Translate this page into:
Adaptation of ethidium bromide fluorescence assay to monitor activity of efflux pumps in bacterial pure cultures or mixed population from environmental samples
⁎Corresponding author at: Department of Botany, Institute of Science, Visva-Bharati (A Central University), Santiniketan, West Bengal 731235, India. bomba.dam@visva-bharati.ac.in (Bomba Dam)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Adaptation of EtBr fluorescence assay for estimation of efflux pump activities. The assay was effective in both Gram negative as well as Gram positive bacteria. The spectrum of the assay was extended to diverse physiologies with efflux pumps. The assay was used for first time directly on cells harvested from natural samples.
Abstract
Quick, simple and reliable assays to screen and understand active efflux pumps operative in pure bacterial cultures or mixed population in environmental samples are important. Although assays are available to monitor efflux activities in pure cultures, none has been used or tested directly on natural samples or cells harvested from them. Here, we have adapted an existing fluorescence assay using ethidium bromide (EtBr) to monitor efflux activities connected to resistance/tolerance to antibiotics (tetracycline), heavy metals (chromium), salt (NaCl) and ionic liquid (1-ethyl-3-methylimidazolium chloride) in different Gram positive and negative bacterial pure cultures. Concentration of 0.5 µg ml−1 of EtBr was found to be optimum for the assay. For all tested physiologies, 1.2–2.2 fold lower EtBr fluorescence (or higher efflux) was detected as compared to control. In addition, the assay was efficiently used for the first time to establish differences in efflux activities in microbial cells harvested from different natural environments. Similar to pure cultures, sediments with higher salinity (7%) compared to lower (3%) ones, and antibiotic contaminated hospital drain samples compared to adjoining garden soil showed up to 1.7-fold and 1.3-fold higher efflux activities respectively. Thus, EtBr fluorescence assay can be used effectively in known or novel bacterial physiologies that involve efflux activities not only in pure cultures, but also in natural environmental microbial groups.
Keywords
Active efflux pump
Ethidium bromide
Fluorescence assay
Environmental sample
Halophiles
Antibiotic resistance
1 Introduction
Bacterial efflux pumps are defensive machineries that actively extrude toxic substances from the periplasm and/or cytoplasm. Efflux of tetracycline by Escherichia coli cells was the first described active pump mediated physiology (Levy and McMurry, 1978). Subsequently, these pumps were found to be utilized for various fundamental cellular functions like maintaining homeostasis of essential metabolites (Eggeling and Sahm, 2003) and resistance/tolerance against antibiotics, heavy metals, and salts (Du et al., 2015; Nies, 2003; Nikaido and Pagès, 2012; Piddock, 2006; Poole, 2007), that help bacteria to survive in extreme environments. Moreover, efflux pumps are now frequently utilized to bioengineer bacterial strains for several biotechnological applications (Fisher et al., 2013; Mingardon et al., 2015; Yang et al., 2015).
Efflux pumps in bacteria belong to five distinct families based on their transmembrane regions, substrate preferences, and energy sources. These are resistance-nodulation-division (RND) family, major facilitator superfamily (MFS), ATP-binding cassette (ABC) superfamily, small multidrug resistance (SMR), and multidrug and toxic compound extrusion (MATE) family (Piddock, 2006; Poole, 2007). RND superfamily is identified only in Gram-negative bacteria, but the rest are present in both groups (Handzlik et al., 2013). ATP hydrolysis is used exclusively by ABC superfamily members to export their substrates, however the other families use proton motive force (Blanco et al., 2016).
For detection and quantification of efflux activities, molecular techniques like quantitative PCR; immunoblotting; radio-labeled and metal-labeled specific substrates; or non-specific fluorescent substrates have been used (Blair and Piddock, 2016; Handzlik et al., 2013; Paixao et al., 2009; Sun et al., 2014; Viveiros et al., 2008). While the first three techniques require prior knowledge of specific target gene/protein/substrate, fluorescent dyes like ethidium bromide (EtBr), Hoechst 33342 or Nile red, can be used as substrate in de novo studies (Olmsted III and Kearns, 1977) (Table 1). Assays utilizing these dyes rely on the principle that fluorescent molecules when used at sub-lethal dose enter cell by passive diffusion, but can be removed only by active efflux system. Therefore, an estimation of relative intracellular concentration of the dye at any point of time can be directly correlated to activity of efflux machinery. In fact these dyes, particularly EtBr, emit very weak fluorescence in aqueous solution (or in media/buffer without viable cells) but the intensity increases with increase in its concentration in the cytoplasm/periplasm of viable cells mostly due to interchelating of the dye to cellular components like DNA (Table 1). Thus, cells with low efflux activity will have higher concentrations of intracellular dyes leading to higher fluorescence and vice versa (Blair and Piddock, 2016). Fluorescence-based assays target the activity of AcrAB-TolC, an RND type multidrug resistant pump in Gram-negative bacteria or ABC and MFS family proteins in Gram-positive bacteria (Coldham et al., 2010; Martins et al., 2011; Paixao et al., 2009; Rodrigues et al,. 2011) (Table 1). All these assays are designed for pure cultures and has never been used directly on environmental samples or cells harvested from them.
Organism
Gram- nature
Target efflux pump family
Fluorescent dye used
Interaction required for fluorescence (molecule)
Objective of the study
Reference
Escherichia coli
- ive
AcrAB-TolC, RND
EtBr
Yes (DNA)
EtBr efflux kinetics by wild type and mutant strains
Paixao et al. (2009)
E. coli
- ive
Non-specific efflux
EtBr
Yes (DNA)
Intrinsic efflux activity using inhibitors
Viveiros et al. (2008)
E. coli
- ive
AcrAB-TolC, RND
EtBr
Yes (DNA)
Role of calcium in multidrug efflux pump
Martins et al. (2011)
Chromohalobacter sp.
- ive
HrdC (TolC homolog)
EtBr
Yes (DNA)
Induction of multidrug efflux pump by salt
Tokunaga et al. (2004)
Staphylococcus aureus
+ ive
NorA, MFS
EtBr
Yes (DNA)
Inhibition of antibiotic efflux by novel inhibitors
Mullin et al. (2004)
Mycobacterium smegmatis
+ ive
LfrA, MFS
EtBr
Yes (DNA)
Drug susceptibility and EtBr efflux of mutant strain
Rodrigues et al. (2011)
S. aureus
+ ive
LmrS, MFS
EtBr
Yes (DNA)
Role of LmrS in antimicrobial efflux
Floyd et al. (2010)
Mycobacterium tuberculosis
+ ive
MDR
EtBr
Yes (DNA)
Understanding efflux mediated drug resistance
Machado et al. (2018)
E. coli, Salmonella enterica serotype Typhimurium
- ive, - ive
TolC, AcrB and AcrF, RND
Hoechst 33342, EtBr
Yes (DNA)
Understanding efflux mediated drug resistance
Coldham et al. (2010)
Acinetobacter baumannii
- ive
AdeABC, RND
Hoechst 33342, EtBr
Yes (DNA)
Understanding efflux mediated drug resistance
Richmond et al. (2013)
S. enterica
- ive
AcrAB-TolC, RND
Hoechst 33342, EtBr, Nile red
Yes (DNA & membrane)
Efflux pump modulators as antibacterial agent
Reens et al. (2018)
E. coli
- ive
AcrAB-TolC, RND
1,2′- dinaphthylamine
Yes (membrane)
Role of phenylalanine residues in transport
Bohnert et al. (2011)
E. coli
- ive
AcrAB-TolC, RND
Nile Red
Yes (membrane)
Effect of different efflux inhibitors
Bohnert et al. (2010)
E. coli, S. enterica serotype
Typhimurium- ive,
- iveAcrAB-TolC, RND
Doxorubicin
No
Understanding efflux mediated drug resistance using mutants
Blair et al. (2015)
E. coli
- ive
EmrAB, RND
Doxorubicin
No
Function of multidrug resistance transporters
Nishino and Yamaguchi (2001)
E. coli,
Pseudomonas aeruginosa,
S. aureus
- ive,
- ive,
+ iveNon-specific efflux
Fluoroquinolones
No
Accumulation of quinolones in different bacteria
Piddock et al. (1999)
P. aeruginosa,
S. aureus
- ive,
+ iveNon-specific efflux
Fluoroquinolone, radio-labeled
No
Method comparision for measurement of quinolones accumulation
Mortimer and Piddock (1991)
E. coli
- ive
TolC, RND
Trimethoprim (TMP) probe
No
Understanding efflux mediated drug resistance using mutant E. coli (ΔtolC)
Phetsang et al. (2016)
E. coli (proteoliposomes)
- ive
AcrAB, RND
1-amino-naphthalene-3,6, 8 trisulfonate
No
Function of multidrug efflux
Zgurskaya and Nikaido (1999)
P. aeruginosa
- ive
MexAB-OprM
Nile Red
Yes (membrane)
Effect of glucose in efflux
Iyer and Erwin (2015)
Enterobacter aerogenes
- ive
Non-specific efflux
1,2′- dinaphthylamine
Yes (membrane)
Inhibition of antibiotic efflux by inhibitors
Brunel et al. (2013)
The current study provides an adaptation of an already existing EtBr fluorescence assay to monitor activity of microbial efflux pumps in several bacterial pure cultures and harvested cells from natural environments. Sediments of saline environments, and hospital drain soil were tested as proof of concept for rapid analysis of environmental samples.
2 Materials and methods
2.1 Bacterial strains and environmental samples
Bacterial strains with well-known active efflux pump physiologies were used to optimize the assay. This includes a Gram negative multidrug resistant (including tetracycline) strain isolated from poultry fecal samples (Banerjee et al., 2018); The second bacterium is a chromium reducing Bacillus cereus strain TCL, isolated from coal mines that use active efflux pump to release the heavy metal from cytoplasm (Banerjee et al., 2019). The third one is a moderately halophilic, Gram negative Halomonas sp. (optimum 6% NaCl requirement) isolated from Sambhar salt lake, Rajasthan. This strain was isolated in course of a recent study to isolate 1-ethyl-3-methylimidazolium chloride ([Emim]Cl) tolerant microorganisms (manuscript communicated elsewhere). These imidazolium based ionic liquids are presently the most preferred solvents for lignocellulosic biomass pretreatment in biofuel production and other industries (Dadi et al., 2006; Khudyakov et al., 2012). In a recent study microbial ionic liquid tolerance was found to be mediated by an MFS efflux system (Ruegg et al., 2014). In fact, Halomonas sp. could also tolerate up to 5.5% [Emim]Cl and active efflux pumps was found to be involved in the process (manuscript communicated elsewhere).
In addition, sediments from saline environments, and soil from antibiotic contaminated hospital drain, both expected to harbor microbial communities with active efflux systems were used. Saline sediments were collected from mangrove forests of Sunderban, West Bengal, India with 2.5% salinity; and those with 3% and 7% salinity from two different locations of Sambhar salt lake, Rajasthan, India. Sambhar lake is the largest inland saline water body in India that remains dry for major part of the year and thus, sediments from different segments vary in their salinity from 2% to 35% (manuscript communicated elsewhere). Antibiotic contaminated drain soils were sampled from two different nearby hospitals and compared to an adjoining garden soil as control.
2.2 Growth conditions
All experiments with bacterial pure culture were performed in heterotrophic complex media, Luria-Bertani (LB) in aerobic shaking condition at 37 °C, except for Halomonas sp. where the LB media was supplemented with 6% NaCl (or as stated in the figure), and 0.2% KCl (Schneegurt, 2012); and pH was adjusted to 8.0 instead of 7.0. Halomonas sp. has higher pH requirements as the bacterium was isolated from Sambhar salt lake with pH 8–9. In addition to NaCl (4%, 6%, 10% and 14%), efflux activity in Halomonas sp. was also tested in sets with [Emim]Cl (1.8%) and optimum NaCl (6%). For B. cereus TCL, 100 µg ml−1 Cr(VI) was used. The strain could very efficiently reduce 200 µg ml−1 of Cr(VI) within 16 h with efficient efflux activity (Banerjee et al., 2019). The commonly used concentration of tetracycline, i.e., 20 µg ml−1, was used for efflux assays, and also for growth, and enumerations. For enumeration of tetracycline resistant bacteria, hospital and garden soils were suspended in PBS buffer and appropriate dilutions spread on antibiotic-supplemented LA plates.
2.3 Processing of environmental samples
Microbial cells in soil or sediments were harvested prior to the assay following a standardized protocol (Sar et al., 2018). Briefly, 5 g sample suspended in 15 ml of 50 mM sodium phosphate buffer was vortexed for 5 min and centrifuged at 1000 g for 10 min to collect the supernatant containing viable cells. Pellet fraction was dissolved again in 15 ml buffer and the steps were repeated to harvest the remaining cells (if any). The pooled 30 ml supernatant was passed through autoclaved filter paper, and the filtrate was centrifuged at 5000 g for 10 min. The pellet fraction containing harvested cells was then used for the assay. When processing sediments from Sambhar salt lake and Sunderban mangroves, NaCl at a concentration equivalent to sample salinity (i.e., 2.5% for Sundarban, and 3%, and 7% for Sambhar) was added to maintain cell stability and viability.
2.4 Efflux assay
20 mg (wet weight) cells from either mid-log grown (OD600 = 0.6) pure culture(s) or harvested from environmental sample(s) were dissolved in 2 ml of 50 mM sodium phosphate buffer and used. To this different substrates that trigger efflux pumps in the respective bacteria or environmental samples was added, like tetracycline (20 μg ml−1) for poultry fecal isolate and hospital drain samples; NaCl for Halomonas sp. (4%, 6%, 10% and 14%) and saline sediments (3%, or 7%); [Emim]Cl (1.8%) for Halomonas sp.; and Cr(VI) (100 μg ml−1) for B. cereus TCL. Finally, glucose (0.4%) and EtBr (final concentration of 0.5 µg ml−1) were added to be used as the energy source (Iyer and Erwin, 2015) and non-specific substrate respectively by the induced efflux pumps. Fluorescence emission was monitored immediately for 30 min in a fluorescence spectrophotometer (Hitachi F-7100, Japan) with excitation and emissions at 520 nm and 590 nm respectively (Ferrari and Peracchi, 2002; Tokunaga et al., 2004). The optimized EtBr fluorescence assay to monitor efflux activities operative in diverse groups of bacteria either in pure form or in cell harvests from environmental samples is represented in a flowchart (Fig. 1).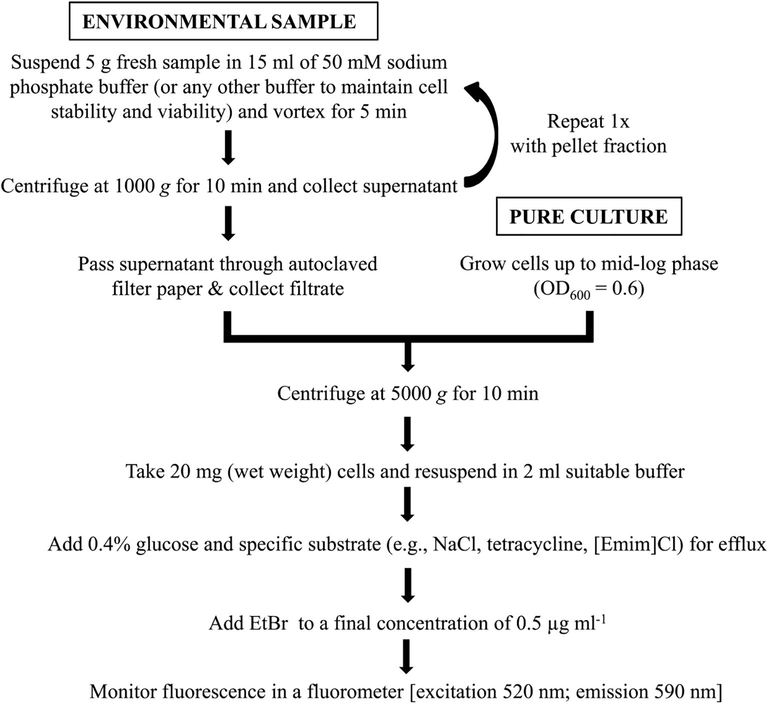
Flowchart presenting steps involved in the EtBr fluorescence assay to monitor activity of efflux pumps in bacterial pure cultures or mixed population from environmental samples.
3 Results and discussion
3.1 EtBr fluorescence assay can be used to monitor diverse efflux activities in bacteria
Fluorescence assay using EtBr is a well-established method to monitor efflux activities in bacteria. The assay was used to establish its efficiency in different efflux pump mediated microbial physiologies like, tetracycline resistance in a Gram-negative isolate from poultry fecal sample; chromium resistance in B. cereus TCL, a Gram positive bacterium from coal mines; and salt and ionic liquid tolerance in Halomonas sp., a Gram-negative, moderate halophile. Their minimum inhibitory concentrations (MIC) (or the lowest concentration with no growth) towards the non-specific fluorescent substrate, i.e., EtBr, is 25, 20 and 9 µg ml−1 respectively. Thus, any concentration of EtBr that is less than half of these values could be used in the respective strains. However, we need to determine the minimum concentration that would work without losing any signal since we want to exploit the assay directly on environmental samples, where MIC determination is not possible. Thus, we checked 0.25, 0.5 and 1.0 µg ml−1 EtBr concentrations in assays with Halomonas sp., which have the lowest MIC. While 0.5 and 1.0 µg ml−1 EtBr resulted in similar fluorescence, use of lower concentrations (0.25 µg ml−1) leads to loss of signal even in control set. Thus, 0.5 µg ml−1 was selected and used for all assays with pure cultures and environmental samples. Similar loss of fluorescence signal was reported with lower EtBr concentrations (<0.50 µg ml−1) in a transporter mutant E. coli K12 strain (Paixao et al., 2009).
Involvement of active efflux pumps in all four tested physiologies, namely, resistance/tolerance to tetracycline (20 μg ml−1), Cr(VI) (100 μg ml−1), NaCl (low: 4%, 6%; high: 10%, 14%), and [Emim]Cl (1.8%) in different pure cultures was established by a 1.2, 1.4, 2.2, and 1.7-fold lower fluorescence (i.e., higher efflux activity) in the respective treated sets compared to the controls (Fig. 2a–d). Among the three strains, efflux activities in Halomonas sp. was higher for both salt and [Emim]Cl tolerance. Active efflux pumps have previously been reported to be associated with tetracycline (Speer et al., 1992) and chromium resistance (Banerjee et al., 2019; Elangovan et al., 2006; Mishra et al., 2012). Even such pumps are reported to be induced by NaCl (Tokunaga et al., 2004). But, effective use of the assay to detect involvement of efflux pumps in [Emim]Cl tolerance suggests that the assay can be used for other unexplored microbial physiologies that involve active efflux pumps. Or in other words, the assay can also be extended to see if a microbial physiology involves active efflux pump or not.![EtBr efflux assay to monitor active efflux pumps operative in bacterial pure cultures. Fluorescence emission from cells of a) a Gram negative multidrug resistant strain in 0 (control) or 20 µg ml−1 tetracycline; b) B. cereus TCL grown in 0 (control) or 100 µg ml−1 Cr(VI); c) Halomonas sp. in 4%, 6%, 10%, and 14% NaCl; and d) Halomonas sp. in 6% NaCl (optimum requirement, control) or 6% NaCl plus 1.8% 1-ethyl-3-methylimidazolium chloride, [Emim]Cl. AU, arbitrary unit. Graphs are representative of three independent time scan measurements.](/content/185/2020/32/1/img/10.1016_j.jksus.2019.06.002-fig2.png)
EtBr efflux assay to monitor active efflux pumps operative in bacterial pure cultures. Fluorescence emission from cells of a) a Gram negative multidrug resistant strain in 0 (control) or 20 µg ml−1 tetracycline; b) B. cereus TCL grown in 0 (control) or 100 µg ml−1 Cr(VI); c) Halomonas sp. in 4%, 6%, 10%, and 14% NaCl; and d) Halomonas sp. in 6% NaCl (optimum requirement, control) or 6% NaCl plus 1.8% 1-ethyl-3-methylimidazolium chloride, [Emim]Cl. AU, arbitrary unit. Graphs are representative of three independent time scan measurements.
3.2 EtBr fluorescence assay is effective to monitor efflux activities in environmental samples
Natural sediments from saline environments, and antibiotic exposed soils were used to validate efficacy of the assay. However, natural samples when used directly for the assay did not yield any fluorescence, quite presumably due to some inhibitors/interferences present therein. Nonetheless, incorporation of a simple cell harvest step yielded the desired results. The harvest protocol relies on the fact that viable cells would remain in solution and has been previously shown to retain up to 94% of the original cell number in different soil samples (Sar et al., 2018). Cells harvested from Sambhar salt lake sediments with 3% salinity produced 1.7-fold higher fluorescence (low efflux) compared to those with 7% salinity (Fig. 3). However, those from sediments of Sunderban mangroves with only 2.5% salinity showed efflux activities similar to the higher (7%) salinity samples of the salt lake. The possible reason might be the higher nutrient availability in mangrove sediments that supports growth of diverse groups of microorganisms with varying efflux activities.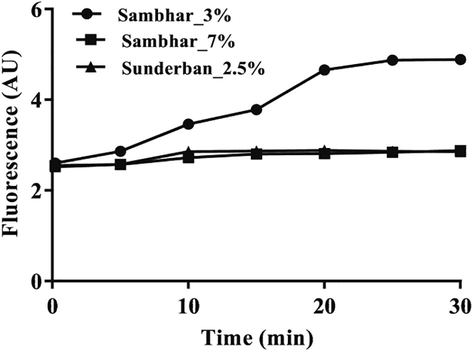
Fluorescence emission from harvested microbial cells of saline sediments of Sambhar lake (3%, and 7% salinity) and Sunderban (2.5% salinity). AU, arbitrary unit. Graphs are representative of three independent time scan measurements.
Similarly, antibiotic exposure in hospital drains is supposed to trigger efflux activities in the residing microbial population. In fact, drain soils from two different hospitals have 1.2 to 1.3-fold lower emission of EtBr fluorescence (i.e., higher activity) than those from a nearby garden soil (Fig. 4a). The observed higher efflux activities in the two hospital drain soils was supported by their higher proportions (6.9% and 7.2%) of tetracycline resistant colonies than the garden soil (0.6%) (Fig. 4b).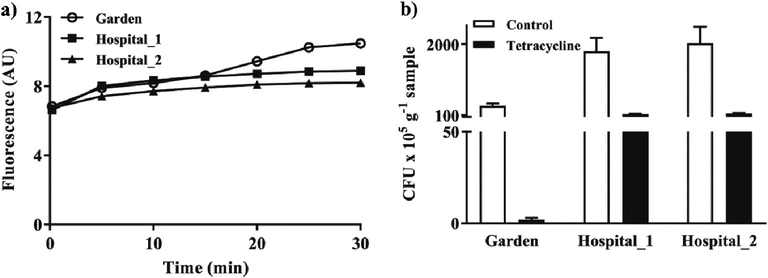
a) Fluorescence emission by microbial cells harvested from antibiotic-exposed drain soil from two hospitals as compared to those from a non-exposed adjoining garden soil. AU, arbitrary unit. Graphs are representative of three independent time scan measurements. b) Microbial load in the three soils on heterotrophic media and one supplemented with tetracycline (20 µg ml−1). Error bars represent standard deviation of three independent replicates.
4 Conclusion
Thus, EtBr fluorescence assay is a quick, reliable and cost-effective method and its spectrum of use was extended to both Gram positive and negative bacterial pure cultures for known or even new efflux mediated physiologies like ionic liquid tolerance. In addition, the assay was demonstrated to be effective in bacterial pure culture as well as mixed microbial populations directly harvested from natural soil/sediment/water samples. And the same holds true for at least two major efflux mediated activities tested, namely salt tolerance and antibiotic resistance. However, it must also be mentioned that the assay cannot be used as an absolute quantification of the tested efflux activity by all microorganisms present in an environment. Rather, it can be used as an efficient tool to compare such activities in different natural sites. Inclusion of efflux pump inhibitors in the assay will further confirm the process as reported earlier (Viveiros et al., 2008; Bohnert et al., 2010; Brunel et al., 2013). In addition, as natural samples contain different types of microorganisms with varying sensitivities towards EtBr, a very low concentration of the dye must be used that would not become toxic to most microbial groups. Thus, this simple assay can be efficiently used for future explorations of microbial efflux mediated activities in unexplored bacterial cultures and natural environmental samples.
Acknowledgements
This work was financially supported by Council of Scientific & Industrial Research (CSIR), India [No. 38(1410)/15/ EMR-II)] and West Bengal Department of Science and Technology (WB-DST), India [No. ST/P/S&T/5G-18/2017]. Srikanta Pal and Sohini Banerjee are thankful to CSIR for their fellowships.
Declaration of Competing Interest
The authors declare no conflict of interest.
References
- A Bacillus strain TCL isolated from Jharia coalmine with remarkable stress responses, chromium reduction capability and bioremediation potential. J. Hazard. Mater.. 2019;367:215-223.
- [Google Scholar]
- Increased productivity in poultry birds by sub-lethal dose of antibiotics is arbitrated by selective enrichment of gut microbiota, particularly short-chain fatty acid producers. Microbiology. 2018;164:142-153.
- [Google Scholar]
- AcrB drug-binding pocket substitution confers clinically relevant resistance and altered substrate specificity. Proc. Natl. Acad. Sci. USA 2015:3511-3516.
- [Google Scholar]
- How to measure export via bacterial multidrug resistance efflux pumps. MBio. 2016;7:e00840-00816.
- [Google Scholar]
- Bacterial multidrug efflux pumps: much more than antibiotic resistance determinants. Microorganisms. 2016;4:14.
- [Google Scholar]
- Optimized nile red efflux assay of AcrAB-TolC multidrug efflux system shows competition between substrates. Antimicrob. Agents Chemother.. 2010;54:3770-3775.
- [Google Scholar]
- Determination of real-time efflux phenotypes in Escherichia coli AcrB binding pocket phenylalanine mutants using a 1, 2′-dinaphthylamine efflux assay. PLoS One. 2011;6:e21196
- [Google Scholar]
- Polyamino geranic derivatives as new chemosensitizers to combat antibiotic resistant Gram-negative bacteria. Bioorg. Med. Chem.. 2013;21:1174-1179.
- [Google Scholar]
- A 96-well plate fluorescence assay for assessment of cellular permeability and active efflux in Salmonella enterica serovar Typhimurium and Escherichia coli. J. Antimicrob. Chemother.. 2010;65:1655-1663.
- [Google Scholar]
- Enhancement of cellulose saccharification kinetics using an ionic liquid pretreatment step. Biotechnol. Bioeng.. 2006;95:904-910.
- [Google Scholar]
- Assembly and operation of bacterial tripartite multidrug efflux pumps. Trends Microbiol.. 2015;23:311-319.
- [Google Scholar]
- New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch. Microbiol.. 2003;180:155-160.
- [Google Scholar]
- A continuous kinetic assay for RNA-cleaving deoxyribozymes, exploiting ethidium bromide as an extrinsic fluorescent probe. Nucleic Acids Res.. 2002;30:e112.
- [Google Scholar]
- Enhancing tolerance to short-chain alcohols by engineering the Escherichia coli AcrB efflux pump to secrete the non-native substrate n-butanol. ACS Synth. Biol.. 2013;3:30-40.
- [Google Scholar]
- LmrS is a multidrug efflux pump of the major facilitator superfamily from Staphylococcus aureus. Antimicrob. Agents Chemother.. 2010;54:5406-5412.
- [Google Scholar]
- Recent advances in multi-drug resistance (MDR) efflux pump inhibitors of Gram-positive bacteria S. aureus. Antibiotics. 2013;2:28-45.
- [Google Scholar]
- Direct measurement of efflux in Pseudomonas aeruginosa using an environment-sensitive fluorescent dye. Res. Microbiol.. 2015;166(6):516-524.
- [Google Scholar]
- Global transcriptome response to ionic liquid by a tropical rain forest soil bacterium, Enterobacter lignolyticus. Proc. Natl. Acad. Sci. USA. 2012;109:E2173-2182.
- [Google Scholar]
- Plasmid-determined tetracycline resistance involves new transport systems for tetracycline. Nature. 1978;276:90.
- [Google Scholar]
- Efflux activity differentially modulates the levels of isoniazid and rifampicin resistance among multidrug resistant and monoresistant Mycobacterium tuberculosis strains. Antibiotics. 2018;7:18.
- [Google Scholar]
- Role of calcium in the efflux system of Escherichia coli. Int. J. Antimicrob. Agents. 2011;37:410-414.
- [Google Scholar]
- Improving olefin tolerance and production in E. coli using native and evolved AcrB. Biotechnol. Bioeng.. 2015;112:879-888.
- [Google Scholar]
- Reduction of chromium-VI by chromium resistant lactobacilli: a prospective bacterium for bioremediation. Toxicol. Int.. 2012;19:25.
- [Google Scholar]
- A comparison of methods used for measuring the accumulation of quinolones by Enterobacteriaceae, Pseudomonas aeruginosa and Staphylococcus aureus. J. Antimicrob. Chemother.. 1991;28:639-653.
- [Google Scholar]
- Inhibition of antibiotic efflux in bacteria by the novel multidrug resistance inhibitors biricodar (VX-710) and timcodar (VX-853) Antimicrob. Agents Chemother.. 2004;48:4171-4176.
- [Google Scholar]
- Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev.. 2003;27:313-339.
- [Google Scholar]
- Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev.. 2012;36:340-363.
- [Google Scholar]
- Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J. Bacteriol.. 2001;183:1455-1458.
- [Google Scholar]
- Mechanism of ethidium bromide fluorescence enhancement on binding to nucleic acids. Biochemistry. 1977;16:3647-3654.
- [Google Scholar]
- Fluorometric determination of ethidium bromide efflux kinetics in Escherichia coli. J. Biol. Eng.. 2009;3:18.
- [Google Scholar]
- Fluorescent trimethoprim conjugate probes to assess drug accumulation in wild type and mutant Escherichia coli. ACS Infect. Dis.. 2016;2:688-701.
- [Google Scholar]
- Multidrug-resistance efflux pumps? Not just for resistance. Nat. Rev. Microbiol.. 2006;4:629.
- [Google Scholar]
- Quinolone accumulation by Pseudomonas aeruginosa, Staphylococcus aureus and Escherichia coli. J. Antimicrob. Chemother.. 1999;43:61-70.
- [Google Scholar]
- A cell-based infection assay identifies efflux pump modulators that reduce bacterial intracellular load. PLoS Pathog.. 2018;14:e1007115
- [Google Scholar]
- Efflux in Acinetobacter baumannii can be determined by measuring accumulation of H33342 (bis-benzamide) J. Antimicrob. Chemother.. 2013;68:1594-1600.
- [Google Scholar]
- Ethidium bromide transport across Mycobacterium smegmatis cell-wall: correlation with antibiotic resistance. BMC Microbiol.. 2011;11:35.
- [Google Scholar]
- An auto-inducible mechanism for ionic liquid resistance in microbial biofuel production. Nat. Commun.. 2014;5:3490.
- [Google Scholar]
- Isolation of high molecular weight and humic acid-free metagenomic DNA from lignocellulose-rich samples compatible for direct fosmid cloning. Appl. Microbiol. Biotechnol. 2018:1-13.
- [Google Scholar]
- Media and conditions for the growth of halophilic and halotolerant bacteria and archaea. In: Advances in understanding the biology of halophilic microorganisms. Springer; 2012. p. :35-58.
- [Google Scholar]
- Bacterial resistance to tetracycline: mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev.. 1992;5:387-399.
- [Google Scholar]
- Bacterial multidrug efflux pumps: mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun.. 2014;453:254-267.
- [Google Scholar]
- Salt-inducible multidrug efflux pump protein in the moderately halophilic bacterium Chromohalobacter sp. Appl. Environ. Microbiol.. 2004;70:4424-4431.
- [Google Scholar]
- Demonstration of intrinsic efflux activity of Escherichia coli K-12 AG100 by an automated ethidium bromide method. Int. J. Antimicrob. Agents. 2008;31:458-462.
- [Google Scholar]
- Improving heterologous polyketide production in Escherichia coli by transporter engineering. Appl. Microbiol. Biotechnol.. 2015;99:8691-8700.
- [Google Scholar]
- Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. USA. 1999;96:7190-7195.
- [Google Scholar]







