pathogenic bacteria. Global J. Environ. Sci. Manage.. 2020;6:47-58.
- [CrossRef] [Google Scholar]
- Assessment of the antioxidant and antibacterial activities of three species of edible seaweeds. J. Food Biochem.. 2015;39:377-387.
- [CrossRef] [Google Scholar]
- Antioxidant and cytotoxic activities of three species of tropical seaweeds. BMC Complement. Altern. Med.. 2015;15
- [CrossRef] [Google Scholar]
- An assessment of the antioxidant and antimicrobial activity of six species of edible Irish seaweeds. Int. Food Res. J.. 2010;17:205-220.
- [CrossRef] [Google Scholar]
- Determination of total phenolic compounds and antioxidant activity of ethanolic extracts of propolis using ATR–FT-IR spectroscopy and chemometrics. Food Anal. Methods. 2018;11:2013-2021.
- [CrossRef] [Google Scholar]
- Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med.. 2016;13:1-6.
- [CrossRef] [Google Scholar]
- Fiori, S.M., Pratolongo, P.D., 2021. The Bahía Blanca Estuary: Ecology and biodiversity, The Bahía Blanca Estuary: Ecology and Biodiversity. Doi: 10.1007/978-3-030-66486-2.
- Seaweeds’ pigments and phenolic compounds with antimicrobial potential. Biomol. Concepts. 2022;13:89-102.
- [CrossRef] [Google Scholar]
- Fucoidan can function as an adjuvant in vivo to enhance dendritic cell maturation and function and promote antigen-specific T cell immune responses. PLoS One. 2014;9:1-10.
- [CrossRef] [Google Scholar]
- Green synthesis and characterization of zinc oxide nanoparticles using bush tea (Athrixia phylicoides DC) natural extract: assessment of the synthesis process. F1000Research. 2022;10:1-22.
- [CrossRef] [Google Scholar]
- A review of the varied uses of macroalgae as dietary supplements in selected poultry with special reference to laying hen and broiler chickens. J. Marine Sci. Eng.. 2020;8
- [CrossRef] [Google Scholar]
- Characterization of carrageenan extracted from hypnea bryoides in Oman. Mar. Drugs. 2019;13:893-899.
- [CrossRef] [Google Scholar]
- An overview on antimicrobial potential of edible terrestrial plants and marine macroalgae rhodophyta and chlorophyta extracts. Mar. Drugs 2023
- [CrossRef] [Google Scholar]
- Natural Dyes for Photonics Applications. In: Gurib-Fakim A., ed. Novel Plant Bioresources: Applications in Food, Medicine and Cosmetics. Vol vol. 1. Wiley-Blackwell; 2014. p. :479-491.
- [Google Scholar]
- Antibacterial activity of some seaweeds from the red sea coast of Egypt. Egyptian J. Aquatic Biol. Fisheries. 2019;23:265-274.
- [CrossRef] [Google Scholar]
- Bio-inspired iron oxide nanoparticles using Psidium guajava aqueous extract for antibacterial activity. Appl. Phys. A Mater. Sci. Process.. 2020;126:1-8.
- [CrossRef] [Google Scholar]
- Antimicrobial potential and seasonality of red algae collected from the southwest coast of India tested against shrimp, human and phytopathogens. Ann. Microbiol.. 2009;59:207-219.
- [CrossRef] [Google Scholar]
- Antimicrobial potential of selected brown seaweeds from Vedalai coastal waters, Gulf of Mannar. Asian Pac. J. Trop. Biomed.. 2011;1:114-120.
- [CrossRef] [Google Scholar]
- Fourier transform infrared spectroscopy for sepia melanin. physics and materials. Chemistry. 2015;3:25-29.
- [CrossRef] [Google Scholar]
- A cytotoxic hydroperoxy sterol from the brown alga, Nizamuddinia zanardinii. DARU, J. Pharmaceutical Sci.. 2013;21:2-5.
- [CrossRef] [Google Scholar]
- Antimicrobial, antioxidant properties and chemical composition of seaweeds collected from Saudi Arabia (Red Sea and Arabian Gulf) Saudi J. Biol. Sci.. 2017;24:162-169.
- [CrossRef] [Google Scholar]
- Biogenic nanoparticles: synthesis, characterisation and applications. Appl. Sci. (Switzerland). 2021;11
- [CrossRef] [Google Scholar]
- Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629-655.
- [CrossRef] [Google Scholar]
- Antimicrobial action of compounds from marine seaweed. Mar. Drugs. 2016;14:1-38.
- [CrossRef] [Google Scholar]
- Inhibitory activity of phenolic acids against Listeria monocytogenes: deciphering the mechanisms of action using three different models. Food Microbiol.. 2019;80:18-24.
- [CrossRef] [Google Scholar]
- Biosorption of heavy metals by a lead (Pb) resistant bacterium, Staphylococcus hominis strain AMB-2. J. Basic Microbiol.. 2019;59:477-486.
- [CrossRef] [Google Scholar]
- Screening for antibacterial activities in some marine algae from the red sea (Hurghada, Egypt) Afr. J. Microbiol. Res.. 2011;5:2160-2167.
- [CrossRef] [Google Scholar]
- Book review: tackling drug-resistant infections globally. Arch. Pharm. Pract.. 2016;7:110.
- [CrossRef] [Google Scholar]
- Marine-based nutraceuticals: an innovative trend in the food and supplement industries. Mar. Drugs. 2015;13:6336-6351.
- [CrossRef] [Google Scholar]
- Antibacterial activities of some marine algae from the Aegean Sea (Turkey) Afr. J. Biotechnol.. 2007;6:2746-2751.
- [CrossRef] [Google Scholar]
- Seaweed extracts as antimicrobial agents in aquaculture. J. Appl. Phycol.. 2015;27:2017-2035.
- [CrossRef] [Google Scholar]
- On the GRAS status of seaweeds. I. Observations on the association between antibacterial activity of ethanolic extracts and metal levels present in selected seaweeds. Bull. Marine Sci. Fisheries, Kochi University. 2002;21:7-12.
- [Google Scholar]
- FT-IR characteristics, phenolic profiles and inhibitory potential against digestive enzymes of 25 herbal infusions. Sci. Rep.. 2022;12:1-11.
- [CrossRef] [Google Scholar]
- Effect of different light qualities on growth, pigment content, chlorophyll fluorescence, and antioxidant enzyme activity in the Red Alga Pyropia haitanensis (Bangiales, Rhodophyta) Biomed. Res. Int.. 2016;2016
- [CrossRef] [Google Scholar]
- A checklist of the benthic marine algae of the Northern Arabian Sea coast of the Sultanate of Oman. Bot. Mar.. 2018;61
- [CrossRef] [Google Scholar]
- Structure-dependent Inhibition of Stenotrophomonas maltophilia by polyphenol and its impact on cell membrane. Front. Microbiol.. 2019;10:1-11.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2024.103431.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







