Translate this page into:
An overview of the molecular and biochemical components of seed dormancy and germination in Brassica napus
⁎Corresponding author. sisohn@korea.kr (Soo-In Sohn)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
Seed dormancy and germination determines the ecological adaptation and sustainability of seed plants. Intensive research efforts have identified the role of several factors in seed dormancy and germination. The major factors are phytohormones, dormancy-specific genes, chromatin factors, developmental and/or structural components of seeds, and exogenous environmental cues, which have been well studied in model plants like Arabidopsis and rice. In general, Brassica napus L. (canola) cultivars and its interspecific hybrids show seed heterogeneity and varying degrees of dormancy, and germination potentials. However, detailed research on understanding the molecular mechanisms of seed dormancy and germination in B. napus is limited as compared to othe model crops like Arabidopsis. Identification of seed dormancy and germination associated QTLs in concert with environmental factors is essential for better management of seed longevity and vigor in economically important oil-seed crops like B. napus. In addition, both maternal and paternal genomes and/or their imprinted genes are often known to influence dormancy and subsequently lead to premature germination in some interspecific hybrids. Thus, in this review we provide an overview on plausible role of genetic, physiological, and environmental factors governing seed dormancy, germination and premature germination traits in B. napus.
Keywords
Phytohormones
Seed dormancy
Brassica napus
Seed germination
Heat stress
Vivipary
1 Introduction
Oilseed rape (Brassica napus L.), also known as canola, is a relatively recent domesticate and the second-largest oilseed crop globally, trailing only soybean (Sohn et al., 2016; Kourani et al., 2022). The eco-physiological adaptation to temperate climates has solidified canola’s position as the dominant oil crop in Europe (Namazkar et al., 2016). However, B. napus is cultivated worldwide for its high-quality oil, vegetables, fodder, and biodiesel production (Raboanatahiry et al., 2021; Sohn et al., 2022a) thus indicating the economic importance. Seed dormancy, a multifaceted adaptive trait, serves as a critical persistence strategy for numerous temperate annual weeds (Baskin and Baskin 1998). Both seed dormancy and germination are inter-related and play crucial roles in seed quality or for its chemical compositions, seedling establishment/eco-physiological adaptation, thereby protecting crop yield (Shu et al., 2015). Research has significantly advanced our understanding of dormancy expression in several species, revealing its modulation by various factors like genotype, progeny placement on the mother plant, and environmental influences (Gutterman 1981; Nimer et al., 1983). Notably, dormancy can be further shaped by post-dissemination environmental conditions (Hazebroek and Metzger, 1990). Based on the maturation stage, B. napus exhibits two types of dormancy. Primary dormancy, characterized by high abscisic acid (ABA) levels, prevents premature germination within the pod and persists from early seed development to seed maturation (Sano et al., 2021; Brown et al., 2022). Secondary dormancy, induced by unfavorable germination conditions after seed dispersal, allows mature seeds to persist in the soil for extended periods (Buijs 2020). B. napus, classified as a non-deep dormant species, exhibits inhibited radicle protrusion within the embryo under conditions unfavorable for germination, such as continuous darkness, freezing temperatures, or low moisture (Pekrun et al., 1997). Brief periods of imbibition under cold stratification or during ripening typically suffice to break seed dormancy in this species (Baskin and Baskin 1998; Brown et al., 2022). Non-deep physiological dormancy presents a dynamic scenario where B. napus seeds can transition between completely dormant, non-dormant, and conditionally dormant states (Sohn et al., 2021; Brown et al., 2022). Conditional dormancy can be broken within constrained environmental settings, enabling seed bank populations to proliferate (Brown et al., 2022). In the non-dormant state, germination is permitted across a broader range of conditions (Baskin and Baskin 1998).
On the other hand, preharvest pod shattering and harvest-related disturbances make B. napus susceptible to seed dispersal (Sohn et al., 2021; Brown et al., 2022). Up to 10,000 seeds/m2 can fall onto the soil surface after harvest (Sohn et al., 2021). While early germination of dispersed seeds allows for control through post-harvest tillage, unfavorable conditions (osmotic stress, oxygen deficiency) in darkness can trigger secondary dormancy and enable seed persistence for years (Sohn et al., 2021; Brown et al., 2022). In fact, a study by Beckie and Warwick (2010) revealed that persistent or dormant seeds can remain viable in the soil for up to 7 years after entering this cycle, creating weed control and contamination issues in subsequent crops. Upon encountering optimal conditions, these dormant seeds can germinate and emerge as volunteer plants, subsequently impacting the yield and quality of newly planted B. napus through cross-pollination (Brown et al., 2022). Various strategies have been investigated for mitigating the persistence of dormant canola seeds in the soil, which can subsequently lead to volunteer oilseed rape emergence in future crops (Schwabe et al., 2019). These strategies, tested in both laboratory and field experiments, include seed coating with KNO3, micronutrients, or gibberellic acid (GA), and pre-harvest spraying of urea ammonium nitrate (UAN) or GA on oilseed rape plants (Schwabe et al., 2019). Interestingly, seed coating treatments were found to reduce secondary dormancy, but paradoxically, they also increased seed persistence in the soil by up to 99 %. Among the coating materials, GA proved most effective in reducing dormancy. However, directly harvested B. napus seeds are generally considered non-dormant and do not require light for germination (Baskin and Baskin 1998; Soltani et al., 2018). Nevertheless, seeds possessing low dormancy or lacking dormancy can lead to preharvest sprouting, which poses significant challenges for cereal crop quality and production (Tuan et al., 2018). Therefore, it is evident that dormancy levels are key trait linked to seedling growth and thereby the yield potential of crop plants.
2 Molecular basis of ABA and GA-dependent signal transduction pathway in seed dormancy and germination
Seed germination in canola is a crucial determinant of crop success, as seedlings must rapidly establish self-sufficiency for survival (Iqbal et al., 2022). This process initiates with water imbibition by the quiescent seed and culminates in the expansion of the embryonic axis within the seed coat (Boter et al., 2019). The well-characterized model plant Arabidopsis thaliana shares over 86 % of its protein-coding regions with B. napus, making it a valuable tool for studying seed germination in the latter species (Cavell et al., 1998). This genetic and physiological similarity has informed much research, directing investigations into B. napus seed germination responses (Zhang et al., 2004). Notably, hormones like gibberellic acid (GA) and abscisic acid (ABA) exhibit antagonistic effects on seed germination, highlighting the complex interplay between internal and external factors (Shu et al., 2018). Despite these shared mechanisms, significant differences exist between the two species in terms of seed size, nutrient content, and dormancy characteristics. These findings emphasize the need for a comprehensive framework encompassing both shared and species-specific regulatory systems governing seed germination in B. napus (Finch-Savage and Footitt 2017; Boter et al., 2019). The intricate interplay of exogenous factors, such as light, temperature, and soil moisture, with endogenous factors like plant hormones (GA, ABA, ethylene, and auxin) and endosperm breakdown, underscores the complexity of the seed germination process (Carrera-Castaño et al., 2020) (Fig. 1). Therefore, a deeper understanding of these interlinked internal and external influences is crucial for optimizing seed germination potential and ultimately, crop yields.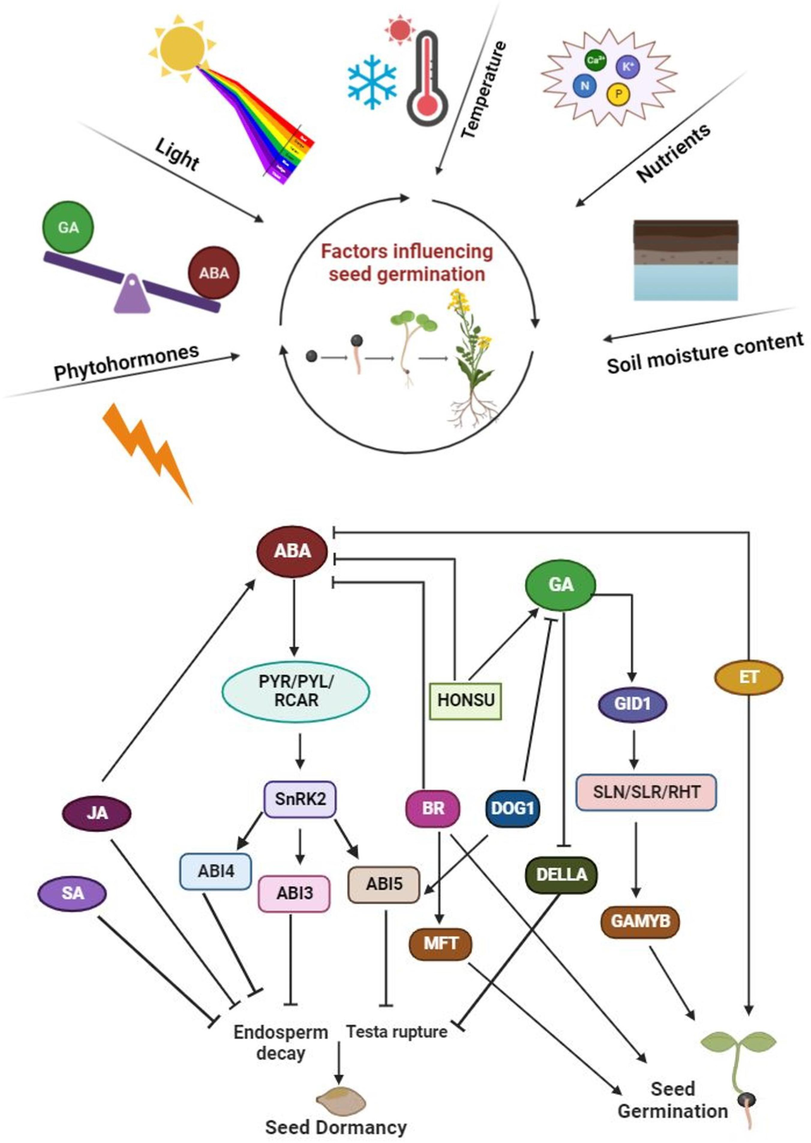
The exogenous and endogenous driving factors of seed germination. a) The external factors influencing seed germination, which are the priming sources for the hormonal gene regulations. Seed germination is affected by different factors such as soil moisture content, nutrients, temperature, light and hormone contents and expression of their respective genes. b) A pathway of genes promoted by different phytohormones for seed germination and dormancy. Repressor of GA biosynthesis, delay of germination 1 (DOG1), activates ABI3 and ABI5 which prevent the testa rupture thereby lead to the dormancy.
The delicate balance between two prominent and antagonistic phytohormones, such as GA and ABA, primarily dictates seed dormancy and germination (Zhou et al., 2015). While ABA suppresses germination by inhibiting radicle elongation, cell wall loosening, and the mobilization of stored proteins required for seedling establishment, GA counteracts these effects by promoting testa degradation and facilitating radicle emergence (Dekkers et al., 2016). The necessity of GA for seed germination is influenced by the ABA concentration within the seeds, as evidenced by the fact that GA-deficient mutants are able to germinate even when ABA is absent. High-dormancy genotypes exhibit greater ABA sensitivity, while germination in low-dormancy genotypes can occur even within the seed due to their inherent ABA insensitivity (Gulden et al., 2004). The concentration of ABA in seeds is regulated not only by the control of ABA biosynthesis but also by its conversion into phaseic acid, which results in ABA inactivation. This conversion is facilitated by ABA-8′ hydroxylases, which are P450 cytochromes encoded by the genes CYP707A1 and CYP707A2 (Kushiro et al., 2004). Several key regulatory proteins governing seed germination in Arabidopsis have been identified by integrating data from mutant studies and omics approaches (Bassel et al., 2011). For successful seed germination, downregulation of ABA signaling is crucial. This process involves the endoplasmic reticulum (ER)-localized bZIP17 protein translocating to the Golgi apparatus via membrane-anchored transcription factor peptidases like S1P (Site-1 Protease) and S2P. Subsequently, bZIP17 reaches the nucleus where it activates the transcription of anti-ABA signaling regulators (Zhou et al., 2015). The main pathway of ABA signaling relies on the PYR/PYL/RCAR-PP2C-SnRKs cascade (Tuan et al., 2018). In the absence of ABA, seed dormancy promoting SnRK2 proteins are inhibited by PP2Cs through dephosphorylation of their kinase activation loop (Ng et al., 2014). However, ABA binding to its receptors, PYR/PYL/RCAR, forms a complex with PP2C, hindering its phosphatase activity and thereby activating SnRK2s (Ng et al., 2014). These activated SnRK2s then stimulate the transcription of ABA-responsive genes through the activation of ABRE-binding protein/ABRE binding factor (AREB/ABF) transcription factors. Among AREB/ABFs, ABA insensitive 3, 4, and 5 (ABI3, ABI4, and ABI5) have been documented to trigger ABA-responsive gene expression, thus controlling seed dormancy (Nambara, and Marion-Poll, 2005). The ABI3 gene, in combination with the seed maturation genes LEAFY COTYLEDON 1 and 2 (LEC1/2) and FUSCA3 (FUS3), makes up the LAFL regulatory network, consisting of LEC1, ABI3, FUS3, and LEC2. ABI3 directly targets several genes, including ABI5, DELLAs, and the bHLH transcription factor PIL5 (PHYTOCHROME INTERACTING FACTOR-LIKE 5) (Tian et al., 2020). ABI 3, ABI5, and DELLAs collaboratively activate SOMNUS, which encodes a C3H zinc finger protein that regulates genes responsible for ABA synthesis and GA catabolism in imbibed seeds, leading to the suppression of germination (Lim et al., 2013). The ABI4 gene encodes a transcription factor of the APETALA2/ethylene response factor family TF that influences various processes by modulating gene expression, including that of ABA degradation genes such as CYP707A1 and CYP707A2 (Chandrasekaran et al., 2020). ABI5 encodes a bZIP transcription factor that binds to ABA Responsive Elements in promoters and interacts with ABI3 through its B1 domain. Additionally, the protein interacts with ABA binding proteins (AFPs), facilitating further protein interactions (Skubacz et al., 2016). In Arabidopsis, AFP2 acts as a negative regulator of primary seed dormancy by inhibiting the expression of DELAY OF GERMINATION 1 (DOG1), a key regulator of seed dormancy. The TF WRKY36 can interact with AFP2 to co-repress TOPLESS RELATED PROTEIN 2 and epigenetically silence DOG1 by decreasing histone acetylation (Deng et al., 2023). Finally, ABA signaling is terminated by the binding of PP2C proteins, namely ABI1 and ABI2, to the ABA receptors (Cutler et al., 2010). The dominant negative mutants abi1-1 and abi2-2 exhibit reduced seed dormancy due to impaired interaction between their mutated PP2C proteins and ABA receptors, confirming the negative role of ABI1 and ABI2 in this process (Zhou et al., 2015). Similarly, the PP2C gene, RD05 reportedly promotes seed germination in Arabidopsis by suppressing ABA signaling (Xiang et al., 2014). Another PP2C protein, HONSU, positively regulates GA signaling while downregulating ABA, thereby favoring seed germination (Kim et al., 2013). Table 1 summarizes key mutants studied in relation to seed dormancy and germination.
S. No
Gene name
Name of the protein encoded
Type of study
Dormancy level
Effect/type of interaction with ABA/GA signaling
References
1.
ABI4
ABA-INSENSITIVE 4
Mutant
Reduced
Positively regulates ABA by inhibiting the gene expression of ABA catabolic genes viz., CYP707A1 and CYP707A2
Shu et al., 2013
2.
ABI4 and ABI5
ABA-INSENSITIVE 4 and 5
Mutant
Reduced
Positively regulates ABA signaling and exhibit similar phenotypes with ABI3 mutants
Finkelstein 1994
3.
NCED
9-cis-epoxycarotenoid dioxygenase
Mutant
Reduced
Positively regulates ABA biosynthesis and increases ABA levels in response to water deficit
Frey et al., 2012; Xu and Cai, 2017
4.
GID1, GID2, and GID3
GA INSENSITIVE
DWARFMutant
Enhanced
Inability of GA to destroy DELLA proteins
Willige et al., 2007
5.
CYP707A2
Cytochrome P450
Mutant
Enhanced
ABA catabolic gene that reduces the levels of bioactive ABA
Matakiadis et al., 2009
6.
WRKY41
WRKY41 transcription factor
Mutant
Reduced
Directly regulates the transcription of ABI3
Ding et al., 2014
7.
DOG1
DELAY OF GERMINATION 1
Mutant
Reduced
Controls seed dormancy by temperature-dependent alteration of the GA metabolism
Graeber et al., 2014
8.
SIZ1
SUMO (small ubiquitin-related modifier) E3 ligase
Mutant
Enhanced
Regulates ABA signalling by inducing the expression of ABI5-dependent signaling genes such as RD29A, Rd29B, AtEm6, RAB18 and ADH1
Miura et al., 2009
9.
NF-YC3, NF-YC4 and NF-YC9
NUCLEAR FACTOR Y C
Mutant
Reduced
Alters ABA and GA signaling
Liu et al., 2016
10.
HONSU
Protein phosphatase 2C (PP2C)
Mutant
Enhanced
A PP2C protein that inhibits ABA signaling
Kim et al., 2013
11.
S2P
Site-2 protease
Mutant
Enhanced
Antagonizes ABA signaling by controlling the activation of the membrane-associated transcription factor bZIP17
Zhou et al., 2015
12.
RDO5
Reduced dormancy 5 (Protein phosphatase 2C)
Mutant
Reduced
Positively regulates seed dormancy without influencing ABA or DOG1 but requires expression of the APUM9 and APUM11 genes that encode RNA binding proteins
Xiang et al., 2014
13.
PIL5
PHYTOCHROME INTERACTING FACTOR 3-LIKE 5
Overexpression
Enhanced
Inhibits seed germination by activating GA catabolic gene (GA2ox2) and repressing GA biosynthesis genes GA3ox1 and GA3ox2
Chen et al., 2008
14.
DAG1
DOF AFFECTING
GERMINATION 1Mutant
Enhanced
Negatively regulates GA biosynthesis and acts downstream of PIL5
Oh et al., 2007
15.
CHO1
CHOTTO1, a putative double APETALA2 repeat transcription factor
Mutant
Reduced
Acts downstream of ABA to inhibit GA biosynthesis during seed germination
Gabriele et al., 2010
16.
DDF1
DWARF AND DELAYED LOWERING 1
Overexpression
Enhanced
Decreases the levels of bioactive GA
Yano et al., 2009
17.
RAF10/RAF11
MAP Kinase Kinase Kinases
Mutant
Reduced
Positively regulates the transcription of ABI3, ABI5 and other ABA related genes
Magome et al., 2004
18.
KYP/SUVH4
Histone methyltransferase
Mutant
Enhanced
Negatively regulates ABA signaling by repressing the transcription of DOG1 and ABI3
Lee et al., 2015
19.
MYB96
MYB DOMAIN PROTEIN 96 (transcription factor)
Mutant
Enhanced
Positively regulates the transcription of ABI4 and some ABA biogenesis genes
Magome et al., 2004
20.
FHY3
FAR1
FAR-RED ELONGATED HYPOCOTYL3, FAR-RED IMPAIRED RESPONSE1
Mutant
Reduced
Positive regulator of ABA signaling by directly activating the transcription of ABI5
Zheng et al., 2012
21.
CSN1
CSN5A
COP9 Signalosome
Mutant
Enhanced
Represses the transcription of RGL2 (repressor of ga1-3 like 2) and ABI5
Tang et al., 2013
22.
HY5
bZIP transcription factor
Mutant
Reduced
Directly induces the transcription of ABI5
Jin et al., 2018
23.
ELF3, PKL, LUX
EARLY FLOWERING 3, PICKLE, LUX ARRHYTHMO
Mutant
Enhanced
Directly represses the transcription of DOG1
Zha et al., 2020
In B. napus, severe hormonal imbalances caused by crosstalk between ABA and auxin, along with downregulation of negative regulators Bna.ARF10 and Bna.GH3.5, have been linked to disrupted seed germination and development (Nguyen et al., 2016). Interestingly, overexpression of the fatty acid desaturase 2 (FAD2) gene promotes hypocotyl elongation and seed germination, suggesting its potential as a downstream component of both GA and light signaling pathways, influencing various physiological traits (Wang et al., 2010). GDSL lipases, which are enzymes involved in seed development, lipid metabolism, stress response, and pathogen defense, also play a role in seed germination. Overexpression of BnGDSL in B. napus increased germination rate while decreasing oil content, highlighting its role in regulating both processes via transcriptional control (Ding et al., 2019). Recent studies have shown that the expression of Ca2+-dependent RSH (CRSH) proteins during B. napus seed maturation leads to the accumulation of alarmones that inhibit plastid and nuclear DNA replication, ultimately preventing seed germination (Turkan et al., 2023). Desiccation plays a crucial role in regulating seed dormancy. During this stage, the expression of CRSH proteins remains low, creating a permissive environment for germination (Turkan et al., 2023). In contrast, seed maturation is characterized by high CRSH levels, leading to the accumulation of alarmones that inhibit DNA replication and prevent germination.
Gibberellic acid (GA) reigns as the principal phytohormone governing seed germination. Its biological activity, however, hinges on a delicate balance between its biosynthesis and inactivation (Vishal and Kumar, 2018). GA20ox and GA3ox enzymes catalyze reactions for GA production, while GA2ox controls its breakdown. Through the activation of GA-responsive genes, it induces cell-wall remodeling enzymes, facilitating embryo expansion and breaching the seed coat. The GA signaling cascade commences with the binding of bioactive GA to its receptor, GIBBERELLIN INSENSITIVE DWARF1 (GID1). This interaction triggers the degradation of DELLA proteins, potent repressors of seed germination via GA-GID1-DELLA complex formation. Further, F-box protein, SCFSLY1/GID2 E3 ubiquitin ligase, interacts with the complex GA-GID1-DELLA, causing DELLA to degrade through the ubiquitin- 26S proteasome pathway (Murase et al., 2008). Following DELLA degradation, GAMYB, a downstream effector of GA signaling, becomes activated. This crucial protein orchestrates numerous downstream processes vital for germination (Ravindran et al., 2017). Interestingly, GAMYB also interacts with GATA family transcription factors, which positively regulate seed dormancy. Additionally, it is negatively regulated by KINASE ASSOCIATED WITH GAMYB1 (KGM), leading to the suppression of hydrolase genes in the aleurone layer, further contributing to dormancy control (Woodger et al., 2003). The pivotal role of GA in germination is underscored by gibberellic acid-deficient mutants like ga1 and ga2. These mutants exhibit profound dormancy and can only germinate in the presence of externally applied GA (Zhang et al., 2020). DELLA proteins, critical antagonists in GA signaling, play a pivotal role in seed dormancy. Notably, mutations in two key DELLA genes, RGA-LIKE2 (RGL2) and SPINDLY (SPY), can bypass the non-germination phenotype of the ga1 mutant, demonstrating their inhibitory effect on germination (Liu et al., 2013). Germination is influenced not solely by the GA response but also by the combined actions of GA and BR (brassinosteroids) signaling. In the BR signaling pathway, two essential basic helix-loop-helix transcription factors, BRASSINOSTEROID ENHANCED EXPRESSION 2 (BEE2) and BEE2 interacting with INCREASED LEAF INCLINATION 1 BINDING bHLH1 (HBI1) play a crucial role in controlling the process of endosperm rupture (Zhong et al., 2021). Moreover, BRI1-EMS-SUPPRESSOR 1 (BES1), which influences BR signaling, has been demonstrated to physically interact with ABI5, reducing its ability to bind to DNA. This interaction diminishes the ABA-mediated inhibition of seed germination by decreasing the expression of ABI5 target genes. Conversely, BIN2, an inhibitor of BR signaling, enhances ABA responses (Zhao et al., 2019).
Reports on the role of auxin in seed dormancy have also emerged, although the precise mechanisms remain unclear. Auxin's role in seed germination varies with concentration, having both stimulatory and inhibitory effects. High levels of auxin have been found to promote seed dormancy through the activation of ABI3, mediated by the AUXIN RESPONSE FACTOR 10 transcription factor and IAA8 (Liu et al., 2013; Hussain et al., 2020). This process results in increased ABI3 expression, although it does not involve direct binding to the ABI3 promoter. Studies on winter-type B. napus genotypes demonstrated that upregulation of auxin biosynthesis pathway genes and exogenous application of indole-3-acetic acid (IAA) increased the seed dormancy (Fei et al., 2009; Lui et al., 2020). Further analysis examining the upstream gene signaling pathways resulting in hormonal modifications may delineate the significant insights into the initiation and regulation of dormancy and germination (Zhong et al. 2021). Ethylene (ET) functions as a crucial antagonist of ABA, promoting seed germination and breaking dormancy. A mutant with a loss-of-function in the ethylene receptor ETHYLENE RESPONSE FACTOR 1 (ETR1), known as reduced dormancy 3, exhibited reduced seed dormancy. ETR1 normally represses ERF12, which forms a repressor complex with TOPLESS that binds to the DOG1 promoter, reducing DOG1 expression and thus seed dormancy (Li et al. 2019). Recently, Guo et al. (2023) reported that EIN2, a key player in ethylene signaling, can inhibit a histone H3 acetylase encoded by HOOKLESS 1 (HLS1) independently of the standard ethylene pathway, preventing the upregulation of ABA signaling genes ABI5 and ABI3, and thus promoting germination.
Mutations in the negative ET regulator Ctr1 (Constitutive Triple Response 1) lead to premature or accelerated germination, underscoring its role in suppressing ET signaling (Liu et al., 2013). Conversely, alterations in positive ET signaling components can induce extreme dormancy. This emphasizes the delicate balance between ET and ABA pathways in regulating seed fate. Secondary seed dormancy in B. napus is partly attributed to the ABA-dependent upregulation of the MFT (MOTHER OF FT and TFL1) gene. Interestingly, brassinosteroids (BRs) counteract ABA's dormancy-promoting effects partly via MFT regulation, creating an anti-ABA signaling feedback loop. This suggests a complex interplay between MFT, ABA, and BRs in controlling seed dormancy dynamics in B. napus (Holdsworth et al., 2008; Lei et al., 2020). The identification of DOG family genes has illuminated the genetic control of dormancy in various plant species (Greaber et al., 2014). In Arabidopsis, the DOG1 gene holds primary responsibility for both primary and secondary seed dormancy (Alonso-Blanco et al., 2003). Interestingly, DOG1 expression exhibits a dynamic pattern, peaking during seed maturation and reaching its minimum during ripening (Greaber et al., 2014). This regulation is further influenced by temperature sensitivity and enhanced ABA sensitivity. Higher levels of DOG1 protein accumulation, encoded by the gene, directly impede seed germination, and conversely, lower levels facilitate it (Greaber et al., 2014). Three copies of DOG1 genes were observed in B. napus (Née et al., 2015), and their transcript and protein expression were elevated after the induction of secondary dormancy, suggesting functional similarity between Arabidopsis and B. napus (Née et al., 2015). Two other genes responsible for inducing dormancy in seeds (the MFT gene) and for regulating germination (TFL1) have been identified in wheat and Arabidopsis (Lui et al., 2020). Genes homologous to MFT and TFL1 have been identified in B. napus and referred to as BnaMFTs. Induction of secondary dormancy using polyethylene glycol resulted in 3.5- fold increased transcripts of BnaMFTs (Lui et al., 2020).
3 Imprinted genes modulate seed dormancy and germination through regulating seed development
Among the different genetic factors, the expression of maternally imprinted genes and/or paternally imprinted genes in endosperm of developing seeds play a role in seed dormancy and germination traits. Interestingly, genomic imprinting reveals high plasticity within and between species indicating the biological significance of imprinted genes rarely conserved among closely related species. There were efforts to identify genomic imprinting in B. napus, which results in parent-of-origin specific differential expression of maternally and paternally inherited alleles (Feng et al., 2009). Recently, 297 imprinted genes, including 283 maternal expressed genes (MEGs) and 14 paternal expressed genes (PEGs) were identified in endosperm at 20 and 25 days after pollination (DAP) through transcriptome analysis (Rong et al., 2021). Interestingly, the imprinted genes were found to be enriched with transposable elements which might enable rapid molecular evolution and preferential expression in endosperms (Yoshida et al., 2018). The clustering of imprinted genes indicates their genome-wide distribution in rapeseed (Liu et al., 2018). Moreover, 11 genes comprised of MEGs (7) and PEGs (4) were observed with differentially methylated regions. Most imprinting in plants occurs in endosperm, therefore, it can be reasoned that endosperm can influence the embryogenesis and seed maturation, thereby influencing the seed dormancy as well as germination efficiencies. Seed coat and its chemical constituents (e.g. flavonoids) known to alter the dormancy levels and germination process. A study comparing the permeability of yellow seed-coated (low in flavonoids and thinner seed coat) and black seed coated (high in flavonoids) rape seed cultivars revealed substantial difference between seed coat of YS and BS, which could affect their germination status (Xuan et al., 2015). Another study dealing with epigenetic regulation of the yellow and black seeded characteristics of B. napus identified extensive methylation changes, responsible for seed-coat variation (Wang et al., 2016). In particular, 10 % of demethylation and 5 % of hypermethylation were detected in yellow rapeseeds compared with black rapeseeds. In fact, two and 13 unique flavonol derivatives were detected in black seeded and yellow seeded rapeseeds. The transcript expression analyses further confirmed that expression of common phenylpropanoid biosynthetic genes (BnPAL and BnC4H), common flavonoid biosynthetic genes (BnTT4 and BnTT6), anthocyanin and proanthocyandin specific genes (BnTT3 and BnTT18), proanthocyandin specific genes (BnTT12, BnTT10, and BnUGT2) and three transcription factor genes (BnTTG1, BnTTG2, and BnTT8) that function in the flavonoid biosynthetic pathway were significantly different between yellow and black seeds (Qu et al., 2013). Altogether, it is clear that the inherent dormancy and germination potential constituted by genomic elements of seeds, in concert with physiological and agronomic factors, determine the seedling adaptability and sustainability.
4 Environmental signals transduced through phytohormones determine seed dormancy levels and their germination efficiencies
Environmental factors like sunlight, temperature, nutrients, and soil water content profoundly influence seed germination by modulating the crucial signaling and metabolic pathways of GA and ABA (Holdsworth et al., 2008). Extensive research in B. napus has identified a network of regulatory genes involved in various stages of germination, encompassing protein biosynthesis, degradation of storage reserves, water transport, cell elongation, oxidative stress response, the ubiquitin system, and the secretory pathway (Ge et al., 2013). Aquaporins (AQPs) play a critical role in water movement across cell membranes during seed imbibition and early embryo growth (Wang et al., 2014). Supporting this, studies have shown that ten AQP genes upregulated during germination and in young B. napus seedlings (Barrero et al., 2010). Light acts as a crucial regulator of numerous plant processes, including photomorphogenesis, phototropism, and flowering, and it also serves as a key determinant of seed germination and dormancy (Lymperopoulos et al., 2018; Saleem et al., 2020). Cryptochromes (CRYs) and phototropins act as photosensory receptors sensitive to blue light (320–500 nm), which has been linked to the inhibition of seed germination (Fig. 2) (Farooq et al., 2022). Through CRY1 (cryptochrome circadian regulator 1), the blue light receptor, blue light stimulates the expression of NCED1 (9-CIS-EPOXYCAROTENOID DIOXYGENASE 1), leading to elevated ABA levels and ultimately inhibiting seed germination (Tuan et al. 2019). Similar findings have been observed in wheat, where blue light upregulates ABA biosynthesis genes and downregulates ABA catabolism genes, preventing germination (Saleem et al., 2020; Yang et al., 2020). In contrast to blue light's inhibitory effect, the initial stages of seed germination are often triggered by phytochromes like PhyB. Furthermore, PhyA mediates the very low fluence response (VLFR) to UV-A-FR light spectra and the R/FR high irradiance response (R/FR-HIR) to accelerate germination when active PhyB is absent (De Wit et al., 2016). Red light, in contrast to far-red light which can exhibit negative effects, has been shown to promote seed germination in various studies (Wang et al., 2012).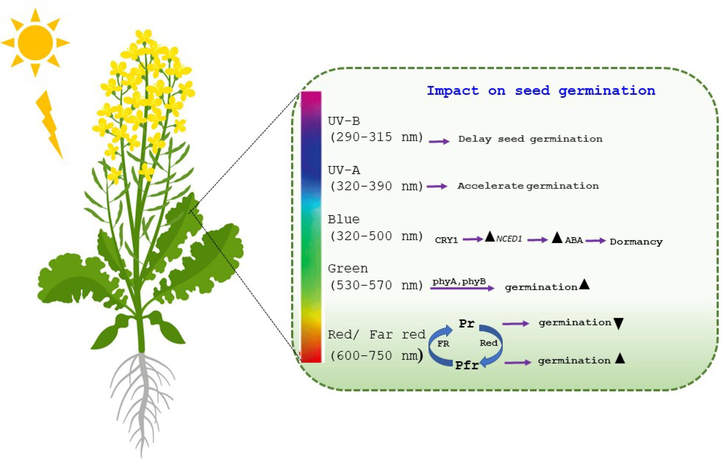
Schematic representation of potential impact of light at different wavelengths during seed germination sourced from literature in different crops. Pr and Pfr represent inactive and active phytochromes.
Temperature deviations from the optimal range (15–30 °C) can drastically impede seed germination through various mechanisms (Carruggio et al., 2021). Excessively high or low temperatures can suppress seed viability, induce thermo-inhibition, or trigger thermo-dormancy, effectively halting the germination process. High temperatures pose a particular threat to seed development by promoting the detrimental reactive oxygen species (ROS) accumulation via lipid peroxidation. This oxidative stress damages crucial cellular components like cell walls, membranes, organelles, and mitochondria, ultimately leading to endosperm decay (Braybrook et al., 2006). FUS3, a gene regulatory protein, plays a pivotal role in modulating the ABA/GA ratio under high temperatures. It negatively regulates GA biosynthesis while enhancing ABA production during post-embryonic development, further contributing to thermodormancy and germination inhibition (Malek et al., 2022). Studies in B. napus have investigated the impact of high-temperature stress (HTS) on triggering thermo-dormancy and thermo-inhibition, as well as the potential of hydropriming as a mitigating strategy. These experiments successfully demonstrated hydropriming's ability to improve germination rates under HTS conditions (Née et al., 2015). While exogenous GA application has shown some success in reducing thermodormancy, it can paradoxically lead to increased seed death and compromised germination in certain B. napus cultivars. Therefore, soil moisture content-mediated priming emerges as a more promising approach for enhancing germination under HTS, albeit with potential trade-offs for seed longevity (Née et al., 2015).
Low temperatures exert a powerful influence on seed dormancy by promoting the accumulation of DOG1 (DELAY OF GERMINATION 1) transcripts (Farooq et al., 2022). This gene plays a crucial role in prolonging dormancy, acting within the zygotic tissues of the embryo rather than the surrounding seed coat. Intriguingly, BnaDOG1 transcript levels have been shown to increase specifically during secondary dormancy in B. napus, highlighting its involvement in this complex dormancy mechanism (Xia et al., 2018). Endosperm decay, a critical step in seed germination, unfolds in three distinct phases (Sachet et al., 2016). Phase I involves imbibition, where the seed absorbs water rapidly while maintaining its original size and shape. Phase II marks the initiation of testa rupture at the micropylar end surrounding the radicle, and Phase III culminates in the rupture of the endosperm and the emergence of the radicle. This coordinated process provides the seedling with essential nutrients and space for initial growth. One of the key prerequisites for successful germination lies in the breakdown of the endosperm cell wall. This task is primarily accomplished by cell-wall-remodeling enzymes (CWREs) such as expansins and xyloglucan endo-transglucosylases (Lee et al., 2015). Gibberellic acid (GA) acts as a vital orchestrator in this process, promoting the expression and activity of CWREs. Conversely, abscisic acid (ABA) plays an antagonistic role, suppressing the loosening of the endosperm and hindering germination. Interestingly, studies have shown that at 24 °C, DOG1 specifically targets expansin 2 (EXPA2), expansin 9 (EXPA9), and xyloglucan endo-transglucosylase 19 (XTH19) genes, key CWREs involved in endosperm breakdown (Sachet et al., 2016; Farooq et al., 2022). This inhibition occurs through DOG1′s differential regulation of GA biosynthesis genes, highlighting its multifaceted role in controlling germination. Hence, achieving the exact balance between GA and ABA signaling becomes crucial for successful seed germination and seedling emergence.
5 Other factors influencing the seed dormancy
Seed dormancy in canola is under the influence of various factors, viz., genetic factors as described previously, physiological, and agronomic factors (Fig. 3). (See Fig. 4).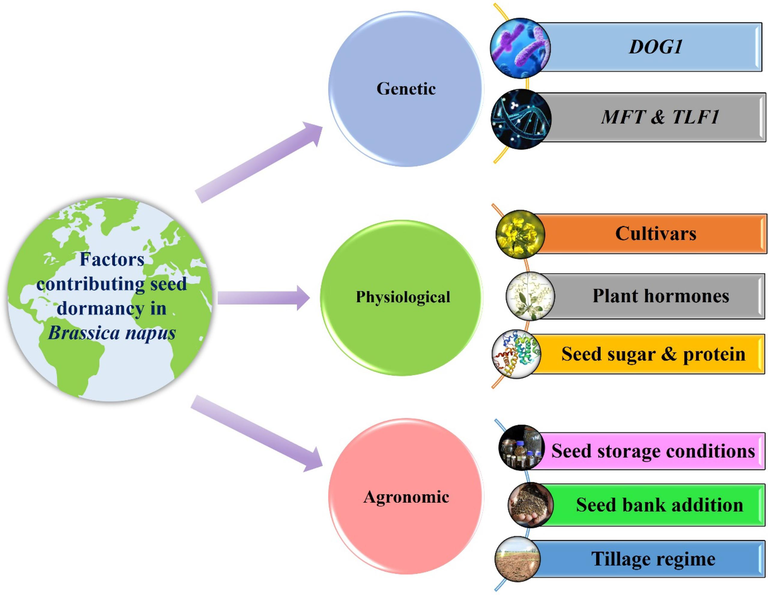
An overview of the factors contributing to seed dormancy in Brassica napus L.
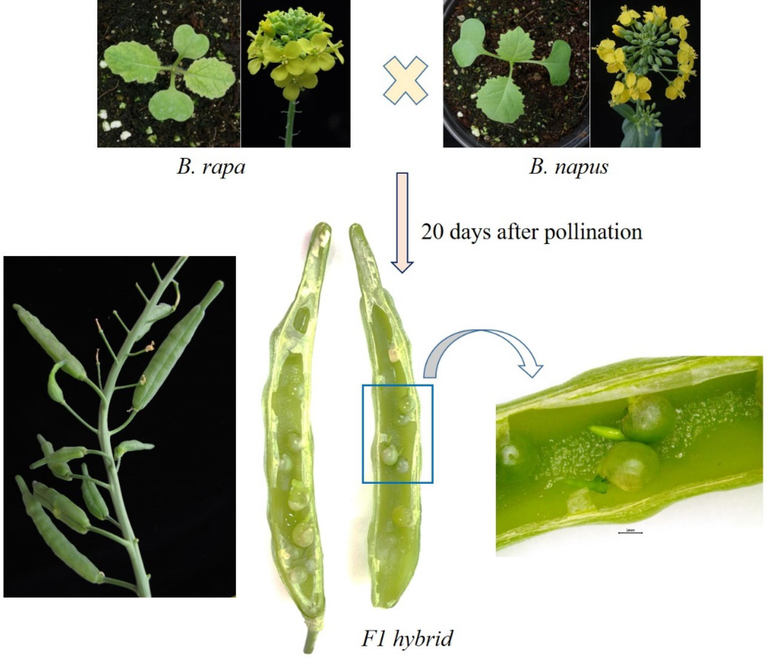
Precocious germination in the interspecific hybrids of B. rapa and B. napus. The F1 hybrids between B. rapa and B. napus was found to have preharvest sprouts after 20 days after pollination.
5.1 Agronomic factors of seed dormancy: Secondary dormancy
5.1.1 Seed storage after harvesting
Environmental factors, particularly storage duration and temperature, significantly influence the development and intensity of secondary dormancy in B. napus (Gulden et al., 2003; Nguyen et al., 2015). Interestingly, even at extremely low temperatures like −70 °C, seed dormancy can be maintained, highlighting its remarkable resilience (Gulden et al., 2004). Cold storage at varying temperatures over extended periods can effectively preserve the degree of secondary dormancy (Momoh et al., 2002). Studies by Vail and Headley (2018) further revealed that temperature cycling between 25 °C and 45 °C induced the most significant changes in dormancy, particularly in spring B. napus genotypes. These findings underscore the importance of considering pre-screening seed storage conditions and potential temperature fluctuations to avoid erroneous interpretation of secondary dormancy levels during subsequent assessments.
5.1.2 Seedbank addition
Seed losses during canola harvest represent a primary pathway for seed contamination and seed bank establishment in previously uninfested fields (Gulden et al., 2003). These losses often occur in two critical phases: first, during seed maturation when fragile pods are susceptible to breakage and seed dispersal, and second, during harvest when seeds escape the combine harvester (Haile and Shirtliffe, 2014). Estimates suggest that losses can reach 5.8 % of the total yield, significantly contributing to seedbank additions. Interestingly, researchers observed variations in dormancy levels among seeds collected at different stages. Seeds harvested before full maturity exhibited higher primary dormancy, while those collected at maturity displayed stronger secondary dormancy (Gulden et al., 2003). This finding reveals the potential mechanism for enhanced resilience of seeds that are lost at various stages of harvest. Reducing seed losses and minimizing their influence on seedbank establishment requires a multifaceted approach. As Raman et al. (2014) pointed out, incorporating pod-shattering tolerant cultivars and implementing appropriate farm management practices can significantly reduce harvest losses and contribute to sustainable canola production.
5.1.3 Tillage regime
Tillage timing and depth can play crucial roles in managing volunteer B. napus populations. Delaying tillage practices until after volunteer emergence can significantly reduce their numbers (Gruber et al., 2005). Additionally, increasing tillage depth to 15 or 25 cm can effectively bury volunteer seeds deeper, further limiting their emergence and survival. Huang et al. (2018) highlight the importance of considering both tillage timing and seed burial depth when managing volunteer populations. Tillage conducted after the autumn season, compared to earlier timings, can significantly reduce seed persistence in the soil seed bank. The optimal tillage regime for managing volunteer B. napus depends on several factors, including climatic conditions and the specific growth patterns of the crop variety being planted (2017). Based on these factors, tillage strategies can be tailored to mitigate soil disturbance and conserve significant resources, all while helping in the efficient management of volunteer populations.
5.2 Physiological factors of seed dormancy: (Physiological dormancy)
5.2.1 Physiological changes during winter and spring
Physiological factors, particularly differences in growth types, influence seed dormancy potential in B. napus. Spring varieties generally exhibit higher dormancy potential compared to winter types (Momoh et al., 2002). This enhanced dormancy in spring genotypes serves as a protective mechanism to prevent premature germination in the fall, which could potentially lead to frost damage. In contrast, winter genotypes possess the ability to germinate during the fall period to experience vernalization, a chilling requirement for subsequent flowering in the spring. Consequently, winter type varieties exhibit a greater sensitivity to environmental cues that regulate dormancy break and timing of flowering (Momoh et al., 2002).
5.2.2 Seed sugar, protein and metabolites
Sugars and proteins are two crucial components of seeds, playing distinct roles in germination and seedling establishment. Glucose acts as a germination inhibitor by influencing ABA signaling pathways, while seed storage proteins contribute to seedling vigor, dormancy, and seed longevity (Dekkers et al., 2004). Cruciferin and napin are the primary seed storage proteins in B. napus, with their relative proportions determined by both genetic and environmental factors. Previous research suggests an inverse relationship between cruciferin and napin content, with napin levels potentially decreasing indirectly due to the selection for low-glucosinolate varieties (Schatzki et al., 2014). If this negative correlation holds true across all B. napus species, modern spring-type low-glucosinolate varieties might be predicted to exhibit higher dormancy levels. However, further studies are necessary to fully explore this association and its underlying mechanisms (Baskin and Baskin, 1998; Schatzki et al., 2014).
Among the several chemical constituents of seed coats/seeds, flavonoids were known to be associated with seed dormancy. In particular, proanthocyanidins (PA) which accumulate during early embroyogenesis until seed maturation, play a crucial role in reinforcing the seed-coat-imposed dormancy. Previous studies suggest that PAs and their precursors inhibit germination by acting as structural barriers and as biochemical inhibitors of GA metabolism. A study investigating flavonoid profiles in yellow-coated and black-coated rapeseeds found that the flavonoid biosynthetic genes, anthocyanin and proanthocyanidin specific genes, as well as regulators in the flavonoid synthetic pathway were relatively repressed in yellow seeds (Chen et al., 2023). In fact, later, it was found that BnTT8 mutation suppressed the expression of phenylpropanoid and flavonoid biosynthetic genes, inhibited proanthocyanidin accumulation in rape seed coats (Zhai et al., 2020). Also, a total of 35 flavonols were identified in the seed coat of B. napus (Qu et al., 2013). The predominant flavonols are kaempferol derivatives, quercetin derivatives, isorhamnetin derivatives, and epicatechin glucosides. However, the role of flavonols in seed dormancy and germination in rapeseeds has yet to be identified.
6 Precocious germination/vivipary is correlated with seed dormancy?
Precocious germination, also referred to as premature germination, pre-harvest sprouting, or vivipary, describes seed germination within the maternal plant during its maturation phase. To prevent premature germination, primary dormancy is a prevalent mechanism in maturing B. napus seeds, although it diminishes significantly in mature seeds (Feng et al., 2009). However, in certain genotypes and under specific environmental conditions, this dormancy can be insufficient, leading to seed germination on the mother plant and potential yield losses. Premature germination is also a quantitative trait influenced by the interplay of genetics, environment, and their interactions (Ren and Bewely, 1998). Excessive moisture and humidity are environmental factors known to promote higher rates of precocious germination (Feng et al., 2009). Additionally, prematurely germinated seeds typically exhibit lower abscisic acid (ABA) concentrations and altered osmotic potential (Johnson-Flanagan et al., 1991). Another significant environmental factor promoting premature germination is heat stress (Brunel-Muguet et al., 2015). This phenomenon likely arises from a decline in the ABA:GA ratio, leading to reduced primary dormancy and increased germination potential (Fahad et al., 2017). While rare in mature B. napus seeds, premature germination may become more frequent if secondary dormancy is compromised, as both primary and secondary dormancy may share similar physiological mechanisms (Feng et al., 2009). Mácová et al., (2022) have found the cultivar-specific premature germination and ABA level reductions in different B. napus cultivars, thus indicating the role of genome composition among others. Premature germination and lower ABA levels in heat stress indicate that seeds did not progress into the dormancy phase but continued directly to germination. Interestingly, the interspecific hybridization between B. napus and B. rapa often exhibits the phenotypic expression of vivipary (Sohn et al. 2022a; Sohn et al. 2022b). Sohn et al. (2022b) observed a higher rate of premature germination in F1 hybrids arising out of B. rapa (maternal) and B. napus (paternal) crosses (Fig. 3). However, the reverse crossing between them (B. napus being maternal parent) do not show vivipary, possibly suggesting the importance of maternal or paternal excess genomes/ploidy levels and also the role of imprinted genes from either parents. However, the precise molecular basis of vivipary except for hormone imbalance in B. napus, is unknown. There was a previous attempt to identify QTLs responsible for vivipary in rapeseed (Feng et al., 2009). They found at least five putative QTLs which were designated as qPHS-2-a, qPHS-2-b, qPHS-2-c, qPHS-2-d and qPHS-7, located in the vicinity of A3G12700, A13C1500, P3C4180 and T2G13420 on LG2 (N11) and P1G8300 on LG7 (N3) rapeseed genome. The accumulated contribution of these five QTLs explained 75.63 % of the phenotypic variance, and individual QTL explained 4.11–50.78 % of the phenotypic variance. Genetic markers are associated with better seed longevity and vigor could lead to the development of targeted agronomic strategies, such as optimized storage conditions or specific treatments during seed maturation that can help to minimize the premature germination.
7 Conclusion and future perspectives
To date, significant progress has been made in elucidating the molecular pathways regulating ABA/GA balance and driving seed dormancy-to-germination transitions in model plant, Arabidopsis. However, research reports on seed germination mechanisms in B. napus is only emerging and it is clear that several QTLs in concert with external and internal cues participating in seed-related traits, including dormancy and germination. Therefore, comprehensive molecular investigations are essential to elucidate the complex dormancy behaviors in B. napus, influenced by various physiochemical factors. Harnessing genetic factors that contribute to sufficient seed vigor can further enhance yield and stability. Genome-wide association studies (GWAS) can identify QTLs and candidate genes associated with seed germination, facilitating gene introgression to expand the genetic and molecular foundation for rapeseed breeding. To unravel the intricate molecular mechanisms underlying seed dormancy and germination, emerging breeding approaches like genomic selection (GS), speed breeding, and Multi-parent advanced generation intercross (MAGIC) populations should be integrated. Additionally, exploring the role of epigenetic factors in B. napus seed dormancy and germination is crucial for a comprehensive understanding of these complex traits. The preharvest sprouting in some of the intersepcific hybrids in rapeseed open up new avenues in research where parental genome conflict theory takes centre place, thus indicating the importance of future studies revolving around the identification of biological significance of materal and paternal imprinted genes or the importance of maternal- or paternal-excess genome contribution towards seed development as it provides the cues for dormancy levels and germiantion pontentials. Similar to primary dormancy, secondary dormancy in rape seeds also play a role in environmental adaptation and seedling vigor. Secondary dormancy is also a result of complex interactive responses among the genome, environment, and agronomic practices. However, considering the fair amount of success achieved with exogenous application of dormancy-breaking substances or treatment that remove the seed-coat imposed dormancy or precise envirnomental monitoring measures, secondary dormancy can be fairly manipulated to meet the requirements. Therefore it is important for future farming practices should consider the temporal and geographic dimensions of environmental sensing, along with the distinct functions and detection mechanisms of environmental factors throughout seed development, storage, and germination. Furthermore, integrating genetic, physiological, and multi-omics data with cutting-edge advancements like artificial intelligence (AI) – machine learning (ML) will speed up the identification of precise molecular basis of these key traits. This significant combination holds the potential to uncover personalized seed treatments tailored to impute specific hormonal needs, biosensors for monitoring hormonal dynamisms, CRISPR mediated gene editing for altering the dormancy related genes for on-demand hormone biosynthesis, and smart agriculture systems for optimizing the success frequency of germination and manage dormancy at scale. These advancements can empower farmers with data-driven insights to improve decision making and eventually enhance agricultural productivity and future food security.
Funding
This study was supported by “Research Program for Agricultural Science & Technology Development and Post-Doctoral Fellowship Program (Project No. PJ01672604)”, National Institute of Agricultural Sciences, Rural Development Administration, Korea.
CRediT authorship contribution statement
Subramani Pandian: Writing – review & editing, Writing – original draft, Visualization, Conceptualization. Jayabalan Shilpha: Writing – review & editing, Writing – original draft, Visualization, Conceptualization. Muthiah Joe Virgin Largia: Writing – original draft, Visualization, Conceptualization. Pandiyan Muthuramalingam: Writing – review & editing, Writing – original draft, Visualization. Muthusamy Muthusamy: Writing – review & editing, Writing – original draft, Visualization. Ravi Jothi: Writing – review & editing, Writing – original draft. Young-Ju Oh: . Soo-In Sohn: Writing – review & editing, Supervision, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Analysis of natural genetic variation at seed dormancy loci of Arabidopsis thaliana. Genetics. 2003;164:711-729.
- [Google Scholar]
- Gene Expression Profiling Identifies Two Regulatory Genes Controlling Dormancy and ABA Sensitivity in Arabidopsis Seeds. Plant J.. 2010;61:611-622.
- [Google Scholar]
- Types of seed dormancy. In Seeds: Ecology, biogeography, and evolution of dormancy and germination. New York: Academic Press, Elsevier Inc.; 1998. p. :27-47.
- Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc. Nat. Acad. Sci.. 2011;108(23):9709-9714.
- [Google Scholar]
- Persistence of an oilseed rape transgene in the environment. Crop Prot.. 2010;29:509-512.
- [Google Scholar]
- An integrative approach to analyze seed germination in Brassica napus. Front. Plant Sci.. 2019;10:1342.
- [Google Scholar]
- Genes directly regulated by LEAFY COTYLEDON2 provide insight into the control of embryo maturation and somatic embryogenesis. Proc. Natl. Acad. Sci. USA. 2006;103:3468-3473.
- [Google Scholar]
- Brown, C., Gulden, R.H., Shirtliffe, S.J. and Vail, S., 2022. A Review of the Genetic, Physiological and Agronomic Factors Influencing Secondary Dormancy Levels and Seed Vigour in Brassica napus L. Can. J. Plant Sci., (ja).
- Heat stress during seed filling interferes with sulfur restriction on grain composition and seed germination in oilseed rape (Brassica napus L.) Front. Plant Sci.. 2015;6:213.
- [Google Scholar]
- A perspective on secondary seed dormancy in Arabidopsis thaliana. Plants. 2020;9(6):749.
- [Google Scholar]
- Conditional seed dormancy helps Silene hicesiae Brullo & Signor. Overcome stressful Mediterranean summer conditions. Plants. 2021;10(10):2130.
- [Google Scholar]
- Collinearity between a 30-centimorgan segment of Arabidopsis thaliana chromosome 4 and duplicated regions within the Brassica napus genome. Genome. 1998;41(1):62-69.
- [Google Scholar]
- Multifaceted signaling networks mediated by abscisic acid insensitive 4. Plant Commun.. 2020;1:100040
- [Google Scholar]
- The Flavonoid Biosynthesis and Regulation in Brassica napus: A Review. Int. J. Mol. Sci.. 2023;24
- [CrossRef] [Google Scholar]
- Integration of Light and Abscisic Acid Signaling during Seed Germination and Early Seedling Development. Proc. Natl. Acad. Sci.. 2008;105:4495-4500.
- [Google Scholar]
- Abscisic Acid: Emergence of a Core Signaling Network. Annu. Rev. Plant Biol.. 2010;61:651-679.
- [Google Scholar]
- Integration of Phytochrome and Cryptochrome Signals Determines Plant Growth during Competition for Light. Curr. Biol.. 2016;26:3320-3326.
- [Google Scholar]
- The Arabidopsis DELAY OF GERMINATION 1 gene affects ABSCISIC ACID INSENSITIVE 5 (ABI5) expression and genetically interacts with ABI3 during Arabidopsis seed development. Plant J.. 2016;85:451-465.
- [Google Scholar]
- A transcription factor WRKY36 interacts with AFP2 to break primary seed dormancy by progressively silencing DOG1 in Arabidopsis. New Phytol.. 2023;238:688-704.
- [Google Scholar]
- Improving seed germination and oil contents by regulating the GDSL transcriptional level in Brassica na-pus. Plant Cell Rep.. 2019;38:243-253.
- [Google Scholar]
- WRKY 41 Controls Arabidopsis Seed Dormancy via Direct Regulation of ABI 3 Transcript Levels Not Downstream of ABA. Plant J.. 2014;79:810-823.
- [Google Scholar]
- Crop production under drought and heat stress: plant responses and management options. Front. Plant Sci.. 2017;8:1147.
- [Google Scholar]
- Underlying biochemical and molecular mechanisms for seed germination. Int. J. Mol. Sci.. 2022;23(15):8502.
- [Google Scholar]
- Metabolic and hormonal processes associated with the induction of secondary dormancy in Brassica napus seeds. Botany. 2009;87:585-596.
- [Google Scholar]
- Feng F., P. L., Hong D., Yang G. 2009. A major QTL associated with pre-harvest sprouting in rapeseed (Brassica napus L.). Euphytica, 169: 57–68.
- Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot.. 2017;68:843-856.
- [CrossRef] [Google Scholar]
- Mutations at Two New Arabidopsis ABA Response Loci Are Similar to the Abi3 Mutations. Plant J.. 1994;5:765-771.
- [Google Scholar]
- Epoxycarotenoid Cleavage by NCED5 Fine-tunes ABA Accumulation and Affects Seed Dormancy and Drought Tolerance with Other NCED Family Members. Plant J.. 2012;70:501-512.
- [Google Scholar]
- The Dof Protein DAG1 Mediates PIL5 Activity on Seed Germination by Negatively Regulating GA Biosynthetic Gene AtGA3ox1. Plant J.. 2010;61:312-323.
- [Google Scholar]
- Spatial and temporal gene expression during seed germination of Brassica napus. Acta Physiologiae Plantarum. 2013;35:2939-2950.
- [Google Scholar]
- DELAY OF GERMINATION 1 Mediates a Conserved Coat-Dormancy Mechanism for the Temperature-and Gibberellin-Dependent Control of Seed Germination. Proc. Natl. Acad. Sci.. 2014;111:E3571-E3580.
- [Google Scholar]
- DELAY OF GERMINATION 1 Mediates a Conserved Coat-Dormancy Mechanism for the Temperature-and Gibberellin-Dependent Control of Seed Germination. Proc. Natl. Acad. Sci.. 2014;111:E3571-E3580.
- [Google Scholar]
- Life cycle and potential gene flow of volunteer oilseed rape in different tillage systems. Weed Res.. 2005;45:83-93.
- [Google Scholar]
- Response to abscisic acid application and hormone profiles in spring Brassica napus seed in relation to secondary dormancy. Can. J. Bot.. 2004;82:1618-1624.
- [Google Scholar]
- Secondary seed dormancy prolongs persistence of volunteer canola in western Canada. Weed Sci.. 2003;51:904-913.
- [Google Scholar]
- Arabidopsis EIN2 represses ABA responses during germination and early seedling growth by inactivating HLS1 protein independently of the canonical ethylene pathway. Plant J.. 2023;115:1514-1527.
- [Google Scholar]
- Influences on seed germinability: phenotypic maternal effects during maturation. Israel Journal of Botany. 1980;29:105-117.
- [Google Scholar]
- Effect of harvest timing on dormancy induction in canola seeds. Weed Sci.. 2014;62:548-554.
- [Google Scholar]
- Environmental control of seed germination in Thlaspi arvense (Cruciferae) Am. J. Bot.. 1990;77:945-953.
- [Google Scholar]
- Molecular networks regulating Arabidopsis seed maturation, after ripening, dormancy and germination. New Phytol.. 2008;179:33-54.
- [Google Scholar]
- Timing and depth of post-harvest soil disturbance can reduce seedbank and volunteers of oilseed rape. Soil Tillage Res.. 2018;175:187-193.
- [Google Scholar]
- The auxin signaling repressor IAA8 promotes seed germination through down-regulation of ABI3 transcription in Arabidopsis. Front. Plant Sci.. 2020;11:111.
- [Google Scholar]
- Canola seed priming and its effect on gas exchange, chlorophyll photobleaching, and enzymatic activities in response to salt stress. Sustainability. 2022;14(15):9377.
- [Google Scholar]
- The COP9 Signalosome Regulates Seed Germination by Facilitating Protein Degradation of RGL2 and ABI5. PLoS Genet.. 2018;14:e1007237.
- [Google Scholar]
- The impact of freezing during maturation on storage products in canola seeds. Physiol. Plant.. 1991;81:301-308.
- [Google Scholar]
- HONSU, a Protein Phosphatase 2C, Regulates Seed Dormancy by Inhibiting ABA Signaling in Arabidopsis. Plant Cell Physiol.. 2013;54:555-572.
- [Google Scholar]
- Genetic and physiological responses to heat stress in Brassica napus. Frontiers in Plant Science. 2022;13
- [Google Scholar]
- The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J.. 2004;23:1647-1656.
- [Google Scholar]
- The Arabidopsis MYB96 Transcription Factor Is a Positive Regulator of ABSCISIC ACID-INSENSITIVE4 in the Control of Seed Germination. Plant Physiol.. 2015;168:677-689.
- [Google Scholar]
- Arabidopsis Putative MAP Kinase Kinase Kinases Raf10 and Raf11 Are Positive Regulators of Seed Dormancy and ABA Response. Plant Cell Physiol.. 2015;56:84-97.
- [Google Scholar]
- Lei, L. I. U., Fan, W. Q., LIU, F. X., Xin, Y. I., Tang, T. A. N. G., Ying, Z. H. O. U., ... & Zhao, X. X. (2020). Increased BnaMFT-transcript level is associated with secondary dormancy in oilseed rape (Brassica napus L.). Journal of Integrative Agriculture, 19(6), 1565-1576.
- ETR1/RDO3 regulates seed dormancy by relieving the inhibitory effect of the ERF12-TPL complex on DELAY OF GERMINATION1 expression. Plant Cell. 2019;31:832-847.
- [Google Scholar]
- ABA-INSENSITIVE3, ABA-INSENSITIVE5, and DELLAs interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell. 2013;25:4863-4878.
- [Google Scholar]
- Genome-wide screening and analysis of imprinted genes in rapeseed (Brassica napus L.) endosperm. DNA Res.. 2018;25:629-640.
- [CrossRef] [Google Scholar]
- Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci.. 2013;110:15485-15490.
- [Google Scholar]
- Increased BnaMFT-transcript level is associated with secondary dormancy in oilseed rape (Brassica napus L.) J. Integr. Agric.. 2020;19(6):1565-1576.
- [Google Scholar]
- Phytochrome and phytohormones: working in tandem for plant growth and development. Front. Plant Sci.. 2018;9:1037.
- [Google Scholar]
- Long-term high-temperature stress impacts on embryo and seed development in Brassica napus. Front. Plant Sci.. 2022;13:844292
- [Google Scholar]
- Dwarf and Delayed-flowering 1, a Novel Arabidopsis Mutant Deficient in Gibberellin Biosynthesis Because of Overexpression of a Putative AP2 Transcription Factor. Plant J.. 2004;37:720-729.
- [Google Scholar]
- Dynamics of seed dormancy and germination at high temperature stress is affected by priming and phytohormones in rapeseed (Brassica napus L.) J. Plant Physiol.. 2022;269:153614
- [Google Scholar]
- The Arabidopsis Abscisic Acid Catabolic Gene CYP707A2 Plays a Key Role in Nitrate Control of Seed Dormancy. Plant Physiol.. 2009;149:949-960.
- [Google Scholar]
- Sumoylation of ABI5 by the Arabidopsis SUMO E3 Ligase SIZ1 Negatively Regulates Abscisic Acid Signaling. Proc. Natl. Acad. Sci.. 2009;106:5418-5423.
- [Google Scholar]
- Variation in the development of secondary dormancy in oilseed rape genotypes under conditions of stress. Weed Res.. 2002;42:446-455.
- [Google Scholar]
- Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459-463.
- [CrossRef] [Google Scholar]
- Concurrent elevation of CO2, O3 and temperature severely affects oil quality and quantity in rapeseed. Journal of Experimental Botany. 2016;67(14):4117-4125.
- [Google Scholar]
- Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol.. 2005;56:165-185.
- [CrossRef] [Google Scholar]
- Secondary dormancy in Brassica napus is correlated with enhanced BnaDOG1 transcript levels. Seed Science Research. 2015;25(2):221-229.
- [Google Scholar]
- Abscisic acid perception and signaling: structural mechanisms and applications. Acta Pharmacol. Sin.. 2014;35:567-584.
- [CrossRef] [Google Scholar]
- A role for seed storage proteins in Arabidopsis seed longevity. J. Exp. Bot.. 2015;66:6399-6413.
- [Google Scholar]
- Disruption of germination and seedling development in Brassica napus by mutations causing severe seed hormonal imbalance. Frontiers in Plant Science. 2016;7:322.
- [Google Scholar]
- Influencia de alguns fatores da planta sobre o grau de dormancia em sementes de mucuna preta. Revista Brasiliera De Sementes. 1983;5:111-119.
- [Google Scholar]
- PIL5, a Phytochrome-Interacting BHLH Protein, Regulates Gibberellin Responsiveness by Binding Directly to the GAI and RGA Promoters in Arabidopsis Seeds. Plant Cell. 2007;19:1192-1208.
- [Google Scholar]
- Induction of secondary dormancy in rape seeds (B. napus L.) by prolonged inhibition under conditions of water stress or oxygen deficiency in darkness. European Journal of Agronomy. 1997;6:245-255.
- [Google Scholar]
- Differential accumulation of phenolic compounds and expression of related genes in black- and yellow-seeded Brassica napus. J. Exp. Bot.. 2013;64:2885-2898.
- [CrossRef] [Google Scholar]
- Rapeseed (Brassica napus): Processing, utilization, and genetic improvement. Agronomy. 2021;11(9):1776.
- [Google Scholar]
- Genome-wide delineation of natural variation for pod shatter resistance in Brassica napus. PLoS ONE. 2014;9:e101673.
- [Google Scholar]
- A novel RGL2–DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA transcription factor. Mol. Plant. 2017;10:1307-1320.
- [CrossRef] [Google Scholar]
- Seed development, testa structure and precocious germination of Chinese cabbage (Brassica rapa subsp. Pekinensis) Seed Sci. Res.. 1998;8:385-398.
- [Google Scholar]
- Genomic imprinted genes in reciprocal hybrid endosperm of Brassica napus. BMC Plant Biol.. 2021;21:1-17.
- [CrossRef] [Google Scholar]
- Leaf gas exchange, oxidative stress, and physiological attributes of rapeseed (Brassica napus L.) grown under different light-emitting diodes. Photosynthetica. 2020;58(3)
- [Google Scholar]
- ABA metabolism and homeostasis in seed dormancy and germination. Int. J. Mol. Sci.. 2021;22(10):5069.
- [Google Scholar]
- Mapping of QTL for the seed storage proteins cruciferin and napin in a winter oilseed rape doubled haploid population and their inheritance in relation to other seed traits. Theor. Appl. Genet.. 2014;127:1213-1222.
- [Google Scholar]
- Overcoming seed dormancy in oilseed rape (Brassica napus L.) with exogenous compounds. Weed Res.. 2019;59(2):119-129.
- [Google Scholar]
- Xyloglucan Metabolism Differentially Impacts the Cell Wall Characteristics of the Endosperm and Embryo during Arabidopsis Seed Germination. Plant Physiol.. 2016;170:1367-1380.
- [Google Scholar]
- ABI4 Regulates Primary Seed Dormancy by Regulating the Biogenesis of Abscisic Acid and Gibberellins in Arabidopsis. PLoS Genet.. 2013;9:e1003577.
- [Google Scholar]
- Dormancy and germination: How does the crop seed decide? Plant Biology. 2015;17(6):1104-1112.
- [Google Scholar]
- APETALA 2-domain-containing Transcription Factors: Focusing on Abscisic Acid and Gibberellins Antagonism. New Phytol.. 2018;217:977-983.
- [Google Scholar]
- The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci.. 2016;7:1884.
- [Google Scholar]
- Characteristics analysis of F1 hybrids between genetically modified Brassica napus and B. rapa. PLoS One. 2016;11(9):e0162103.
- [Google Scholar]
- A review of the unintentional release of feral genetically modified rapeseed into the environment. Biology. 2021;10(12):1264.
- [Google Scholar]
- Characteristics and Fitness Analysis through Interspecific Hybrid Progenies of Transgenic Brassica napus and B. rapa L. ssp. International Journal of Molecular Sciences. 2022;23(18):10512.
- [Google Scholar]
- Interspecific Hybridization of Transgenic Brassica napus and Brassica rapa—An Overview. Genes. 2022;13(8):1442.
- [Google Scholar]
- A review of the relationship between primary and secondary dormancy, with reference to the volunteer weed oilseed rape (Brassica napus) Weed Res.. 2018;59:5-14.
- [Google Scholar]
- Far-red elongated hypocotyl3 and far-red impaired response. Transcription Factors Integrate Light and Abscisic Acid Signaling in Arabidopsis. Plant Physiol.. 2013;163:857-866.
- [Google Scholar]
- Direct and indirect targets of the arabidopsis seed transcription factor abscisic acid insensitive3. Plant J.. 2020;103:1679-1694.
- [Google Scholar]
- Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci.. 2018;9:668.
- [Google Scholar]
- Molecular mechanisms underlying abscisic acid/gibberellin balance in the control of seed dormancy and germination in cereals. Front. Plant Sci.. 2018;9:668.
- [Google Scholar]
- Localization and expression of CRSH transcript, level of calcium ions, and cell cycle activity during Brassica napus L. seed development. Ind. Crop. Prod.. 2023;195:116439
- [Google Scholar]
- Vail, S., and Headley, C. 2018. Seed storage conditions and secondary dormancy. Unpublished raw data.
- Regulation of seed germination and abiotic stresses by gibberellins and abscisic acid. Front. Plant Sci.. 2018;9:838.
- [Google Scholar]
- Overexpression of FAD2 promotes seed germination and hypocotyl elongation in Brassica napus. Plant Cell, Tissue and Organ Culture (PCTOC). 2010;102:205-211.
- [Google Scholar]
- Comparative proteomics analysis reveals the mechanism of pre-harvest seed deterioration of soybean under high temperature and humidity stress. J. Proteom.. 2012;75:2109-2127.
- [Google Scholar]
- Comparison of DNA methylation in the developing seeds of yellow- and black-seeded Brassica napus through MSAP analysis. Euphytica. 2016;209:157-169.
- [CrossRef] [Google Scholar]
- Wang, Y., Zhao, Z., Liu, F., Sun, L. and Hao, F., 2020. Versatile roles of aquaporins in plant growth and development. International journal of molecular sciences, 21(24), p.9485.Ge, F. W., Tao, P., Zhang, Y., & Wang, J. B. (2014). Characterization of AQP gene expressions in Brassica napus during seed germination and in response to abiotic stresses. Biologia plantarum, 58(2), 274-282.
- The DELLA Domain of GA INSENSITIVE Mediates the Interaction with the GA INSENSITIVE DWARF1A Gibberellin Receptor of Arabidopsis. Plant Cell. 2007;19:1209-1220.
- [Google Scholar]
- A Mak-like kinase is a repressor of GAMYB in barley aleurone. Plant J.. 2003;33:707-717.
- [CrossRef] [Google Scholar]
- Integrating proteomics and enzymatic profiling to decipher seed metabolism affected by temperature in seed dormancy and germination. Plant Sci.. 2018;269:118-125.
- [Google Scholar]
- Reduced Dormancy5 encodes a protein phosphatase 2C that is required for seed dormancy in Arabidopsis. Plant Cell. 2014;26:4362-4375.
- [Google Scholar]
- Function of Brassica napus BnABI3 in Arabidopsis gs1, an allele of AtABI3, in seed development and stress response. Front. Plant Sci.. 2019;10:67.
- [Google Scholar]
- Comparison of vitality between seedlings germinated from black-coated and yellow-coated seeds of a turnip rape (Brassica rapa L.) subjected to NaCl and CdCl2 stresses. Plant Growth Regul.. 2015;76:61-70.
- [CrossRef] [Google Scholar]
- The role of light in regulating seed dormancy and germination. J. Integr. Plant Biol.. 2020;62:1310-1326.
- [Google Scholar]
- CHOTTO1, a Putative Double APETALA2 Repeat Transcription Factor, Is Involved in Abscisic Acid-Mediated Repression of Gibberellin Biosynthesis during Seed Germination in Arabidopsis. Plant Physiol.. 2009;151:641-654.
- [Google Scholar]
- Genome-wide analysis of parent-of-origin allelic expression in endosperms of brassicaceae species, brassica rapa. Plant Cell Physiol.. 2018;59:2590-2601.
- [CrossRef] [Google Scholar]
- Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol. J.. 2020;18:1153-1168.
- [CrossRef] [Google Scholar]
- Practical Methods for Breaking Seed Dormancy in a Wild Ornamental Tulip Species Tulipa thianschanica Regel. Agronomy. 2020;10:1765.
- [Google Scholar]
- Resynthesizing Brassica napus from interspecific hybridization between Brassica rapa and B. oleracea through ovary culture. Euphytica. 2004;140:181-187.
- [Google Scholar]
- BES 1 hinders ABSCISIC ACID INSENSITIVE 5 and promotes seed germination in Arabidopsis. New Phytol.. 2019;221:908-918.
- [Google Scholar]
- A Novel Role for Histone Methyltransferase KYP/SUVH4 in the Control of Arabidopsis Primary Seed Dormancy. New Phytol.. 2012;193:605-616.
- [Google Scholar]
- A transcriptional hub integrating gibberellin–brassinosteroid signals to promote seed germination in Arabidopsis. J. Exp. Bot.. 2021;72:4708-4720.
- [Google Scholar]
- Membrane-associated transcription factor peptidase, site-2 protease, antagonizes ABA signaling in Arabidopsis. New Phytol.. 2015;208:188-197.
- [Google Scholar]







