Translate this page into:
Actinomycetes mediated targeting of drug resistant MRSA pathogens
⁎Corresponding author. kkb@vit.ac.in (Krishnan Kannabiran)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The present study was to check the anti-Methicillin Resistant Staphylococcus aureus (MRSA) activity of actinomycetes isolated from terrestrial soil samples. The cell free supernatant and the ethyl acetate extract prepared from the potential isolate were studied for anti-MRSA activity. The culturing conditions of the potential isolate were optimized and characterized by polyphasic approach. The molecular taxonomy and phylogenetic studies of the potential isolate showed that it belonged to the genus Streptomyces. The partial 16S rDNA nucleotide sequencing analysis of the isolate yielded 1367 nucleotides and it was submitted to the GenBank under the accession number MF974561. Blast search of the nucleotide sequence of the isolate in the GenBank database showed 100% sequence similarity with Streptomyces gancidicus. The ethyl acetate (EA) extract showed 20 and 21 mm zone of inhibition against two MRSA strains (ATCC 43300) and (ATCC700699), respectively. Partial purification and GC-MS analysis of EA extract showed the presence of 1,1-Dichloropentane (DCP) (76%) as a major compound.
Keywords
Actinomycetes
Anti-MRSA activity
Cross-streak method
MRSA
Well diffusion method
1 Introduction
Antibiotic resistance by bacterial pathogens is considered as a major threat to mankind and infectious diseases are responsible for increased mortality worldwide (Lee et al., 2016). To overcome this problem several approaches have been explored and several classes of drugs are being tried constantly to manage and to control pathogenesis by these organisms. Drugs from natural sources are considered to be very effective to control these pathogens. Currently natural sources are explored for novel antibiotics and new chemical entities. Natural sources have provided several new chemical compounds and continued to be a good source for many active chemical compounds. There are still unexplored or under explored regions, which need to be explored for novel bioactive compounds. More and more drugs are needed from natural sources to combat drug resistant pathogens. Drug resistant pathogens are the major cause for the spreading of bacterial diseases, making the treatment ineffective and leading to the death of the patient. The mechanism of the resistance by pathogens is mainly due to the production of β-lactamases, up regulated efflux pumps and target site mutations (Sakoulas and Moellering, 2008). Methicillin resistant Staphylococcus aureus (MRSA) is one of the superbugs which are responsible for causing life threatening diseases in nosocomial settings. Presence of a large stretch of foreign DNA in MRSA isolates mec element and the presence of mecA gene encoding 76KDa penicillin-binding protein (PBP2a) is the major difference between MRSA and methicillin-sensitive S.aureus (MSSA). Clinical isolates exhibits methicillin resistant by expressing an altered penicillin binding protein 2 (PBP2) that has a lower penicillin binding affinity. MRSA strains express a foreign PBP2a responsible for the property of methicillin resistance. The rise of resistance worldwide has urged researchers to find novel antibiotics from natural sources for controlling the spread of infection. Marine microbes have contributed more than 10,000 compounds having several biological activities (Gad, 2017).
Actinomycetes are Gram positive filamentous bacteria, well known for its high G + C content and exist in wide variety of natural habitats of both marine and terrestrial environment (Yuan et al., 2010). Among the actinomycetes group, Streptomyces species are aerobic and spore forming bacteria belonging to the order Actinomycetales under phylum Actinobacteria. It is the predominant producer of various secondary metabolites and many of the commercially available antibiotics was produced by them (Kämpfer, 2012). Streptomyces was continues to be the good sources for several commercially important antibiotics, anticancer compounds, enzymes, enzyme inhibitors etc. (Goodfellow et al., 1988). The purified compound G60H from Streptomyces sp. was found to be effective against several S.aureus ATCC 25923, S.aureus ATCC 43300, S. aureus 38, S. aureus 39, S. aureus 636, S. aureus S1 and S. aureus R2 (Driche et al., 2015). In the present study Streptomyces sp. VITBKA3 isolated from terrestrial soil sample was studied for anti-MRSA activity.
2 Materials and methods
2.1 Isolation of actinomycetes
Terrestrial soil samples were collected from different locations of Kaliammanpet, Gudiyattham Taluk, Vellore district, Tamil Nadu, India. Soil sample was collected using sterile spatula in polyethylene bag and transported to the laboratory for further process. Soil samples were pretreated, serially diluted and seeded onto the Actinomycetes isolation agar (AIA) medium in different dilutions using spread plate technique. After inoculation the plates were kept at room temperature for 5–7 days. After 7 days specific actinomycetes colonies were sub-cultured by quadrant streaking on AIA and cultured for 5–7 days at 28 °C.
2.2 Screening of isolates
Cross streak method was used for primary screening of pure actinomycetes isolates for antibacterial activity by perpendicular streak method on Mueller Hinton Agar (MHA) medium. MRSA (ATCC 43300), MRSA (ATCC700699), Vancomycin resistant enterococci (VRE) (ATCC 51299), Staphylococcus aureus, Proteus vulgaris, Bacillus cereus and Escherichia coli were used as test organisms. Secondary screening was performed by agar well diffusion method.
2.2.1 Characterization of actinomycetes isolates
The potential actinomycetes isolates were characterized by morphological, biochemical and molecular taxonomic methods. The microscopic characterization was done by using cover slip culture method. The spore structure and surface morphology was analyzed using SEM.
2.2.2 Fermentation and preparation of EA extract
Seed culture was prepared using Tryptone yeast extract broth and inoculated in 1L Erlenmeyer flask was kept in rotary shaker for 7 days for incubation at 28° C. After 7 days, equal volume of ethyl acetate was mixed with ISP1broth containing potential isolate in 1L Erlenmeyer flask were kept at 28 °C for 24 h in shaker. After incubation the supernatant was separated using separating funnel. The supernatant collected was concentrated in rotary evaporator and the ethyl acetate (EA) extract was prepared as described earlier (Liu et al., 2011).
2.2.3 Assay of antibacterial activity
Agar well diffusion method was used for testing the antibacterial activity of actinomycetes. The test organisms were seeded on MHA medium and the wells were made using sterile well borer. The cell free supernatant and the EA extract of the potential isolate were loaded onto the wells separately and incubated at 37 °C for 24 h. After incubation the inoculated plates were observed for the clear zone of inhibition. All values are from three independent experiments.
2.3 Molecular characterization
The potential actinomycetes isolate, VITBKA3 cultured on ISP1 media was used for the extraction of genomic DNA by using Q1Amp DNA mini kit (Qiagen, Germany). The 16S rDNA was amplified by PCR using the universal forward and reverse primers, 8F (50-AGAGTTTGATC(A/C)TGGCTCAG-30) and 1392R (50-ACGGGCGGTGTGTACA-30). The PCR product obtained was purified by Nucleospin PCR cleanup gel extraction kit (Macherey—Nagel GmbH and Co. KG, Germany). The DNA obtained was sequenced by ABI3730xl sequencer (Applied Biosystems, USA). The 16S rDNA nucleotide sequence obtained was searched through the GenBank database for its similarity using BLAST search. Phylogenetic tree was constructed by neighbor joining method using MEGA software.
2.4 GC-MS analysis
The EA extract of the isolateVITBKA3 was analyzed in GC-MS JEOL (GCMATE II GC-MS, Agilent Technologies 6890N Network GC system for GC). The column (HP5) used was fused silica 50 m × 0.25 mm I.D. The column temperature were maintained at 100 °C for 20 min, 235 °C for 3 min and injector temperature was maintained at 240 °C. The carrier gas used was helium and the split ratio is 5:4. The sample (1 μl) was evaporated in a split less injector at 300 °C. Approximate run time will be around 30 min. The compounds present in the EA were then identified by Mass Spectra (MS) coupled with GC. The molecular mass and structure of the compounds were obtained by matching with the reference compounds listed in the National Institute of Standards and Technology (NIST) library.
3 Results
Terrestrial soil samples yielded fifty actinomycetes isolates. Starch casein agar (SCA) and AIA medium were used for the isolation of actinomycetes. When comparing to Starch casein agar medium Actinomycetes isolation agar was found to be the best medium for the isolation of actinomycetes.
3.1 Antibacterial activity of actinomycetes isolates
Out of 50 isolates, VITBKA3 showed Anti-MRSA activity against ATCC 43,300 and ATCC700699. The anti-MRSA activity was confirmed by both primary screening (cross-streaking method) followed by secondary screening (well diffusion method) according to the guidelines given by Clinical and Laboratory Standards Institute (CLSI, 2012). The cell free supernatant and the EA extract (100 µg) of VITBKA3 showed activity against both the MRSA ATCC strains (Table 1). The cross streak method of the isolate VITBKA3 showing activity against MRSA ATCC 43,300 and 700,699 is shown in Fig. 1. The zone of inhibition exhibited by EA extract of VITBKA3 is shown in Fig. 2.
Bacterial strains
Zone of inhibition (mm)*
Cell free supernatant
Ethyl acetate extract (100 µg)
MRSA (ATCC 43300)
20.3 ± 0.57
18.3 ± 1.52
MRSA (ATCC700699)
19.5 ± 0.57
0
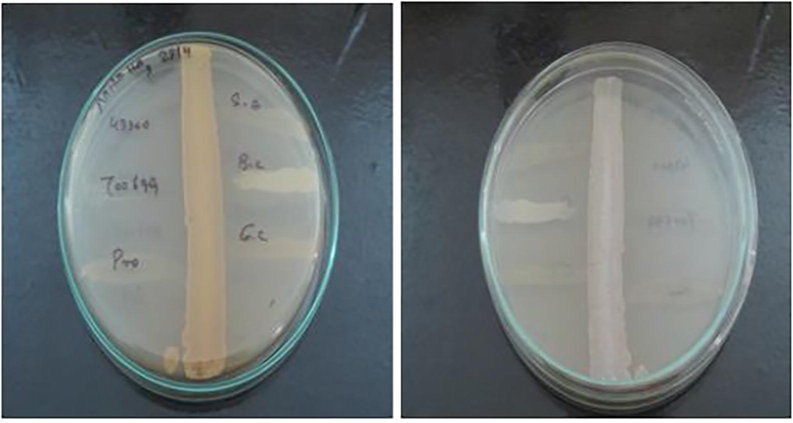
Anti-MRSA activity of VITBKA3against MRSA strain 43300 and 700699 by cross streak method.
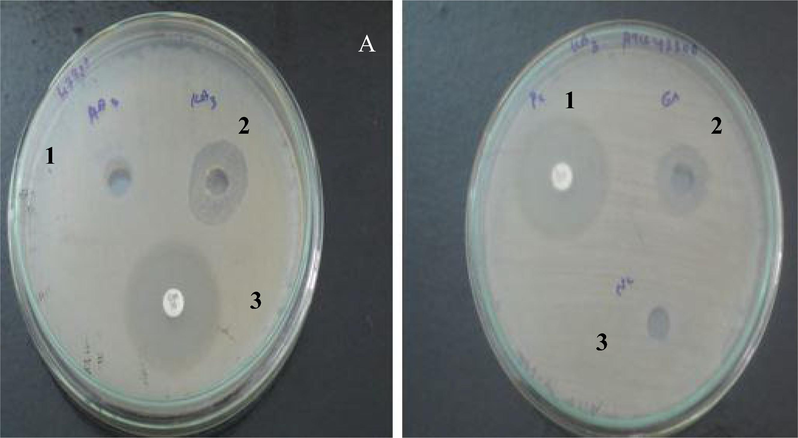
Anti-MRSA activity of VITBKA3 against MRSA strain (ATCC 43300).
A) Cell free supernatant of VITBKA3
B) Ethyl acetate extract of VITBKA3
1-VITAD3
1-Positive control (ciprofloxacin 10 µg)
2-VITBKA3
2-VITBKA3
3-Positive control (ciprofloxacin 10 µg)
3-Negative control
3.2 Morphological and biochemical characterization of the isolate
Morphological analysis using SEM showed that the spores of the isolate was coil like chains of cocci with smooth spore surface (Fig. 3) and also non-motile in nature. The isolate utilized all the carbon and nitrogen sources tested for its growth. AIA was found to be the optimal media for maximal growth. The growth was maximal at pH 7.0 and 28 °C with 3% NaCl concentration. The biochemical characteristics of the isolate are presented in Table 2. All the characteristics of the potential isolate indicate that it may belong to the genus Streptomyces. + – Positive, – – Negative, ++ – positive in 7 days, +++ – positive in 3 days.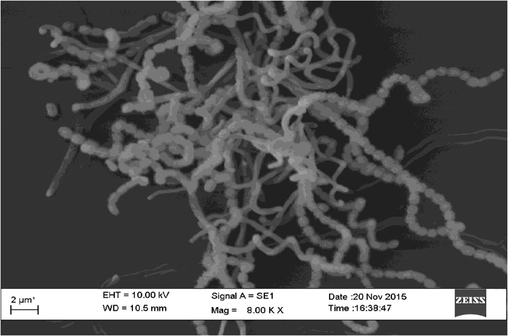
SEM image of the actinomycetes isolate VITBKA3.
Characteristics
VITBKA3
Indole test
–
Methyl red
–
Vogesproskauer
–
Citrate utilization test
–
Urea degradation
+
Triple sugar iron agar test
K/A (Alkaline slant, Acid butt)
Catalase test
+
Oxidase test
+
Mannitol motility
Non-motile
Carbon sources
Mannitol
+++
Inositol
+++
Cellulose
+
Salicin
+++
Arabinose
+++
Raffinose
+++
NaCl concentration
1%
+++
2%
+++
3%
+++
4%
++
5%
+
3.3 Molecular characterization
The taxonomy and phylogeny of the potential isolate indicate that the isolate comes under the genus Streptomyces and designated as Streptomyces sp. VITBKA3.The partial 16SrDNA sequence (1367 nucleotides) showed 100% sequence similarity with Streptomyces gancidicus. The partial 16S rDNA sequence was deposited in GenBank under the accession number MF974561. The phylogenetic tree shows the relationships between isolated actinomycetes and related strains. Percentage bootstrap values based on 1000 resampled data sets are shown at the nodes. The scale bar indicates 0.5 nucleotide substitution per nucleotide position (Fig. 4).
Phylogenetic tree of Streptomyces sp.VITBKA3.
3.4 GC-MS analysis
The partial characterization of secondary metabolites present in EA extract of VITBKA3 was done by GC-MS and the chromatogram obtained is shown in Fig. 5. The GC-MS spectrum showed the presence of 1, 1, Dichloropentane (76%) with the retention time of 2.79 and 5.04 as a major compound (Table 3) and 1,2,3, trimethyl cyclohexane (13%) with the retention time of 20.54.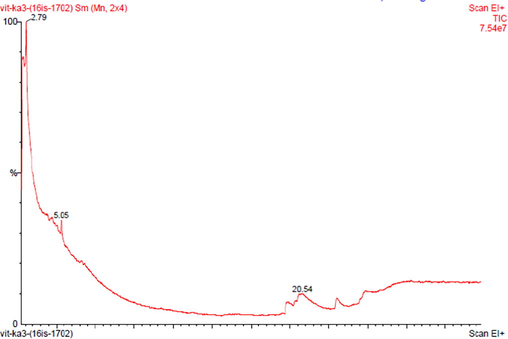
GC-MS spectrum of the ethyl acetate extract of Streptomyces sp.VITBKA3.
Retention time
Compound name
Structure
Area (%)
2.79 and 5.04
1,1-Dichloropentane
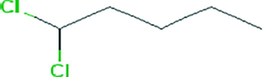
76
20.54
1,2,3,Trimethyl cyclohexane
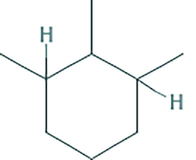
13
4 Discussion
The emergence of drug resistant bacterial pathogens has become a huge global threat all over the world making the treatment ineffective. Development of an effective control measures for the treatment and management of drug resistance is of current importance. In order to look for antagonistic agent terrestrial actinomycetes isolates were screened for antibacterial activity against the multi-drug resistant MRSA strains. Primary screening of Streptomyces sp. VITBKA3 indicated that it has the ability to inhibit the growth of two MRSA strains. The secondary screening (disk diffusion method) of EA extract of VITBKA3showed significant zone of inhibition against tested MRSA strains suggesting that the isolate produces bioactive compound. Many Streptomycesstrains were found to produce various secondary metabolites against various microbial pathogens. But few of them are only reported to have anti-MRSA activities. The results of GC-MS analysis of EA extract of Streptomyces sp. VITBKA3 showed the presence of 1, 1, Dichloropentane (76%) with the retention time of 2.79 and 5.04 as a major compound and 1, 2, 3, trimethyl cyclohexane (13%) with the retention time of 20.54. Several secondary metabolites produced by marine actinomycetes have been reported to be active against drug resistant bacterial pathogens such as MRSA and VRE strains (Ceylan et al., 2008). Few reports are available on anti-MRSA activity of actinomycetes and its compounds. Abyssomicin C, anti-MRSA compound from actinomycetes was reported by Bister et al. (2004). Laidlomycin obtained from Streptomyces sp. CS684 has been shown to be effective against MRSA and VRE strains (Yoo et al., 2007). Novel antibiotics, neocitreamicins I and II with activity against MRSA and VRE strains have been reported from Nocardia (Peoples et al., 2008). A partially purified compound (100 mg/ml) extracted from Streptomyces sp. showed 22 mm zone of inhibition against MRSA strains has been reported (Higginbotham and Murphy, 2010). Etamycin extracted from actinomycetes strain CNS-575 was reported to be active against hospital and community-associated MRSA (HA-and CA-MRSA) (Haste et al., 2010). Dichloromethane extracted from actinomycetes isolates (I-400A, B1-T61, M10-77) has been reported to exhibit strong antibacterial activity against MRSA (ATCC 43300) and vancomycin-resistant E. faecalis (ATCC 51299) (León et al., 2011). Antimicrobial compound 2, 4-dichloro-5-sulfamoyl benzoic acid (DSBA) extracted from marine Streptomyces sp. VITBRK2 have been reported to be active against MRSA and VRE strains (Rajan and Kannabiran, 2014). To overcome the drug resistance superbugs newer antibiotics are very much required. Hence, further probing of Streptomyces sp. VITBKA3 strain would certainly yield anti-MRSA compound.
Acknowledgements
The authors are thankful to the management of VIT for providing the necessary facilities to carry out this study.
Conflict of interest
The authors declare that there is no conflict of interest.
References
- Abyssomicin C—A Polycyclic Antibiotic from a Marine Verrucosispora Strain as an Inhibitor of the p-Aminobenzoic Acid/Tetrahydrofolate Biosynthesis Pathway. Angew. Chem. Int. Ed.. 2004;43(19):2574-2576.
- [Google Scholar]
- Isolation of soil streptomyces as source antibiotics active against antibiotic-resistant bacteria. Eur. Asia J. BioSci.. 2008;2:73-82.
- [Google Scholar]
- Clinical and Laboratory Standards Institute, 2012. M100-S22. Performance standards for antimicrobial susceptibility testing: 22nd informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA.
- A new Streptomyces strain isolated from Saharan soil produces di-(2-ethylexyl) phthalate, a metabolite active against methicillin-resistant Staphylococcus aureus. Ann. Microbiol.. 2015;65:1341-1350.
- [Google Scholar]
- Phylogenetic diversity and Anti-MRSA activity of halotolerant actinobacteria in great salt plains. Oklahoma 2017
- [CrossRef] [Google Scholar]
- Actinomycetes in Biotechnology. London: Academic Press Inc; 1988. p. :1-88.
- Activity of the streptogramin antibiotic etamycin against methicillin-resistant Staphylococcus aureus. J. Antibiot.. 2010;63(5):219.
- [Google Scholar]
- Identification and characterization of a Streptomyces sp. isolate exhibiting activity against methicillin-resistant Staphylococcus aureus. Micrbiol. Res.. 2010;165:82-86.
- [Google Scholar]
- Kämpfer, P., 2012. Genus Streptomyces: the Actinobacteria. In: Goodfellow, M., Kämpfer, P., Busse, H.-J., Trujillo, M.E., Suzuki, K., Ludwig, W., Whitman, W.B. (Eds.), Bergey's Manual of System.
- Anti-multi drug resistant pathogen activity of siderochelin A, produced by a novel Amycolatopsis sp. KCTC 29142. Kor. J. Microbiol.. 2016;52(3)
- [Google Scholar]
- Study of marine actinomycetes isolated from the central coast of Peru and their antibacterial activity against Methicillin-Resistant Staphylococcus aureus and Vancomycin-Resistant Enterococcus faecalis. Rev. Peru Med. Exp. Salud Publica.. 2011;28(2):237-246.
- [Google Scholar]
- Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis.. 2011;52(3):18-55.
- [Google Scholar]
- Neocitreamicins I and II, novel antibiotics with activity against methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococci. J. Antibiot.. 2008;61(7):457.
- [Google Scholar]
- Extraction and identification of antibacterial secondary metabolites from marine Streptomyces sp. VITBRK2. Int. J. Mol. Cell Med.. 2014;3(3):130.
- [Google Scholar]
- Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin. Infect. Dis.. 2008;46:360-367.
- [Google Scholar]
- Production and biological activity of laidlomycin, anti-MRSA/VRE antibiotic from Streptomyces sp. CS684. J. Microbiol.. 2007;45(1):6-10.
- [Google Scholar]
- Actinopolymorpha cephalotaxi sp. nov, a novel actinomycete isolated from rhizosphere soil of the plant Cephalotaxus fortunei. Int. J. Syst. Evol. Microbiol.. 2010;60(1):51-54.
- [Google Scholar]







