Translate this page into:
Aberrant DNA methylation of CDKN2B gene in the Type 1 diabetes mellitus and analysis of epigenetic markers
⁎Corresponding author at: V.K. Gopalakrishnan Professor, School of Medicine, Bule Hora University Institute of Health, Bule Hora University, BuleHora, Ethiopia. Department of Biochemistry, Centre for Plant Tissue Culture and Central Instrumentation Facility, Karpagam Academy of Higher Education, Coimbatore 641 021, Tamil Nadu, India. vkgopalakrishnan@gmail.com (V.K. Gopalakrishnan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Introduction
Aberrant DNA methylation is one of the important molecular markers in acute lymphoblastic leukemia (ALL).
Objectives
To determine Aberrant DNA methylation profiles of the CDKN2A and CDKN2B genes in ALL samples from T1DM cases.
Results
Aberrant hypermethylation of CDKN2B gene and cytogenetic abnormalities were analyzed in >60% of subjected cases. Total cholesterol, triglycerides and low-density lipoprotein cholesterol were not significantly varied between experimental and control (p >0.05). Expression analysis of human peripheral blood mononuclear cells revealed decreased mRNA levels of CDKN2B. The decreased level of p21 and CDK4 expression in the T1DM cases than control. The number of monocytes, lymphocytes and neutrophils significantly varied in T1DM samples and control (p < 0.05).
Conclusions
Aberrant methylation is one of the important mechanisms involved in the expression of CDKN2B genes and is an important prognostic marker for the early detection of ALL risks.
Keywords
DNA methylation
Aberrant
Expression
Diabetes mellitus
Prognostic markers
1 Introduction
Epigenetics is termed as mitotically heritable changes in the expression of genes and these changes do not affect the DNA sequences. These are considered non-genetic factors that generally interact with associated genes; however, they can be influenced by inherited genetic variation (Jerram et al., 2017). DNA methylation is regulated by the catalytic activity of DNA methyltransferases and the impaired function of DNA methyltransferases affects the normal DNA methylation process. DNA methylation is one of the important epigenetic modifications, and it is very important for silencing various imprinted genes, silencing retrotransposons and X-chromosome inactivation. It is involved in the expression of genes during individual tissues development and differentiation. Moreover, aberrant methylation is associated with disease response. DNA methylation is involved in the development of various diseases including cardiovascular diseases, diabetes, atherosclerosis, and tumours and is a reversible process (Kandi and Vadakedath, 2015).
Type 1 diabetes (T1D) results in decreased production of insulin and epigenetics plays a major role in disease susceptibility. In cancer, methylation silences various types of genes and it is generally considered that methylation of a promoter is very important for the process of gene silencing (Srivastava and Lodhi, 2022). The CDKN2A/B locus, at chromosome 9p21, associates high diabetes risk across various geography and ethnicities. In malignancies, CDKN2B (p15) (tumour suppressor gene) is frequently silenced (Xia et al., 2021). Methylation of CDKN2B gene was used as an important biomarker for acute myeloid leukaemia and myelodysplastic syndrome. The epigenetic changes do not alter the genetic code, however, it involved in the induction of gene activity and involved in pathogenesis in leukemia cases. Methylation of CpG islands considerably reduced the transcription of specific genes and it affects the expression of tumour suppressor genes. Hypo methylation has been frequently determined in malignant cells and is involved in genetic instability, increased proto-oncogene expression, and the complete role of methylation patterns in cancer is not elucidated (Rodrigues et al., 2010). There are several environmental and genetic factors associated with T1DM. Hence, analysis of human leukocyte antigen response is very important and the immune response is associated with T1DM pathogenesis (Pugliese, 2017). Previous functional studies linked the upregulation of CDKN2A and CDKN2B genes and increase the risk of T2DM (Hannou et al., 2015). The present study analyzes the immune systems function in T-ALL cases and expression changes of CDKN2A and CDKN2B genes.
2 Materials and methods
2.1 Samples
In this study, a total of 71 diabetic cases and 23 non-diabetic controls were evaluated between July 2020 and June 2021. All the participants involved in this study were confirmed using biochemical tests. The patients that had >110 mg/dL glucose concentration in the blood were considered diabetic and those patients who had <110 mg/dL glucose in the serum were considered as a non-diabetic group. T1DM was analyzed from the fasting plasma and the value ≥110 mg/dL or HbA1C value ≥6.5% was considered a normal or hyperglycaemic condition. The gender, age were collected from the clinical records and the mean age of the diabetic patients was 63 ± 2.3 years, including 53 males and 41 females. The biochemical analysis of serum was determined using clinical diagnostic kits and the result was expressed as mg/dL. Genomic DNA was isolated from the seven ALL patients (5 males and 2 females) and four control cases (2 males and 2 females). Bone marrow sample was obtained from ALL control and experimental subjects. The selected ALL cases were associated with diabetic disorder. All selected diabetic and non-diabetic cases were provided written informed consent.
2.2 DNA methylation of CDKN2A and CDKN2B genes
2.2.1 Extraction of DNA and bisulphite conversion
The blood sample was used for the extraction of genomic DNA using a Genomic DNA Kit (Qiagen, Germany) according to the manufactures instructions. The concentration and purity of the extracted DNA were performed by a NanoDrop spectrophotometer (Thermo Scientific). The quantified DNA has diluted appropriately for all experiments. About 400 ng DNA was used for bisulphite conversion using an EZ DNA Methylation–Gold Kit as described by the manufacture’s protocol (Zymo Research, USA). Bisulfite-converted DNA samples were finally stored at −20 °C until use.
2.2.2 Methylation–specific polymerase chain reaction
In this study, methylation of CDKN2A and CDKN2B genes was evaluated by methylation specific primers, as described earlier (Chen et al., 2017). Polymerase chain reaction products were used for the determination of the methylation process. The sequencing process revealed the confirmation of bisulphite conversion. The primer sequences of CDKN2A methylated primers were 5′–GTA GGG TTT AGA GTC GTT TCG A–3′ (forward) and 5′–AAC TAC AAA CTA AAA CCC ACG C-3′ (reverse). The unmethylated primers for CDKN2A were 5′–CGT AGG GTT TAG AGT TGT TTT GA–3′ (forward) and 5′–AAC TAC AAA CTA AAA CCC ACA CA–3′ (reverse). CDKN2B methylated primers were 5′–GCG TTC GTA TTT TGC GGT T–3′ (forward) and 5′–CGT ACA ATA ACC GAA CGA CCG A–3′ (reverse). Whereas, 5′–TGT GAT GTG TTT GTA TTT TGT GGT T–3′ (forward) and 5′–CCA TAC ATA ACC AAA CAA CCA A–3′ (reverse) primers were used for the amplification of unmethylated CDKN2B. The final PCR reaction volume was 20 μL, comprising forward and reverse primers (0.5 µL), bisulphate–converted DNA (1.6 µL), PCR mix (10 μL), DNase/RNase–free Millipore water (Chen et al., 2017).
2.3 Cytokines analysis and enzyme‑linked immunosorbent assay (ELISA)
ELISA test was performed for the determination of cytokines from the human plasma. The plasma blood was collected and centrifuged at 3000 rpm for 5 min. The centrifuged plasma was stored at 4 °C and blood parameters were determined. For ELISA assay, heparinised blood was used and the level of cytokines was analyzed using ELISA method (R&D Systems, UK).
2.4 Gene expression analysis by quantitative real‑time PCR (qPCR)
Gene expression analysis was performed on the diabetic and non-diabetic ALL and normal cases during the study period. For this study, human peripheral blood mononuclear cells were initially separated from the blood of individuals using a commercial kit and RNA was obtained (Qiagen, USA). The amount of RNA in the sample was quantified using a Nanodrop spectrophotometer (Thermoscientific) and 1.0 – 1.5 μg RNA was transcribed and cDNA was obtained using a cDNA kit. Then the DNA was amplified using a PCR machine. The expression analysis of CDKN2A and CDKN2B was performed. Endogenous control was used to normalize the mRNA levels.
2.5 Western blotting analysis
Human peripheral blood mononuclear cells (PBMCs) were aseptically lyzed in ice cold conditions using lysis buffer (Tris-HCl buffer, pH 7.5, 0.05 M, Tween 20 – 2%; NaCl- 0.2 M, and NP-40 0.2%) containing protease inhibitors. The protein sample was prepared and loaded on 12% SDS-PAGE and Western blotting analysis was performed as described earlier. The primary antibodies such as anti-CDK4, anti-p21 and anti-β-Actin and secondary antibodies (anti-mouse IgG-HRP and anti-rabbit IgG-HRP) were applied and the results were analysed (Vinue et al., 2019).
2.6 Statistical analysis
Data were described in the study as the mean ± standard deviation. The difference between experimental and control samples was compared by Student’s t-test. Analysis of variance was performed and the p-value <0.01 was considered significant.
3 Results
3.1 Clinical history of patients
The general characteristics of the study population were described in Table 1. The patients with >75 were highly susceptible to a diabetic than younger ages (<40). The relationship between the ages of diabetic patients was significant (p < 0.001). The blood sugar level was comparatively lower in females than males. About 5.3% of diabetic cases were associated with other complications.
Parameters
Diabetic
Non-Diabetic
p-value
Sex
Male
41
13
<0.01
Female
30
10
<0.01
Male (41–50 Years)
7
2
<0.001
Male (51–60 Years)
21
5
<0.0001
Male (61–70 Years)
8
5
<0.045
Male (71–80 Years)
5
1
<0.001
Female (41–50 Years)
5
1
<0.001
Female (51–60 Years)
7
5
>0.05
Female (61–70 Years)
13
2
<0.001
Female (71–80 Years)
5
2
>0.043
3.2 Clinical characterization of diabetic and normal cases
The biochemical parameters such as blood pressure, total cholesterol, HDL-C, LDL-C and triglyceride contents were determined among diabetic and normal cases. Diabetic cases were associated with increased systolic pressures than normal cases. Based on high pressure, the human subjects were categorized into two, viz, 41–60 years and 61–80 years. The amount of HDL-C was lower in diabetic cases than in control groups. Total cholesterol, triglycerides and LDL cholesterol were not significantly varied between the experimental and control (p >0.05). The biochemical parameters of diabetic patients and normal cases were described in Table 2. HDL-C – High density lipoprotein cholesterol, LDL-C- Low-density lipoprotein cholesterol. The value represents the average of three independent experiments.
41–60 years
61–80 years
Characters
T1DM
Control
T1DM
Control
Total Cholesterol (mg/dL)
169
174.2
173.2
176.4
HDL-C (mg/dL)
51.2
53.8
52.5
53.8
LDL-C (mg/dL)
109
110.2
112.2
115.9
Triglycerides (mg/dL)
69.7
73.3
74.1
75.6
3.3 DNA methylation of CDKN2B and CDKN2A genes
The methylation result of the CDKN2B was described in Fig. 1. A total of seven samples from ALL males and three from ALL females were methylated. CDKN2B gene was common among the patients and methylation was observed in male bone marrow tissues (M03, M09, M25, M33, M34; M39 and M42) and female bone marrow samples (F49, F54 and F63). DNA methylation was not observed in the selected specimens associated with CDKN2A.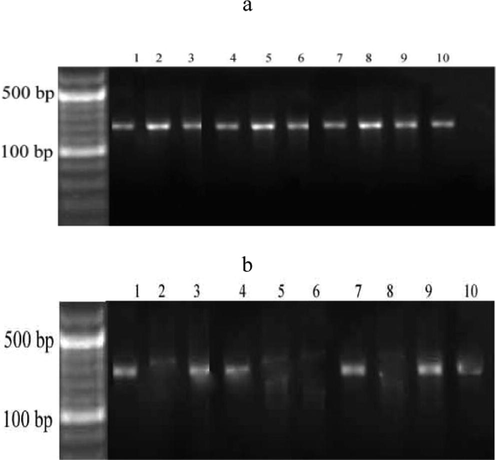
Methylation pattern of tumour suppressor gene, CDKN2B (a and b). A total of ten leukemia samples from seven males (1 = M03, 2 = M09, 3 = M25, 4 = M33, 5 = M34; 6 = M39 and 7 = M42) and three females (8 = F49, 9 = F54 and 10 = F63) were selected and methylation changes were analyzed.
3.4 Expression of CDKN2B levels in T1DM cases and biomarker analysis
Expression analysis revealed variation in the T1DM cases and control individuals. Expression analysis of human peripheral blood mononuclear cells revealed fewer mRNA levels of CDKN2B and the result was described in Fig. 2. Analysis revealed variations in the regulation of CDK4 and p21 protein expressions. These cell cycle inhibitors showed a decreased level of p21 and CDK4 expression in the T1DM cases than control. CDKN2A gene was negatively correlated with mRNA expression in diabetic cases associated with ALL-disease and normal individuals. The percentage of lymphocytes and monocytes was high in normal samples than in diabetic samples. However, neutrophils (%) were high in the diabetic group than control (Fig. 3). The circulating CD3 + marker level was high than CD4 + and was statistically significant (p < 0.01) (Fig. 4). The expression of CDK4 and p21 was analyzed and the results revealed variation in the expressed level in control and T1DM samples. Fig. 5a and b revealed mRNA expression in control and T1DM subjects. The cytokine levels in diabetic and control subjects were analyzed. The cytokine markers were high in normal cases than T1DM samples (Fig. 6).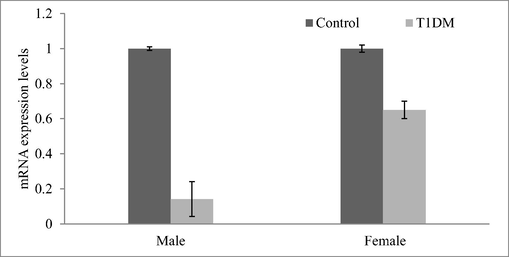
mRNA levels of CDKN2B from the tissues extracted from the T1DM patients and control individuals.
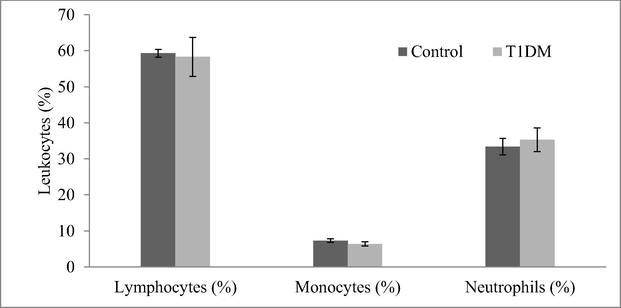
Analysis of leukocytes (%) in diabetic and control individuals. The number of monocytes, lymphocytes and neutrophils was determined in diabetic cases and control individuals. A student t-test was performed and the value < 0.01 was considered significant.
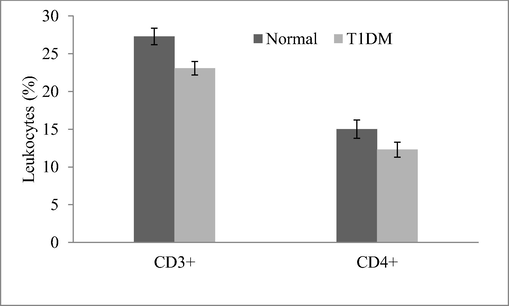
Analysis of circulating marker (CD3 + and CD4 + ) in ALL cases associated with the diabetic disease. The student t-test was performed and the value < 0.01 was considered significant.
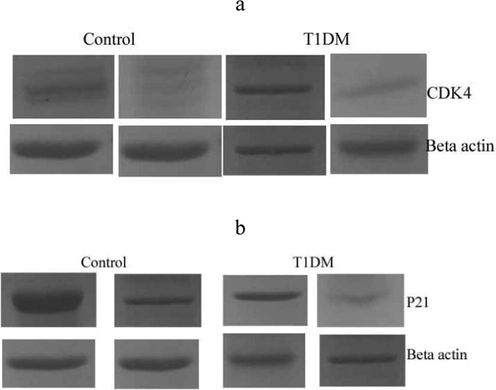
Analysis of human peripheral blood mononuclear cells from diabetic and control subjects. Protein expression of CDK4 (a) and p21 (b) from diabetic and control subjects. The expressed level of proteins was normalized to β-actin levels.
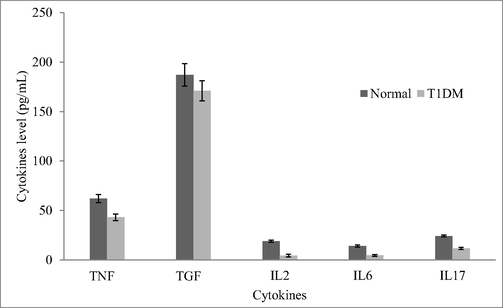
Analysis of circulating cytokine levels in diabetic and control subjects. The circulating cytokine level was assayed and the amount was expressed pg/mL blood.
4 Discussion
In humans, an abnormal methylation can indirectly affect the activity of genes with the specific disruption of the protein synthesis process by enhancing the possibility for mutational changes resulting in reduced chromosomal stability and dysfunction (Liu et al., 2022). Methylation changes of the CDKN2B were determined in this study and CDKN2B gene was more methylated than the CDKN2A gene. Methylation changes in the promoter region affect the functions of the cell growth regulator to inactivate the complexes (cyclin D/CDK4 and D/CDK6) in the G1 phase of the cell cycle. Inactivation of CDKN2B and CDKN2A was reported in neoplasia revealing methylation or homozygous deletion in the promoter region (Wong et al., 2000). In ALL and AML, hypermethylation of CDKN2B was reported. Our study was consistent with earlier ALL experiments, where >50% of samples were involved to have promoter-based methylation of CDKN2B. In certain studies, the percentage of CDKN2B methylation has been considerably less (<30%) (Roman-Gomez et al., 2004). Methylation of CDKN2B has also been reported in squamous cell carcinoma, diffuse large B-cell lymphoma, hepatocellular carcinoma and glioblastoma (Wemmert et al., 2009; Wong et al., 2003). In ALL patients, earlier reports showed various methylation frequencies for the CDKN2B gene and methylation was based on the selected methods (Chim et al., 2003). The unmethylated phase of the CDKN2B gene was observed in this study, which was in agreement with the study of Nephew and Huang (2003).
The expression changes of CDKN2B mRNA were determined in methylated samples, however, CDKN2A mRNA expression was not observed in this study. In the unmethylated specimens, the expression level and methylated CpG (%) were analyzed. The expression of the CDKN2B gene was considerably high than the CDKN2A gene. The fact of epigenetic gene silencing is that the rate of loss of protein biosynthesis is not almost uniform throughout the development of tumour cells and was distinct from genetic changes (Jones and Baylin, 2002). It has been reported that various factors are involved to bind CpG-containing sequences and these molecules failed to bind the methylated CpG sequences, therefore transcription process is significantly affected (Wong et al., 2000).
T1DM increased the risk of various diseases however the cause of risk has not been completely reported. In our study, we analyzed the changes in mRNA expression of the CDKN2B gene in circulating leukocytes of diabetes patients associated with ALL cases. The risk associated with CDKN2B inhibitor in ALL cases and the level of CD3 + and CD4 + marker changes in the T1DM cases revealed lower circulating levels in the blood. The T-cell function and circulating CD3 + and CD4 + reduced mRNA transcripts related to CD3 + and CD4 + markers. The number of cytokines such as TNFα, TGFβ, IL2, IL6 and IL17 were determined in control and diabetic cases associated with ALL. The amount of these cytokines reduced in T1DM patients than in normal cases revealed the lower expression of various transcription factors. The expression of the CDKN2B gene has been associated with various disease risks. CDKN2A and CDKN2B genes are linked with carbohydrate metabolism, β-cell islet biology and insulin secretion (Bochenek et al., 2013; Kong et al., 2018). T1DM cases showed decreased levels of cytokines in the blood sample and affected the expression of mRNAs associated with cytokines. Likewise, T1DM patients showed decreased amounts of cytokines TNFα, TGFβ, IL2, IL6 and IL17. The expression of the CDKN2B gene was correlated with mRNA expression and methylation in ALL cases. However, the association between the CDKN2B gene and methylation changes were not very clear. Recently, Song et al. (2020) reported non-coding RNA (CDKN2B-AS1) associated with various diseases. Aberrant expression of CDKN2B-AS1 linked with hepatocellular carcinoma, ovarian cancer, breast cancer, laryngeal squamous cell carcinoma and osteosarcoma, glioma, lung cancer, esophageal squamous cell carcinoma and gastric cancer. Recent studies also reported the potent association of CDKN2B-AS which is present within the p14, p15 and p16 gene cluster at 9p21 locus with cancers, cardiovascular diseases and diabetes (Golchin et al., 2017). The CDKN2A/B genomic locus is mainly linked with various metabolic diseases and human cancers.
5 Conclusions
In this study, we analyzed a total of 71 samples from diabetic cases diagnosed ALL positive and 23 samples were non-diabetic individuals associated with ALL. The expression pattern of CDKN2 was determined in this study and plays a significant role in the regulation of cytokines. The level of CDKN2 expression and translation influenced the regulation and apoptosis of the cell cycle pathway. Inactivation of CDKN2 may lead to the development and progression of cancers. The amount of circulating cytokines level decreased in ALL cases than in control individuals. A deeper knowledge of intracellular pathways, gene expression, cell signalling, and molecular mechanisms will helpful for the identification of new biomarkers, as well as novel therapeutic targets in the management of ALL patients with T1DM.
Acknowledgement
The authors thank Bule Hora University for the support and the technical analysis. The authors thank Department of Biochemistry and Cancer Research Center, FASCM, Karpagam Academy of Higher Education for the support. Researchers Supporting Project number (RSP2023R393), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- The large non-coding RNA ANRIL, which is associated with atherosclerosis, periodontitis and several forms of cancer, regulates ADIPOR1, VAMP3 and C11ORF10. Human Mol. Genetics. 2013;22(22):4516-4527.
- [Google Scholar]
- FOXF2 promoter methylation is associated with prognosis in esophageal squamous cell carcinoma. Tumor Biol.. 2017;39(2):1010428317692230
- [Google Scholar]
- Epigenetic inactivation of INK4/CDK/RB cell cycle pathway in acute leukemias. Ann. Hematol.. 2003;82(12):738-742.
- [Google Scholar]
- Analysis of two CDKN2B-AS polymorphisms in relation to coronary artery disease patients in North of Iran. Int. J. Mol. Cell. Med.. 2017;6(1):31.
- [Google Scholar]
- Functional genomics of the CDKN2A/B locus in cardiovascular and metabolic disease: what have we learned from GWASs? Trend. Endocrinol. Meta.. 2015;26(4):176-184.
- [Google Scholar]
- The fundamental role of epigenetic events in cancer. Nat. Rev. Genetics. 2002;3(6):415-428.
- [Google Scholar]
- Effect of DNA methylation in various diseases and the probable protective role of nutrition: a mini-review. Cureus.. 2015;7(8)
- [Google Scholar]
- ANRIL: a lncRNA at the CDKN2A/B locus with roles in cancer and metabolic disease. Front. Endocrinol.. 2018;9:405.
- [Google Scholar]
- DNA methylation in polycystic ovary syndrome: Emerging evidence and challenges. Reproduct. Toxicol.. 2022;111:11-19.
- [Google Scholar]
- Epigenetic gene silencing in cancer initiation and progression. Cancer Lett.. 2003;190(2):125-133.
- [Google Scholar]
- Epigenetic alterations of p15INK4B and p16INK4A genes in pediatric primary myelodysplastic syndrome. Leuk. Lymphoma. 2010;51(10):1887-1894.
- [Google Scholar]
- Promoter hypermethylation of cancer-related genes: a strong independent prognostic factor in acute lymphoblastic leukemia. Blood. 2004;104(8):2492-2498.
- [Google Scholar]
- CDKN2B-AS1: an indispensable long non-coding RNA in multiple diseases. Curr. Pharmaceut. Design. 2020;26(41):5335-5346.
- [Google Scholar]
- DNA methylation malleability and dysregulation in cancer progression: Understanding the role of PARP1. Biomolecules. 2022;12(3):417.
- [Google Scholar]
- Changes in CDKN2A/2B expression associate with T-cell phenotype modulation in atherosclerosis and type 2 diabetes mellitus. Transl. Res.. 2019;203:31-48.
- [Google Scholar]
- p15 promoter methylation-a novel prognostic marker in glioblastoma patients. Int. J. Oncol.. 2009;34(6):1743-1748.
- [Google Scholar]
- The study of p16 and p15 gene methylation in head and neck squamous cell carcinoma and their quantitative evaluation in plasma by real-time PCR. Eur. J. Cancer. 2003;39(13):1881-1887.
- [Google Scholar]
- Aberrant p15 promoter methylation in adult and childhood acute leukemias of nearly all morphologic subtypes: potential prognostic implications. Blood, J. Am. Soc. Hematol.. 2000;95(6):1942-1949.
- [Google Scholar]
- Dominant role of CDKN2B/p15INK4B of 9p21. 3 tumor suppressor hub in inhibition of cell-cycle and glycolysis. Nat. Commun.. 2021;12(1):2047.
- [Google Scholar]







