Translate this page into:
Antibiotic resistance in Salmonella: Targeting multidrug resistance by understanding efflux pumps, regulators and the inhibitors
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
An increased number of research articles in terms of infectious disease and multidrug resistance challenges reveals that antibiotic resistance is becoming a serious health issue. Multidrug resistance in Salmonella is acquired through efflux pumps extruding the antibiotics from the bacterial cells. Efflux pumps in bacteria help in developing resistance and tolerating antibiotics. A collective effort of efflux pumps, regulators, and other cellular components helps in multidrug resistance, pathogenicity, biofilm formation, etc. As a result, efflux pumps have become an important drug target for novel drugs and their inhibitors may help to fight the resistance to existing antibiotics. Efflux pumps recognize the substrates, and prevent their accumulation inside the cells, thus interrupting their mode of action. Drug resistance in Salmonella is regulated by the expression of transcriptional regulator families. In this review, the different efflux pumps, their regulators, and inhibitors are summarized to target the antibiotic resistance pattern of Salmonella. In addition, a special focus is made to elucidate the roles of Salmonella efflux pumps in virulence and biofilm formation. The detailed study of efflux pump inhibitors may also help to explore and identify novel antibiotic components or additives that enhance the drug efficacy in multidrug-resistant strains.
Keywords
Antibiotic resistance
Efflux pump
Inhibitors
Regulators
Salmonella
1 Introduction
Salmonella is the main cause of diarrhea annually 550 million humans are infected, by 2020 million are under the age of 5. Many cases are deadly and life-threatening. The increase in antibiotic resistance to Salmonella has become a serious threat leading to severe problems in healthcare. According to WHO, pathogens, for new antibiotics are urgently needed. One of the highest attention needed pathogens is Salmonella, which is resistant to fluoroquinolone (Table 1). Recently, a report has shown that tetra and penta-resistance are found in Salmonella (Xiang et al., 2020). The development of resistance from multidrug (MDR), and extensive resistance (XDR) leading to pan drug resistance (PDR) may cause a post-antibiotic era in which bacterial infections cannot be controlled by antibiotics. Generally, pathogenic bacteria have mechanisms to face many risks surrounding them, the most important of which are antibiotics. These mechanisms include (i) efflux pumps, which remove the drugs from bacterial cells to the outside and causing losing their effectiveness by lowering their concentration to non-toxic levels. (ii) Antibiotic inactivation by bacterial enzymes that modify or destroy antibiotics structure. (iii) Alteration of target site via spontaneous mutation, which usually occurs in the cell envelope by modifying the chemical structure of their molecular targets. (vi) Preventing drug entry through modifying the frequency, size, and selectivity of porin channels, which are found in the bacterial envelope (Fig. 1) (Abdi et al., 2020).

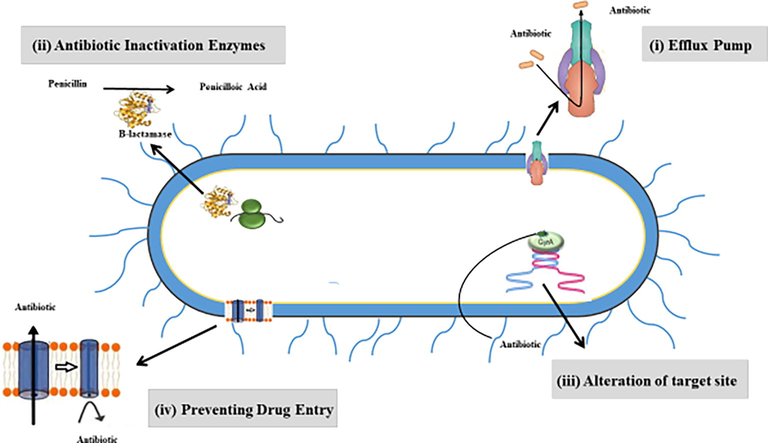
Schematic diagram highlighting the main antimicrobial resistance mechanisms in pathogenic bacteria which include: i) drug efflux systems, which expel the antimicrobials outside the bacterial cell and leading to reducing their effectiveness to non-toxic levels. ii) Antibiotic inactivation enzymes, which modify or destroy the structure of antibiotics. iii) Alteration of target site, which usually occurs in the cell envelope via spontaneous mutation by chemical modification of their molecular targets. iv) Preventing drug entry through modifying the frequency, size, and selectivity of porin channels, which found in the bacterial envelope and plays a crucial role of antibiotic entry into the bacterial cell.
The cell wall of bacteria is a complex multilayered structure that possesses complex proteins, and efflux pumps, spanning the inner membrane or cytoplasmic membrane and the outer membrane partitioned by periplasmic space. Therefore, the bacterial cell wall plays a major role in antibiotic resistance in pathogenic bacteria by preventing antibiotics and other toxic compounds, from entering the bacterial cell (Fisher and Mobashery, 2020). One of the main properties of efflux pumps is the recognition and extrusion variety of structurally diverse substrates outside the bacterial cell. This may lead to preventing the intracellular accumulation of these substances inside bacterial cells and leading to reducing their concentrations to non-toxic levels. In addition, efflux pumps prevent these substrates from reaching their sites of action inside the bacterial cell (Kornelsen and Kumar, 2021). However, the efflux pump and outer membrane barrier in bacteria are nonspecific defense mechanisms and inhibition studies in efflux pumps have proved to decrease resistance development in bacteria. The drugs enter the bacterial cell through the outer membrane via porin channels or lipopolysaccharide and reach the cytoplasm then the efflux pump expels out the harmful chemicals from the periplasm to the medium. Generally, bacteria possess five families of bacterial efflux pumps, which, are well-defined via biochemical, microbiological, and structural studies. These efflux pumps include ATP binding cassette (ABC) family, multidrug and toxic compound exporters (MATE), small multidrug resistance (SMR), resistance nodulation division proteins (RND), and major facilitator superfamily (MFS) were characterized by sequence homology (Pagès et al., 2005).
2 Pathogenicity of Salmonella
Salmonella (Enterobacteriaceae) Gram-negative rods, facultative anaerobe associated with foodborne illness, causing Salmonellosis (Jajere, 2019). Two broader species namely Salmonella enterica and S. bongori has been described and more than 2600 serovars belonging to the former species are reported as animal and human pathogen. Salmonella enterica serovar Typhimurium causes food poisoning known to be the second-largest cause of bacterial diarrhea (Sutkuviene et al., 2013). Multidrug-resistant Salmonella infections lead to a twofold higher mortality rate than normal strains responsible for 1–4% worldwide. Upon entry to the host body, Salmonella raises the acid tolerance response (ATR) to tolerate low pH in the stomach as well as the internal pH homeostasis of the bacterial cell. Salmonella enterica as an intracellular pathogen enters the M cells to cause gastrointestinal diseases or systemic diseases by crossing epithelial cells with specialized antigen sampling cells (M cells) depending on the strains or serovars (Buckley et al., 2006). The host cells undergo cytoskeletal changes that help in the entry of Salmonella. The capability to enter the host cells and cause infections are mediated by Salmonella Pathogenicity Islands (SPI) in specific virulent phenotypes. These genes are attained through horizontal gene transfer (Zha et al., 2019). The entry is mediated by the genes present in Salmonella Pathogenicity Island (SPI-1 contains at least 29 genes), by infecting the macrophages, stimulating mucosal inflammation, and disseminating from lymph nodes to phagocytes, spleen, and liver (Lou et al., 2019). Type secretion system (T3SS) that functions like a molecular syringe encoded by SPI1 and SPI2, helps in the effector protein secretion required for invasion of epithelial cells in the intestine that increases secretion and inflammation. The effector proteins help in translocating the proteins required for intracellular survival, multiplication, and causing infections (Srikanth et al., 2011). T3SS (Inv/Spa) helps in delivering proteins such as SopE, SipA, SipC, and StpP from Salmonella cytosol into host cells that help in cytoskeletal reshuffling (Groisman and Mouslim, 2000). Further, for the survival and growth of Salmonella SPI-2, 3 and 4 are required. During the enteric phase of disease inflammation and chloride secretion are mediated by SPI-5 (Marcus et al., 2000). Chronic infections of the gallbladder are caused by Salmonella that invades the liver or from the biliary tract. Further, it forms biofilms in gallstones and causes reinfection of the gastrointestinal tract, and transmits through the fecal route to new hosts (Crawford et al., 2010). Persistent Salmonella infection may cause irritable bowel syndrome and cancer by impairing the mucosal barrier, T3activation leading to host immune disorders and unbalancing the microbial community (Zha et al., 2019).
3 Multi-drug resistance in Salmonella
Fluoroquinolones were included from 1980 in Europe and further in 1995 in the USA for the treatment of salmonellosis in veterinary medicine (Bager and Helmuth, 2001). The resistance development in Salmonella had been reported by chromosomal mutation and exclusively by epidemic dissemination of mutant strain by the clonal spread. In recent years, ceftriaxone and azithromycin became the secondary drug of choice for treating typhoid due to the emergence of fluoroquinolone-resistant strains (Klemm et al., 2018). For animals and poultry, antibiotics are incorporated into feeds and water to control infections. The feeding rate varies and is subjected to under-dosing of drugs leading to an increase of selective pressure on Salmonella strains and inducing the development of resistant strains. Further, even for clinical Salmonellosis, these drugs such as fluoroquinolones are not widely used for treatment again unintentionally force Salmonella to acquire resistance (Dekker et al., 2019). In contrast, Salmonellosis in human are treated with fluoroquinolones, and the resistance is acquired by the mutation in the quinolone determining region (QRDR) of gyrA, gyrB, parC, and parE in chromosomal DNA. The plasmid-mediated quinolone resistance (PMQR) is coded by qnr determinants namely qnrA, qnrB, qnrC, qnrD, and qnrS and aac(6′)-ib-cr and quinolone extrusion qepA and oqxAB (Cui et al., 2019). Carbapenems and colistin are considered as the drug of choice for multidrug resistant Salmonella, however, carbapenemase and plasmid-mediated colistin resistance mcr-1- Salmonella emerged as a threat to these drug options (Liu et al., 2016). Recently developed multidrug resistant in Salmonella Typhimurium may cause life-threatening diseases and cannot be treated with common antibiotics. Salmonella entrica serovar Typhimurium DT104 showed multidrug resistance to ampicillin, chloramphenicol, florfenicol, streptomycin, sulfonamides, and tetracycline by the genes (43 kb) present in Salmonella genomic island I when compared with the Salmonella enterica serovar Typhimurium LT2 (Baucheron et al., 2004). Moreover, MDR strains of Salmonella have shown resistance to colistin, carbapenem, and tigecycline (Dawan and Ahn, 2020). New therapeutic strategies such as targeting the efflux pump and other resistance mechanisms are needed to fight against the developing multidrug resistance in Salmonella. Sequential antibiotic therapy and antibiotic rotation that reduces side effects have gained attention in past years. However, these approaches may favor the cross resistance to different groups of antibiotics (Vestergaard et al., 2016). The cross resistance evaluation in different strains of Salmonella Typhimurium under sequential antibiotic treatment showed that effective therapeutic strategies, appropriate selection, and antibiotic therapy helps in preventing cross resistance. (Dawan and Ahn, 2020).
4 Efflux system in Salmonella
The efflux pumps porins and the active role is documented. The multiple target gene mutation in Salmonella contributed largely to the multiple drug resistance by activation of efflux pumps. Numerous efflux pump encoding genes were identified in Salmonella sp. Ten or more genes in chromosomal and plasmid DNA encoding for efflux pump were identified (Nishino, 2016). The drug-resistant genes responsible for drug transportation and the functioning of efflux pumps have been documented. Most of these transporters have a wide substrate range to withstand a large set of antibiotics within the small genome of bacterial cells (Nishino et al., 2007). Additionally, it helps in quorum sensing regulated virulence (Szemeredi et al., 2020) biofilm formation, colonization, cell invasion, adhesion, and heavy metal resistance (Ebbensgaard et al., 2020). ArcAB is an important efflux transporter and OmpF porin is considered a major influx that selectively allows antibiotics inside the cells (Piddock, 2019). Efflux pump AcrAB-ToIc is overexpressed during the exposure to antimicrobial agents by using transcriptional activators such as RamA, MarA, SoxRS, and Rob. In the absence of RamA and presence of soxR transcriptional activators Salmonella can respond to the selective antibiotic pressure and develop mutation (Grimsey et al., 2020). This overexpression may lead to mutations showing multidrug resistance. However, Salmonella susceptible to antibiotics and chemical agents were reported to have defects or mutations in the ArcAb operon (Alav et al., 2020). The amino acid residue substation in the AcrB drug-binding pocket formed an alternate substrate binding specificity and increased the susceptibility to other antibiotics (Blair et al., 2015). The mutations in local repressor genes, global regulatory gene and transport gene promotor region, and upstream insertion elements in transporter genes are found responsible for the overexpression of efflux pumps (Piddock et al., 2010).
The drug efflux system in Salmonella consists of ten pumps (Fig. 2) belonging to four different exporter families. ToIc is the major outer membrane channel helping in the extrusion of toxic antibiotics from the cells. Several research has been done to identify the expression of these pumps and their physiological properties in multidrug resistant strains of Salmonella. There are some more proteins belonging major facilitator superfamily (MFS) that help in the extrusion of solutes and other toxic components from the cells in which the proper physiological properties are well studied (Kumar et al., 2020). Therefore, detailed research is needed to reveal the contribution of pumps in developing multidrug resistance.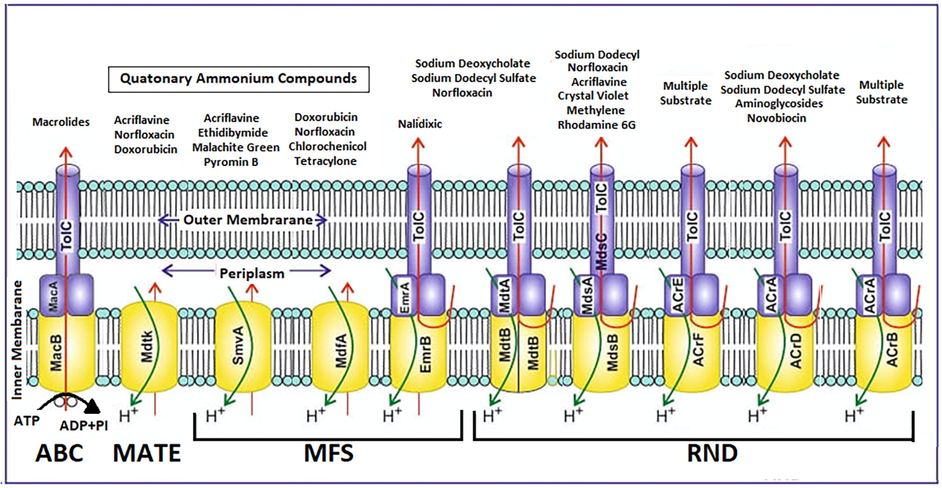
Schematic diagram describing representative structures of the main efflux pump systems of Salmonella including transporters, efflux pumps, and substrates. These transporters are the adenosine triphosphate (ATP)-binding cassette (ABC) superfamily, the multi-drug and toxic compound extrusion (MATE) family, the major facilitator superfamily (MFS) family, and the resistance nodulation-cell division (RND) family. All these transporters use electrochemical gradients to drive their transport processes, except the ABC transporters, which utilize ATP to drive substrate transport processes.
4.1 Multidrug and toxic compound exporters (MATE)
This is a secondary transport system that utilizes an electrochemical gradient of cations for drug transportation. MdtK is the MATE type efflux pump in Salmonella Typhimurium, and it is substrate-specific and pumps out the antimicrobial drug such as aminoglycosides, fluoroquinolones, cation drugs from the cells (Nishino et al., 2009). It can transport drugs with unrelated chemical structures and is called multidrug exporters (Kuroda and Tsuchiya, 2009). It has 12 transmembrane proteins and the protein structure remains similar to MSF, however, there are differences in the aminoacid sequences (Farhat et al., 2020). It utilizes sodium gradients to eject the toxic compounds from the cells.
4.2 ATP binding cassette (ABC)
ABC family are the transporters responsible for the uptake and efflux of components by the cells. Nucleotide-binding and transmembrane components of different or sometimes the same proteins form this domain. The permeases with six transmembrane regions are connected as dimers in the cytoplasmic membrane from the pores for transport (Nishino et al., 2009).
5 Resistance nodulation division proteins (RND)
In Salmonella RND proteins are located in the inner membrane and with the help of other proteins and outer membrane channel plays a major role in the drug efflux system with the tripartite system (AcrAB-ToIC). It transports substrates such as antibiotics, dyes, detergent and host metabolites. The substrates for AcrAB-ToIC are acriflavine and ethidium bromide is captured by AcrAB and extruded across the outer membrane through the ToIC channel. Even this RND helps in the virulence of Salmonella, and the virulence is affected if some components of AcrAB-ToIC are altered. The absence of RND protein components makes Salmonella susceptible to drugs and overexpression of these genes leads to multidrug resistance (Higgins et al., 2020). Therefore the studies on these proteins are gaining importance to fight the multidrug resistance and decrease the virulence of Salmonella (Blair and Piddock, 2009).
6 Major facilitator superfamily (MFS)
This comprises membrane proteins that are involved in symport, antiport, or uniport of various substrates and solutes such as sugars, ions, drugs, and metabolites by using proton motive force or electrochemical gradient. Further, the superfamily consists of two families DHA12 and DHA14 with 12 and 14 transmembrane proteins (Li and Nikaido, 2004). These transporters are considered a largest group among the exporter families found in bacteria. It transports H+/drug or solutes from the cell by using proton motive force (PMF) as an energy source (Andersen et al., 2015). EmrAB is an important pump in Salmonella for developing drug resistance. EmrA, is a membrane fusion protein that transports the substrates across the inner and outer membrane. This pump has been reported to develop resistance to triclosan, novobiocin, sodium deoxycholate, and nalidixic acid (Rensch et al., 2014). MdfA is another pump in the MFS family conferring resistance to tetracycline, norfloxacin, and doxorubicin (Yamasaki et al., 2016). This pump also helps in the alkaline adaptation by transporting Na+ or K+/H+ in addition to drug/H+ transport (Lewinson et al., 2004). Some genes responsible for resistance in Salmonella are encoded in the integrin sites of SGI-1. The floR (gene for chloramphenicol/florfenicol resistance), tetG (tetracycline resistance), and qnr (fluoroquinolone resistance) belong to the MF superfamily (Braibant et al., 2005). For tetracycline resistance TetA, TetB and TetG are responsible for Salmonella and belong to the family DHA12. This also confers tigecycline resistance (Akiyama et al., 2013). TetA gene is also considered a supplementary gene to the ramR that regulates the ramA responsible for multidrug resistance in Salmonella (Tawyabur et al., 2020).
7 Regulators modulating the resistance
Utilizing multiple transporters for multidrug resistance by the bacterial cell depends upon the regulation of the transporter expression. Multidrug transporter is expressed under a specific and rich transcriptional control. Multiple transcriptional regulators repress or activate the genes for controlling the expression of AcrAB-ToIC and MDR pumps in Salmonella (Nikaido et al., 2011). Cellular activity, growth factors, and environmental conditions may transform the expression of these regulators. A list of regulators is given in Table 2. Efflux pump inhibition methods such as dissipation of proton motive force, chemical inhibition, or deletion of efflux pump are activated by RamA, which increases the expression of acrAB (Lawler et al., 2013). Thereby cells may increase the efflux pump activity that compensates for low activity by using the expression of regulators to overcome the inhibitors (Marshall et al., 2020). Even for expression of AcrAB efflux pump multilevel regulations are needed, in which AcrR is essential with the help of other regulators such as SoxS, Rob, and MarA. Mutations in acrR overexpress the acrAB in Salmonella lead to multidrug resistance For transcriptional activators marA and soxS expression acrB, acrF and acrD are jointly regulated. Integration of IS1 or IS 10 elements in the acrEF operon in the upstream region increases the acrEF expression (Olliver et al., 2005). Earlier acrAB acrD and acrEF genes were known to regulate multidrug transport. Later positive regulation of multidrug efflux pump by mdtABC and arcD genes with the help of BaeR signal transduction system was identified (Nishino et al., 2007).
Transcriptional
regulatorRegulation
Reference
TetR
Tetracycline resistance
Miruka et al., 2011
AraC/Xyls
Regulates both transcriptional activators and repressors
Shaheen et al., 2020
PhoPQ
Signal transduction systems
Dalebroux and Miller, 2014
Mer-R
Mercury resistance
Brown et al., 2003
Mar-R regulator
Multiple drug resistance
Hünnefeld et al., 2019
LeuO
LysR-type regulators
Dillon et al., 2012
The resistance of Salmonella is regulated by transcriptional regulator families. For example TetR for tetracycline, AraC/Xyls that regulate both transcriptional activators and repressors, signal transduction systems (PhoPQ), mer-R for mercury resistance, Mar-R regulator for multiple drug resistance, LysR-type regulators (LeuO) and histone-like protein regulators involved in repression of new genes that are responsible for antibiotic resistance (Shaheen et al., 2020). Nonantibiotic compounds such as dephostatin help in reducing both virulence and Polymyxin resistance by disrupting the signaling of SsrA-SsrB and PmrB-PmrA two-component regulatory system and enhancing the susceptibility to colistin (Tsai et al., 2020). The EmrAB genes responsible for the functioning of a pump in the major facilitator family is regulated by EmrR (Andersen et al., 2015). The MFS efflux pump genes such as tetA, tetB, and tetG are controlled by the TetR (Colclough et al., 2019). The expression of AcrAB-ToIC MDR efflux pumps is regulated by multiple transcriptional regulators by repressing or activating the promotor of efflux pump genes based on environmental or cellular activity. The transcriptional activator RamA, which increases the acrAB expression is sensitive to inhibition of efflux activity by deletion of genes or using chemicals, or by indulging proton motive force (Nikaido et al., 2011). This may be the response to inhibitors to increase the action of the pump to pump the chemicals from the cells.
8 Virulence and efflux pumps
The efflux pumps present in bacteria are not only helping in extruding the drugs from the cells but also helps in maintaining the physiological activities of the cells including virulence (Blanco et al., 2016). In Salmonella efflux pumps are essential to overcome the lethal effects of bile salts and host defense mechanisms (Pasqua et al., 2019). Mutant strains of Salmonella lacking one or more efflux pumps are unable to adapt to the host environment with bile resistance (Urdaneta and Casadesús, 2018). In a recent study the genomes of African iNTS isolates of Salmonella enterica serovar Typhimurium, were analyzed and found the multilateral pump MacAB with ToIC, which is upregulated during macrophage infection. This also helps Salmonella to colonize in the gut and survive in presence of antimicrobial peptides. Earlier studies on the inactivation of acrB, acrAB, and acrEF resulted in attenuation of Salmonella Typhimurium (Blair et al., 2015). The compounds that are inhibiting the efflux pump not only favor an alternative approach to multidrug resistance treatment, but also opens helps in anti-virulence and anti-biofilm that offers clinical benefit.
9 Role of efflux pump in Salmonella in biofilm formation
Biofilms are also formed with the help of an efflux pump. It helps in transport of extra polymeric substances and quorum quenching molecules supportive to make biofilms. It regulates the genes responsible for biofilm formation and extrudes metabolic intermediates, harmful chemicals, and antibiotic molecules. Further, it stimulates the aggregation of cells in the formation of biofilms. Biofilm is one of the major factor contributing toward the tolerance and resistance to antibiotic resistance by directly extruding the antibiotics or indirectly forming the biofilm (Reza et al., 2019). Inhibition of the efflux system by using efflux pump inhibitors and interrupting the biofilm formation may reverse the antibiotic resistance in bacteria. It potentiates antibiotic efficiency. Salmonella forms biofilms on abiotic and biotic surfaces including gall stones and epithelial cells and causes reinfection. These biofilms formed by Salmonella on food products are one of the major causes of food borne illness. It is more difficult to treat these biofilms due to their high tolerance to antibiotics than the planktonic cells. The MIC and MBC values of the antibiotics become 100–1000 higher due to biofilm formation and endure in host making the treatment difficult. Biofilm formation are controlled by many factors including genes csqD, adrA, and bapA for matrix formation and adhesion (Gonzalez-Escobedo and Gunn, 2013). Efflux pumps in Salmonella plays an important role in formation of biofilm. The biofilm ability as decreased after the deletion of efflux pumps including acrD, acrEF, emrAB, macAB, mdfA, mdsABC, mdtBC, mdtK and tolC from Salmonella enterica serovars (Baugh et al., 2012). AcrD transports the regulatory molecules controlling the genes in biofilm formation. The RND acrD is also involved in transcriptomic variations according to the habitat associated with environmental sensing, stress tolerance, pathogenicity and metabolic pathways. MacAB-ToIC has been reported to involve in efflux of protoporphyrin, an intermediate biomolecule that chelates transition metals to form metallophorphyrins for various biological functions (Turlin et al., 2014). It confirms the functional role of efflux pumps in biofilm formation. MdsABC also helps in Salmonella to defend chemicals, to overcome oxidative stress and macrophage mediated killing that prevents biofilm formation. The over expression of MdsABC led to increase in 1-palmitoyl-2-stearoyl-phosphatidylserine (PSPS) that helps in host cell death and survival of Salmonella in phagocytic cells (Song et al., 2015). MATE superfamily MdtK extrudes antibiotics and some antimicrobial peptide components from PhoP/PhoQ system which possess antimicrobial activity. Dipeptides efflux function was also reported in Salmonella (Kapach et al., 2020). MdtK is 90% similar in both Salmonella and E.coli and found in all Enterobacteriaceae groups (Nishino et al., 2006). MdtK pump may play an important role in the efflux of dipeptides in cells of Salmonella by preventing the accumulation of antimicrobial dipeptides through quorum sensing. The formation of curli, the cell surface protein responsible for matrix formation in biofilm is also exaggerated in efflux mutants, where the transport of activators and regulators for csgB and csgD curli genes were affected (Barnhart and Chapman, 2006). The four families of efflux pumps reported in Salmonella are important in biofilm formation. However, RND family is predominant and molecules on quorum sensing in Salmonella were not reported.
10 Inhibitors of efflux pumps
Exploring the efflux pump inhibitors helps to overcome the problem of multidrug resistance in bacteria. The outer membrane (OM) permeability barrier with the help of an efflux pump is the non-specific protection mechanism that increases antibiotic resistance. Studies on components inhibiting the efflux mechanism can increase the antibiotic susceptibility thereby preventing the development of resistance to antibiotics (Table 3). Novel approaches for identifying the natural components and inhibitors of efflux pump allow the antibiotics to accumulate inside the bacterial cell for effective treatment (Farhat et al., 2020). Several efflux pump inhibitors have been reported to date and the mechanism of these inhibitors are not completely understood. Some molecules act as a competitive to the substrates and export it and some depolarize the plasma membrane making it susceptible to the antibiotic. Further, the efflux pump inhibitors restore the activity of the drug inside the cells and inhibit the growth of multidrug resistant strains. The compounds or molecules help in inhibiting the efflux pump regulate the overexpression of the efflux pump help the antibiotic attach to the target receptors interrupt the assembly of efflux pumps, competitive or non-competitive inhibition of antibiotic binding, hindering the pores of efflux pumps and inhibiting the energy source of efflux pump activity (Aparna et al., 2014). Heterocyclic nitrogen containing compounds that could act as EPI were tested in Salmonella Typhimurium with an overexpressed AcrAB-ToIC efflux pump. The strains in which efflux pump acrAB inactivated were hyper-susceptible to antibiotics such as ciprofloxacin chloramphenicol, and erythromycin. There was a 2–4-fold decrease in the MIC values in presence of trimethoprim. Theobromine and norepinephrine separately tested with ciprofloxacin also showed synergetic activity in Salmonella strains (Piddock et al., 2010).
Compound
Target efflux pump
Substrate
Reference
Compound (OU33858)
MacAB
Macrolids
Yamagishi et al., 2020
Trimethoprim
AcrAB-ToIC
Ciprofloxacin
Piddock et al., 2010
Cathinone
Carbonyl cyanide m chlorophenylhydrazone, norepinephrine and trimethoprim
AcrAB-ToIC
Ciprofloxacin
Rushdy et al., 2013
Tetraphenylphosphonium and ethidium cations
AcrAB-ToIC
Polymyxin B and Gramicidin D
Sutkuviene et al., 2013
Phe-Arg-β-naphtylamide
AcrAB-ToIC OqxAB
Tigecycline
Chen et al., 2017
Polyaminoisoprenyl derivatives
AcrAB-ToIC
Erythromycin
Doxycycline
Nalidixic acid
ChloramphenicolLieutaud et al., 2020
Hexadecanoic acid
Ethyl ester
Cyclopropane
1-(1-hydroxy-1-heptyl) 2-methylene-3- pentyl from Artemisia tournefortiana
AcrAB-ToIC
Ciprofloxacin
Khosravani et al., 2020
Chlorpromazine
AcrAB-ToIC
Chloramphenicol, tetracycline, nalidixic acid and triclosan
Bailey et al., 2008
Trans-cinnamaldehyde,
β- resorcyclic acid
carvacrol
thymol
eugenol
Ampicillin
Chloramphenicol
Streptomycin
Sulfamethoxazole
TetracyclineJohny et al., 2010
Pyranopyridine
AcrAB-ToIC
Fluoroquinolones,
Β-lactams
Chloramphenicol
Minocycline
Erythromycin
Linezolid(Aron and Opperman, 2016
11 Conclusion
Multidrug efflux pump in Salmonella helps in antibiotic resistance leading to untreatable bacterial infections. Understanding the role of efflux pumps their regulators and inhibitors may help in identifying novel target and control resistance in Salmonella. Identifying the efflux inhibitor molecules that can enhance the effect of antibiotics may also restore the efficacy of drugs to fight against multidrug-resistant strains of Salmonella. Modern technologies and molecular approaches may help to discover drug targets and novel molecules that can fight against the development of antibiotic resistance and treat Salmonella infections.
Acknowledgement
The author extends his appreciation to the Deanship of Research, Shaqra University, Kingdom of Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Acinetobacter baumannii efflux pumps and antibiotic resistance. Infect Drug Resist.. 2020;13:423-434.
- [Google Scholar]
- The tetA gene decreases tigecycline sensitivity of Salmonella enterica isolates. Int. J. Antimicrob. Agents. 2013;42(2):133-140.
- [Google Scholar]
- Multidrug efflux pumps from enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int. J. Environ. Res. Public Health.. 2015;12(2):1487-1547.
- [Google Scholar]
- Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation. PLoS ONE. 2014;9(7) e101840
- [Google Scholar]
- Optimization of a novel series of pyranopyridine RND efflux pump inhibitors. Curr. Opin. Microbiol.. 2016;33:1-6.
- [Google Scholar]
- Epidemiology of resistance to quinolones in Salmonella. Vet. Res.. 2001;32(3/4):285-290.
- [Google Scholar]
- RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob. Agents Chemother.. 2008;52(10):3604-3611.
- [Google Scholar]
- AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar typhimurium DT104. Antimicrob. Agents Chemother.. 2004;48(10):3729-3735.
- [Google Scholar]
- Loss of or inhibition of all multidrug resistance efflux pumps of Salmonella enterica serovar Typhimurium results in impaired ability to form a biofilm. J. Antimicrob. Chemother.. 2012;67(10):2409-2417.
- [Google Scholar]
- AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc. Natl. Acad. Sci. U.S.A.. 2015;112(11):3511-3516.
- [Google Scholar]
- Expression of homologous RND efflux pump genes is dependent upon AcrB expression: implications for efflux and virulence inhibitor design. J. Antimicrob. Chemother.. 2015;70(2):424-431.
- [Google Scholar]
- Structure, function and inhibition of RND efflux pumps in Gram-negative bacteria: an update. Curr. Opin. Microbiol.. 2009;12(5):512-519.
- [Google Scholar]
- Bacterial multidrug efflux pumps: Much more than antibiotic resistance determinants. Microorganisms. 2016;4(1):14.
- [Google Scholar]
- Structural and functional study of the phenicol-specific efflux pump floR belonging to the major facilitator superfamily. Antimicrob. Agents Chemother.. 2005;49(7):2965-2971.
- [Google Scholar]
- The MerR family of transcriptional regulators. FEMS Microbiol. Rev.. 2003;27(2–3):145-163.
- [Google Scholar]
- The AcrAB-TolC efflux system of Salmonella enterica serovar Typhimurium plays a role in pathogenesis. Cell. Microbiol.. 2006;8(5):847-856.
- [Google Scholar]
- Efflux pump overexpression contributes to tigecycline heteroresistance in Salmonella enterica serovar Typhimurium. Front. Cell. Infect. Microbiol.. 2017;7:37.
- [Google Scholar]
- TetR-family transcription factors in Gram-negative bacteria: Conservation, variation and implications for efflux-mediated antimicrobial resistance. BMC Genom.. 2019;20(1):731.
- [Google Scholar]
- Gallstones play a significant role in Salmonella spp. gallbladder colonization and carriage. Proc. Natl. Acad. Sci. (U.S.A).. 2010;107(9):4353-4358.
- [Google Scholar]
- Prevalence and characterization of fluoroquinolone resistant Salmonella isolated from an integrated broiler chicken supply chain. Front. Microbiol.. 2019;10:1865.
- [Google Scholar]
- Salmonellae PhoPQ regulation of the outer membrane to resist innate immunity. Curr. Opin. Microbiol.. 2014;17(1):106-113.
- [Google Scholar]
- Assessment of cross-resistance potential to serial antibiotic treatments in antibiotic-resistant Salmonella Typhimurium. Microb. Pathog.. 2020;148:104478.
- [Google Scholar]
- Fluoroquinolone-resistant Salmonella enterica, Campylobacter spp., and Arcobacter butzleri from local and imported poultry meat in Kumasi, Ghana. Foodborne Pathog. Dis.. 2019;16(5):352-358.
- [Google Scholar]
- LeuO is a global regulator of gene expression in Salmonella enterica serovar Typhimurium. Mol. Microbiol.. 2012;85(6):1072-1089.
- [Google Scholar]
- The role of efflux pumps in the transition from low-level to clinical antibiotic resistance. Antibiotics. 2020;9(12):855.
- [Google Scholar]
- Efflux pumps as interventions to control infection caused by drug-resistance bacteria. Drug Discov. Today. 2020;25(12):2307-2316.
- [Google Scholar]
- Constructing and deconstructing the bacterial cell wall. Protein Sci.. 2020;29(3):629-646.
- [Google Scholar]
- Identification of Salmonella enterica serovar typhimurium genes regulated during biofilm formation on cholesterol gallstone surfaces. Infect. Immun. 2013;81(10):3770-3780.
- [Google Scholar]
- Overexpression of RamA, which regulates production of the multidrug resistance efflux pump AcrAB-TolC, increases mutation rate and influences drug resistance phenotype. Antimicrob. Agents Chemother.. 2020;64(4)
- [Google Scholar]
- Molecular mechanisms of Salmonella pathogenesis. In Antimicrob. Agents Chemother.. 2000;13(5):519-522.
- [Google Scholar]
- Association of virulence and antibiotic resistance in salmonella—statistical and computational insights into a selected set of clinical isolates. Microorganisms. 2020;8(10):1-17.
- [CrossRef] [Google Scholar]
- The MarR-Type regulator MalR s involved in stress-responsive cell envelope remodeling in Corynebacterium glutamicum. Front. Microbiol.. 2019;10:1039.
- [Google Scholar]
- A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and adaptation and antimicrobial resistance including multidrug resistance. Vet. World. 2019;12(4):504-521.
- [Google Scholar]
- Effect of subinhibitory concentrations of plant-derived molecules in increasing the sensitivity of multidrug-resistant Salmonella enterica serovar typhimurium DT104 to antibiotics. Foodborne Pathog. Dis. 2010;7(10):1165-1170.
- [Google Scholar]
- Loss of the Periplasmic Chaperone Skp and Mutations in the efflux pump AcrAB-TolC play a role in acquired resistance to antimicrobial peptides in Salmonella typhimurium. Front. Microbiol.. 2020;11:189.
- [Google Scholar]
- Phytochemical composition and anti-efflux pump activity of hydroalcoholic, aqueous, and hexane extracts of Artemisia tournefortiana in ciprofloxacin-resistant strains of Salmonella enterica Serotype Enteritidis. In Iran J. Public Health. 2020;49(1)
- [Google Scholar]
- Emergence of an extensively drug-resistant Salmonella enterica serovar typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio. 2018;9(1)
- [Google Scholar]
- Multidrug resistance efflux pumps in Acinetobacter spp.: An update. Antimicrob. Agents Chemother 2021:AAC-00514.
- [Google Scholar]
- Functional and structural roles of the major facilitator superfamily bacterial multidrug efflux pumps. Microorganisms. 2020;8(2):266.
- [Google Scholar]
- Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta - Proteins and Proteomics. 2009;1794(5):763-768.
- [Google Scholar]
- Genetic inactivation of acrAB or inhibition of efflux induces expression of ramA. J. Antimicrob. Chemother.. 2013;68(7):1551-1557.
- [Google Scholar]
- Alkalitolerance: A biological function for a multidrug transporter in pH homeostasis. Proc. Natl. Acad. Sci. USA. 2004;101(39):14073-14078.
- [Google Scholar]
- New Polyaminoisoprenyl antibiotics enhancers against two multidrug-resistant gram-negative bacteria from Enterobacter and Salmonella species. J. Med. Chem.. 2020;63(18):10496-10508.
- [Google Scholar]
- Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: A microbiological and molecular biological study. Lancet Infect. Dis.. 2016;16(2):161-168.
- [Google Scholar]
- Salmonella pathogenicity island 1 (SPI-1) and its complex regulatory network. Front. Cell. Infect. Microbiol.. 2019;9:270.
- [Google Scholar]
- Salmonella pathogenicity islands: Big virulence in small packages. Microbes Infect.. 2000;2(2):145-156.
- [Google Scholar]
- Tetracycline efflux pump in different Salmonella enterica isolated from diarrhea patients in two rural health centers in Western Kenya. Arch. Clin. Infect. Dis.. 2011;6(1)
- [Google Scholar]
- Regulation of the AcrAB multidrug efflux pump in Salmonella enterica serovar Typhimurium in response to indole and paraquat. Microbiology. 2011;157(3):648-655.
- [Google Scholar]
- Nishino, K., 2016. Antimicrobial drug efflux pumps in Salmonella. In efflux-mediated antimicrobial resistance in bacteria Springer International Publishing. pp. 261–279.
- Virulence and drug resistance roles of multidrug efflux systems of Salmonella enterica serovar Typhimurium. Mol. Microbiol.. 2006;59(1):126-141.
- [Google Scholar]
- Regulation of multidrug efflux systems involved in multidrug and metal resistance of Salmonella enterica serovar Typhimurium. J. Bacteriol. Res.. 2007;189(24):9066-9075.
- [Google Scholar]
- Regulation and physiological function of multidrug efflux pumps in Escherichia coli and Salmonella. Biochim. Biophys. Acta - Proteins and Proteomics. 2009;1794(5):834-843.
- [Google Scholar]
- Overexpression of the multidrug efflux operon acrEF by insertional activation with IS1 or IS10 elements in Salmonella enterica serovar typhimurium DT204 acrB mutants selected with fluoroquinolones. Antimicrob. Agents Chemother.. 2005;49(1):289-301.
- [Google Scholar]
- Inhibitors of efflux pumps in Gram-negative bacteria. Trends. Mol. Med.. 2005;11(8):382-389.
- [Google Scholar]
- The MFS efflux pump EmrKY contributes to the survival of Shigella within macrophages. Sci. Rep.. 2019;9(1):1-11.
- [Google Scholar]
- Natural and synthetic compounds such as trimethoprim behave as inhibitors of efflux in Gram-negative bacteria. J. Antimicrob. Chemother.. 2010;65(6):1215-1223.
- [Google Scholar]
- The 2019 Garrod Lecture: MDR efflux in Gram-negative bacteria—how understanding resistance led to a new tool for drug discovery. J. Antimicrob. Chemother.. 2019;74(11):3128-3134.
- [Google Scholar]
- Salmonella enterica serovar Typhimurium multidrug efflux pumps EmrAB and AcrEF support the major efflux system AcrAB in decreased susceptibility to triclosan. Int. J. Antimicrob. Agents. 2014;44(2):179-180.
- [Google Scholar]
- Effectiveness of efflux pump inhibitors as biofilm disruptors and resistance breakers in Gram- negative (ESKAPEE) bacteria. In Antibiotics. 2019;8(4)
- [Google Scholar]
- Contribution of different mechanisms to the resistance to fluoroquinolones in clinical isolates of Salmonella enterica. Braz. J. Infect. Dis.. 2013;17(4):431-437.
- [Google Scholar]
- Transcriptional regulation of drug resistance mechanisms in Salmonella: where we stand and what we need to know. World J. Microbiol. Biotechnol.. 2020;36(6):85.
- [Google Scholar]
- MdsABC-mediated pathway for pathogenicity in Salmonella enterica serovar typhimurium. Infect. Immun.. 2015;83(11):4266-4276.
- [Google Scholar]
- Salmonella effector proteins and host-cell responses. Cell Mol. Life Sci.. 2011;68(22):3687-3697.
- [Google Scholar]
- Evaluation of the efficiency of synthesized efflux pump inhibitors on Salmonella enterica ser. Typhimurium cells. Chem. Biol. Drug Des.. 2013;82(4):438-445.
- [Google Scholar]
- Ketone- and cyano-selenoesters to overcome efflux pump, quorum-sensing, and biofilm-mediated resistance. Antibiotics. 2020;9(12):1-17.
- [Google Scholar]
- Isolation and characterization of multidrug-resistant Escherichia coli and Salmonella spp. from healthy and diseased turkeys. Antibiotics. 2020;9(11):770.
- [Google Scholar]
- Targeting two-component systems uncovers a small-molecule inhibitor of salmonella virulence. Cell Chem. Biol.. 2020;27(7):793-805.
- [Google Scholar]
- Protoporphyrin (PPIX) efflux by the MacAB-TolC pump in Escherichia coli. Microbiol. Open. 2014;3(6):849-859.
- [Google Scholar]
- Antibiotic combination therapy can select for broad-spectrum multidrug resistance in Pseudomonas aeruginosa. Int. J. Antimicrob. Agents. 2016;47(1):48-55.
- [Google Scholar]
- World Health Organization, 2017. WHO publishes list of bacteria for which new antibiotics are urgently needed. https://who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed.
- Investigation of a salmonellosis outbreak caused by multidrug resistant Salmonella typhimurium in China. Front. Microbiol.. 2020;11:801.
- [Google Scholar]
- An efflux inhibitor of the MacAB pump in Salmonella enterica serovar Typhimurium. Microbiol. Immunol.. 2020;64(3):182-188.
- [Google Scholar]
- Phenotype microarray analysis of the drug efflux systems in Salmonella enterica serovar Typhimurium. J. Infect. Chemother.. 2016;22(11):780-784.
- [Google Scholar]
- Salmonella infection in chronic inflammation and gastrointestinal cancer. Dis.. 2019;7(1):28.
- [Google Scholar]







