Translate this page into:
A brief overview to systems biology in toxicology: The journey from in to vivo, in-vitro and –omics
⁎Corresponding author. manraj@upm.edu.my (Manraj Singh Cheema)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Toxicology aims to comprehend and envisage the adverse outcomes of chemicals and xenobiotics on the biological system. Predicting toxicity on the biological system is a complex and immense process. In-vivo, in-vitro, pre-, clinical and –omics level experimental approaches have been utilized to describe the toxicological impact of these chemicals and this has generated a vast wealth of data. Hence, there now exist a need for a system that can interrelate and provide accurate and robust extrapolation of these data across various systems. Therefore, it is essential to re-shift our notion from empirical, animal-based testing to a mechanistic understanding. Systems biology is one such system that can extrapolate and interrelate these vast biological system data. Systems biology is a computational and mathematical modelling approach developed to understand interrelationships between networks of biological systems. The use of systems biology to answer toxicology-related questions is termed as systems toxicology. In this review we will look at the standard and classical toxicology experimentations and how we can use mechanistic data (systems biology) to answer toxicology-related questions using systems toxicology and what are the future opportunities in systems toxicology. The advancement of systems toxicology heralds new dawn of technologies that will aid in our quest to better comprehend and envisage the adverse outcomes of chemicals and xenobiotics on the biological system.
Keywords
In-vivo and in-vitro
Network biology
-Omics
Systems biology
Systems toxicology
Toxicology
- ADME
-
Absorption, distribution, metabolism and excretion
- AOP
-
Adverse outcome pathway
- cDNA
-
Complementary DNA
- DILI
-
Drug-induced liver injury
- DNA
-
Deoxyribonucleic acid
- NMR
-
Nuclear magnetic resonance
- PPI
-
Protein-protein interactions
- QST
-
Quantitative systems toxicology
- TetR
-
Tet repressor proteins
- UPS
-
Ubiquitin-proteasome system
Abbreviations
1 Introduction
Toxicity is demarcated as the adverse sequence of events that are triggered by exposure towards biological, physical or chemical agents (Klaassen and Amdur, 2013) and is manifested in various forms; from mild malfunctions to serious organ impairment and even death (Klaassen and Amdur, 2013). These reversible or irreversible events are influenced by the poison’s absorption, distribution, metabolism and excretion (ADME) properties, as well as the interactions of the toxicant or its metabolites with cellular macromolecules (Hodgson, 2004).
Toxicology encompasses the study of these events at entire biological system, ranging from molecules to complex ecosystems and requires extensive inter-disciplinary approaches. The question is whether such complex interaction(s) can be studied and mapped. The answer may lie in systems toxicology, where principles and methods of other disciplines; biology, chemistry, molecular biology, computer science and informatics are employed to better understand the resilience of biological system towards toxicants insults. The progressive advancement in genomics and systems-orientated perspective on biology has herald the shifting from empirical testing to a mechanistic understanding of toxicants perturbation. Understanding this systems-orientated perspective in biology involves gathering large sets of high-content technologies data and/or data from mining related literature, articles and databases, followed by recommending possible mathematical models that might be associated to this data set. Thereafter, numerical predictions are obtained from accurate computer simulation generated from these mathematical models and finally, the quality assessment of the models and numerical prediction with actual experimental data.
Hence, this review article aims to provide a guide and framework to new and novice researchers that are interested in applying systems biology in toxicological perspective. It provides an overview of the progression in the field of toxicology; from the in-vivo and in-vitro studies to vast ranges of tools such as the –omics as well as high data and information content screening, bioinformatics and systems biology to determine the changes on cells, tissues, and organisms upon chemical or xenobiotic exposure. Extra emphasis will be given on the latter portion.
Search strategies utilising the following bibliographic databases; Scopus, PubMed, Science Direct and EBSCOHost (Medline, Cumulative Index to Nursing and Allied Health Literature; CINAHL and Academic Search Complete) were developed using the selected keywords such as “in vivo”, “in vitro”, “animal testing”, “network biology”, “systems biology”, “mechanistic data”, “systems toxicology”, “-omics”, “toxicogenomics*”, “transcriptomics*”, “proteomic*”, “metabolomic*” and “synthetic biology”. Boolean operators, wildcards, exact, truncation and other commands were utilised whenever appropriate. The electronic databases searching across all databases was conducted on October 2019 and no limit was set for the evaluated time. Manual searches of reference lists of relevant articles was also performed and partial grey literature search was conducted using Google Scholar.
2 In-vivo and in-vitro toxicology
In-vivo toxicological studies are those in which various compounds and their entities are tested on whole living organisms; usually, non-human animals and plants whereas in-vitro toxicity studies is the scientific analysis on the effect of these compounds and their entities on a cultured mammalian or bacterial cells.
In the context of toxicology, the in-vivo and in-vitro models are often referred to as and continues to be the gold standard in the toxicity evaluation of chemicals (Pereira and Tettamanti, 2011). Data generated and gathered from these study models are usually extrapolated to the human biology system context to provide ‘safe’ exposure data for human usage and consumption but equally, it proves not to be without flaws and limitations (Pereira and Tettamanti, 2011; Fielden and Kolaja, 2008; Hartung and Daston, 2009). One of the biggest challenges in animal testing have been false negatives which by itself can be interpreted as could be nothing or even worse something even bigger (Hartung and Daston, 2009). Animal behavior, physiology and environment differ greatly from human and this may lead to false or skewered results (Hartung and Daston, 2009; Fielden and Kolaja, 2008). Though the in-vitro model addresses this issue of the animal to human extrapolation factor but fails to represent the physiology of a whole biological system (Hartung and Daston, 2009). Most cell system is representative of only one cell type, often monoclonal with no cell–cell interactions and this fails to mimic the normal human biological processes. The lack of the capability to undergo biotransformation is one of the best-known limitations of the in-vitro and the cancer origin of most cell culture adds to this problem (Hartung and Daston, 2009).
The rapid growth of understanding of mechanistic toxicology and its basis are not sufficiently simulated in the current animal testing and there is now a need to integrate mechanistic investigations in toxicity evaluation and this may be provided by the in-vitro model. The integration and combination of both of these models would probably have a better representation of the accuracy of toxicity estimations. One possible avenue that addresses this interaction is the –omics (Guttmacher and Collins, 2003). The rise of the genomic paved new avenues of exploration in better understanding the relatedness between the in-vivo and in-vitro models. It was the missing link that connects these two models through the use of various experimental approaches.
3 –Omics in toxicology
The integration of in-vivo and in-vitro studies with –omics studies would provide comprehensive and in-depth toxicological analysis. With Watson and Crick’s discovery of DNA structure in 1953 (Watson and Crick, 1953), appreciating the need to understand the genetic code’s translation to the function of certain gene and protein that may impact on the functioning and physiology of cells, organ and organism as a whole paved a new frontier for toxicity estimations (Hamadeh et al., 2002). Though this concept sounds basic and simple, however, the capability to fully comprehend the ability to translate the code to function is an enigma still challenging most of today’s scientists (Hamadeh et al., 2002; Afshari, Hamadeh, and Bushel, 2011). It requires many technology advances, collaborative sciences and integration of data information. Having said that, considerable progress has been made in regards to the integration of –omics with toxicology that creates a platform in assessing the cellular, tissue, organ and organisms’ wellbeing through the translation and interpretation of molecular distresses. The progression of these molecular techniques is then used in a whole-genomic capacity to the studies of the toxic effect of compounds is known as toxicogenomics ((Nuwaysir et al., 1999); Fig. 1). Toxicogenomics is the understanding of the molecular and cellular effects of toxicants in a whole biological system. It encompasses multidisciplinary sciences including informatics, biotechnology and engineering with traditional toxicological research (Hamadeh et al., 2002; Afshari, Hamadeh, and Bushel, 2011; Nuwaysir et al., 1999; Pennie et al., 2000).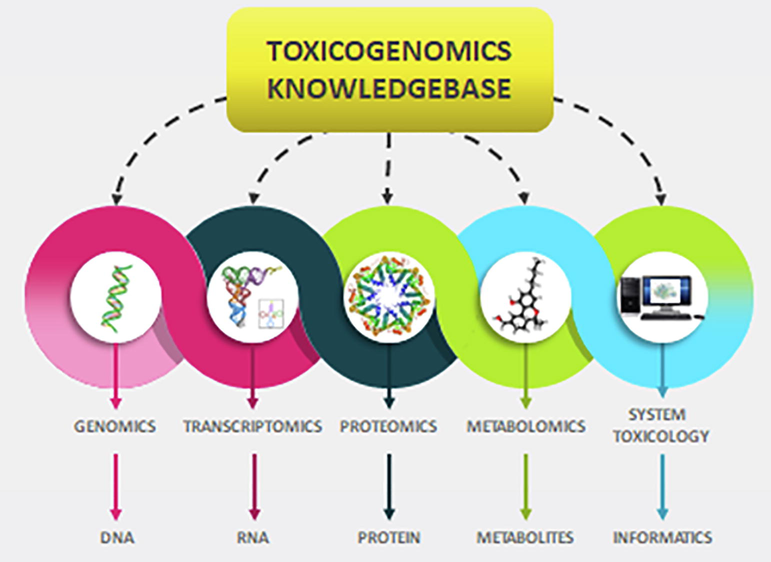
Schematic diagram showing flow of toxicogenomics knowledgebase. Toxicogenomics is the amalgamation of genomics (genome), transcriptomics (transcriptome), proteomics (proteins) and metabonomics (metabolites) knowledge in order to attain a comprehensive understanding of the effect of a particular chemical/toxicant on the biological system. Systems toxicology is created when networks of conventional toxicology along with quantitative analysis of large networks representing molecular and functional changes that are taking place across various levels of biological organisations are integrated. The knowledgebase serves as a library and computational system that carry out tasks to create new information and understanding by employing and integrating data, information and latest knowledge.
Typical toxicogenomics experiments usually observe the changes of transcripts across a genome ensuing cells and/or tissues to a chemical/compound insult (Fig. 2). Toxicogenomics data analysis can track several different paths which include discovery and comparison of classes and mechanistic analysis (Afshari, Hamadeh, and Bushel, 2011).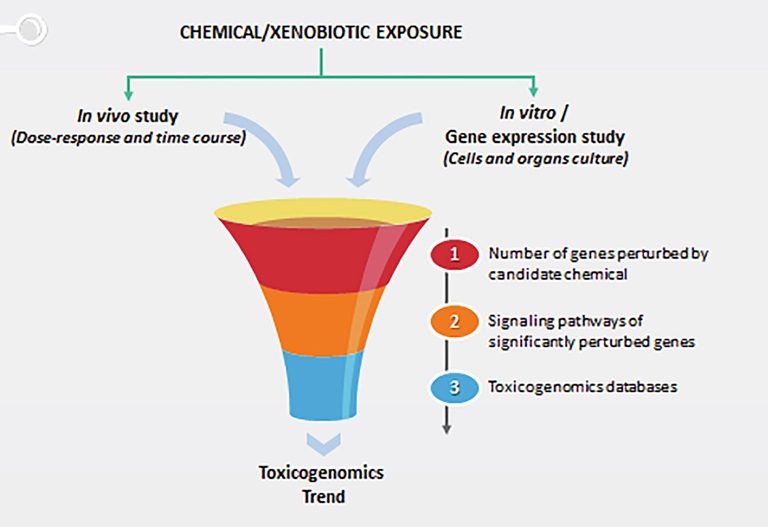
A flow scheme example of toxicogenomics. In-vivo and in-vitro models are exposed to varying chemical/xenobiotics doses and cells, tissues or organs are collected at different time points and thereafter subjected to various methods of experimental analysis. Experimental analysis is carried out to (1) determine numbers of genes altered in every sample in each organ in response to candidate chemical/xenobiotic and (2) mapping these observed changes into annotated pathways. Steps (1) and (2) offers preliminary assessment of potential pathways and mechanisms in response to candidate chemical/xenobiotic. Thereafter, (3) the potential pathways and mechanisms can then be compared and plotted against libraries of data to identify similarity of chemical/xenobiotic action/response to other previously determined chemical/xenobiotic. In addition, this allows the flexibility to analyse dose/time profiles either individually or as a trend.
Adapted from Afshari et al., 2011.
This standard method of evaluating RNA, proteins and metabolites is known as ‘–omic’ technologies (Fig. 1). These technologies are so-called based on their ability to characterize most if not all members of a family of molecules in a single analysis. With these new tools and technologies, complete assessments of functional activity of biochemical pathways, genetic sequences differences among individuals and species can be obtained. These methods of high-throughput and multi-endpoint analysis comprise of gene expression arrays which enable the expression of all human genes to be simultaneously measured on a single “chip” (Hamadeh et al., 2002; Afshari, Hamadeh, and Bushel, 2011; Nuwaysir et al., 1999; Pennie et al., 2000; Aardema and MacGregor, 2002; Waters and Fostel, 2004; Pognan, 2004; Hayes and Bradfield, 2005; Smetanová et al., 2015). Similarly, robust and powerful methods for protein analysis (proteomics: the large-scale study of proteins structure and functions; (Pognan, 2004; Anderson and Anderson, 1998; Blackstock and Weir, 1999; James, 1997; Wilkins et al., 1996) and for analysis of cellular small molecules (metabonomics: the study of the cellular metabolites and its intermediates formed and degraded under pathophysiological stimuli or genetic modification; (Nicholson, Lindon, and Holmes, 1999; Nicholson et al., 2002; Holmes, Wilson, and Nicholson, 2008); Fig. 1).
These technologies allows gathering of genomic-sequence data rapidly and its gene and protein footnote, thus allows for rapid analysis of gene-expression to better understand a toxicant mode-of-action on the biological system (Waters and Fostel, 2004).
Examples of such technologies are cDNA and oligonucleotide microarrays (Vermeeren and Michiels, 2011), protein chips (Yang et al., 2011) and molecular profiling by nuclear magnetic resonance (NMR; (Nelson, 2003) that permits simultaneous expression measurements of numerous genes, proteins, metabolites and its intermediates that accelerates toxicant pathways discovery, mode of actions and chemical and drug targets.
Therefore, there is a possibility of the data generated could be stored in a global depository through informatics (toxicoinformatics) and collectively analyzed to be able to offer a complete view of the genetic and biochemical machinery of the cell function and eventually the biological system. The integration of transcriptomics, proteomics and metabonomics data could add value to the expansion of toxicogenomics knowledgebase (Fig. 1) and eventually evolve to become systems biology (Waters and Fostel, 2004; Hartung et al., 2017; Kitano, 2002). This is due to the fact that current gene, protein and metabolite expression profile are just simple ‘snapshots’ at a particular time whereas in contrast, systems toxicology (as with systems biology) attempts to delineate the interaction of all of the elements that occurs in a specific biological system upon toxicant exposure, thus enabling to understand the toxicological responses on a mechanistic level (Kitano, 2002; Hartung et al., 2017).
The main challenge in toxicogenomics is the coordination and standardization across fields and disciplines that would ensure a better universal representation of data (Waters and Fostel, 2004). One possible solution is the progression and advancement of the systems biology that exploits computational and mathematical modelling of complex biological systems that would standardize the data from the –omics era.
4 Systems toxicology
Systems biology unifies studies of biological system at the molecular level (Waters and Fostel, 2004). It involves and encompasses perturbing system, monitoring molecular expression, integrating response data and modelling and displaying the molecular structure and network function of the system. When this concept is applied into toxicology context, it is then known as systems toxicology (Hartung et al., 2017; Kitano, 2002; Kongsbak et al., 2014; Slikker et al., 2007; Mc Auley et al., 2015; Plant, 2015). Systems toxicology describe the -omics as well as ‘classical’ endpoints evaluation of a biological system, including perturbation by toxicants and stressors, monitoring molecular expression and toxicological parameters and repetitively integrating response data to model and represent the archetypically toxicological system.
The wealth of in-vivo, in-vitro and –omics data is still not fully utilized as it does not lead to an accurate predictive power on the biological system and the robustness of these systems may be questionable. The understanding of the effects are usually done in a single system and broader perspective are merely determined by extrapolation. An accurate prediction and comprehensive integration capabilities are therefore required and systems toxicology aims to address this gap by incorporating computational approaches used in system biology in addressing toxicology-related problems. The computational approaches ranges from relational databases (repository of curated information and screening tools) and even potential digital organism (Plant, 2015) (Fig. 3).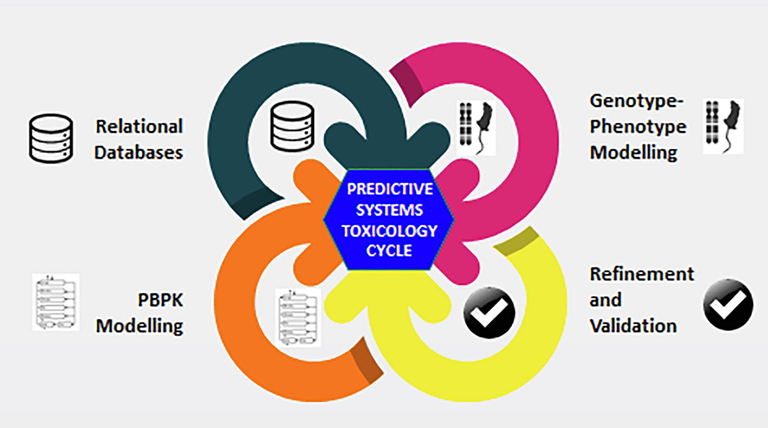
Safety assessment within the systems toxicology cycle. Systems toxicology approaches (PBPK modelling, relational databases, genotype-phenotype modelling) are further strengthened with advance experimental data (AOPs, exposomic, toxicoproteomics, toxicometabolomics data). Continuous cycle (note the arrows that are continuous and inter-related) of refinement and validation as well as revisiting assessment, review and addressing experimental gaps will further heighten the model’s predictive power that will provide great confidence for safety assessment.
Adapted from Plant, 2015.
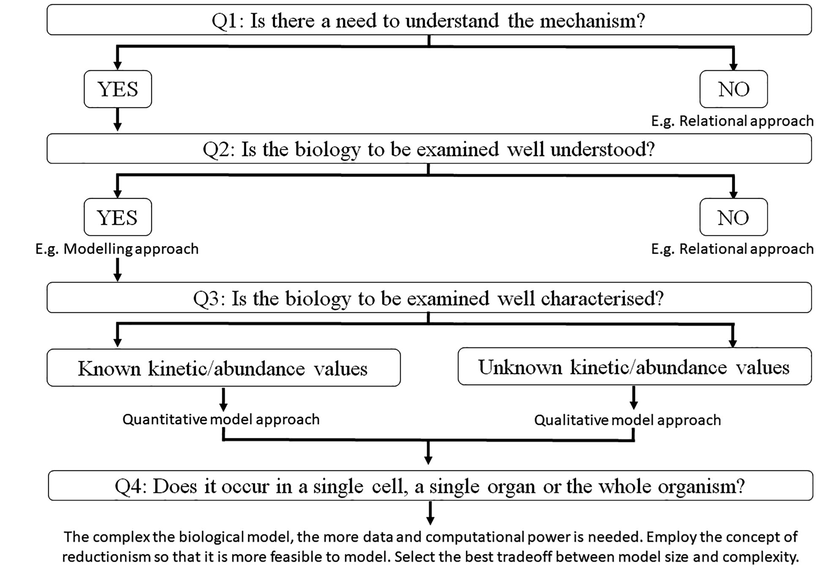
To novice researchers, systems toxicology may sounds complex and confusing due to countless available approaches. Using article by Plant, 2015 as a guide, this issue will be addressed here along with the available databases to assist researchers. The ultimate determinant of a successful application of systems toxicology is knowing and deciding the researcher requirements from this approach. These following questions will aid in selecting the best possible approach (Plant, 2015) (Fig. 4).
-
Is there a need to understand the mechanism?
-
Is the biology to be examined well understood?
-
Is the biology to be examined well characterised?
-
Does it occur in a single cell, a single organ or the whole organism?
-
Decision tree to define most appropriate approach(es) in systems toxicology. Firstly it is important to decide the aim of employing systems toxicology approach and the available data. There are many possible decision trees but are generally addressed by these 4 questions. A relational approach is most suitable is the answer to both Question 1 (Q1) and Question 2 (Q2) is a no. Relational approach allows for prediction of associations between network components without the need of mechanistic understanding. Modelling approach is applicable if the answer to Q2 is a yes, with Question 3 (Q3) and Question 4 (Q4) deciding which type of model can be used. Quantitative models are able to simulate biology in high precision while qualitative models are able to estimate the behaviour of the system but with low precision and accuracy. The determinant of which models to be used depends on data availability. The decision on Q4 depends on the tradeoff between model size and complexity as increase in model complexity requires tremendous biological data to make the model that produces a more accurate biology and vice versa.
The relational approach may be ideal if the answer to question I is a no. Relational approach (Table 1) analyses the association between predicted network components without requiring the comprehension of mechanistic footings. This may include protein–protein and/or chemical-protein interactions predictions by evaluating the toxicity of a chemical depending upon its structural fragments (Plant, 2015).
The relational approach would still be ideal if the answer to question II is also a no. If the answer is yes, then the best approach is a modelling approach, but this depends on question III and IV answers (Plant, 2015). To determine whether a quantitative or qualitative model approach will be employed, the answer to question III will be the determining factor. Quantitative approach (network connectivity and with kinetic/abundance values are known) allows the highest level of biology simulation accuracy in terms of predicting dose- and time- courses (Plant, 2015). Qualitative approach (known network connectivity but unknown kinetic/abundance values) forecast the system’s behavior with no fixed numbers. The decision to select quantitative or qualitative model approach heavily depends on the data availability (Table 2; (Plant, 2015).
DATABASES
WEBSITE
Network topology
BioCyc
https://biocyc.org/
KEGG
https://www.genome.jp/kegg/
MetaCyc
https://metacyc.org/
Enzyme nomenclature and annotation
ExplorEnz
https://www.enzyme-database.org/
HAMAP
https://hamap.expasy.org/
IntEnz
https://www.ebi.ac.uk/intenz/
TCDB
http://www.tcdb.org/
Protein abundance
PAXDB
https://pax-db.org/
Enzyme kinetics
BRENDA
https://www.brenda-enzymes.org/
SABIO-RK
https://www.h-its.org/projects/sabio-rk-biochemical-reaction-kinetics-database/
Cellular signalling pathway
NCATS BioPlanet
https://tripod.nih.gov/bioplanet/
Finally, the answer to question IV is vital in determining the complexity within the model by providing a degree of reductionism. As the model complexity increases (i.e. increase in biology representation/reproducibility), it becomes progressively strenuous to both the biological data that is necessary to make the model and the computational power necessary to run the model (Plant, 2015). Therefore, a compromise is required between the complexity of the biological system and sub-systems and model size, whereby higher complexity will reduce the model size and vice versa (Table 3).
RESOURCES
WEBSITE
Network
BiGG
http://bigg.ucsd.edu/
Cytoscape
https://cytoscape.org/
ERGO
https://www.igenbio.com/ergo
KEGG mapper
https://www.genome.jp/kegg/mapper.html
MetaCyc
https://metacyc.org/
MetaNetX
https://www.metanetx.org/
Model SEED
https://modelseed.org/
Pathway tools software
PDID
http://biomine.cs.vcu.edu/servers/PDID/index.php
STITCH
http://stitch.embl.de/
Transpath
http://genexplain.com/transpath/
yAPOPTOSIS
http://www.ycelldeath.com/yapoptosis/
The network-based focus of systems toxicology and the availability of large information data sets and modelling approaches have improved understanding of toxicological events and effects at multiple biological levels. Systems toxicology approaches have been utilized in risk assessment (Aguayo-Orozco, Taboureau, and Brunak, 2019; Hayes et al., 2019; Lanzoni et al., 2019; Sturla et al., 2014), safer design of chemicals and identification of safer alternatives (Voutchkova et al., 2010), drug design, discovery and development (Bloomingdale et al., 2017; Blomme and Will, 2016), toxicity prediction (Kiani et al., 2016) mostly in kidney (Ramm et al., 2019; Te et al., 2016), liver (Te et al., 2016; Longo et al., 2019; Battista et al., 2018) and heart (Li et al., 2018). Selecting optimal drug cocktails combination relies on the usage of such computational approaches as it necessitates a distinctive understanding of network biology behavior (Folger et al., 2011; Hopkins, 2008). In environmental toxicology, the establishment of adverse outcome pathway (AOP) concept and modelling approaches are used for the identification of hazards and defining risk assessments for the high number of environmental chemicals (Villeneuve et al., 2014a, 2014b). Moreover, systems toxicology is being employed to have a representative and collective understanding of diseases and treatments. For instance, systems toxicology highlighted a threshold dose suggestive of a long-term biphasic effect of formaldehyde exposure that leads to carcinogenicity (Martin et al., 2019). By using systems toxicology, an identification information framework of paracetamol/acetaminophen overdose was applicable for healthy and high-risk individuals as well as being able to define the quantitative estimation of liver injury probability was made possible (Mason et al., 2019). The same approach was also used to better comprehend the mechanisms of drug-induced liver injury (DILI) and provide evidences for reducing its risk (Peng et al., 2019). Implementation of quantitative systems toxicology (QST) modelling, mechanisms accountable several macrolide antibiotics with varying degree of toxicity; despite being within a class of drugs and having structural similarity were successfully elucidated (Woodhead et al., 2019). A systems toxicology including physiology, histology and molecular measurements approach was used to ascertain the effect of modified-risk tobacco products aerosols in comparison to conventional cigarette showed a reduction to almost sham exposure levels (Phillips et al., 2019; Smith et al., 2016).
Though systems toxicology provides to an extent a representative and collective data of toxicants acting on an organism has a whole, there are still several challenges and limitations to be addressed. By resolving these limitations, it will help in influencing how effectively these disciplines are constantly relevant in toxicology research. One main limitation is the calibration of the computational models since this aspect is important especially in toxicology as risk evaluation is of utter importance and extremely sensitive models weaken their potential effectiveness (Mc Auley et al., 2015; Plant, 2015). An optimization is required to make this model much more robust and recently research had ongoing to address this key limitation. Another limitation is the consideration of spatial scale that deals from the smallest cellular structure to the large whole organism. Many projects have embarked to resolve the problem but it remains in its infancy (Mc Auley et al., 2015; Plant, 2015).
To resolve this issue, a review provided a comprehensive listing of over 900 key in silico data resources relevant to ADME properties, animal, biological databases, chemical identity and properties, clinical trials, -omics, patents-related databases, pathways, pharmacovigilance, protein–protein interactions (PPIs), toxicology (including nano-material toxicity) amongst others. The paper highlights and suggested the need for a common platform for mapping and integration of databases to provide better accessibility and translatability (Pawar et al., 2019).
5 Future opportunities for systems toxicology
The advancement in systems toxicology is increasing exponentially and paved vast area of expansion of advancement that includes toxicoproteomics (Nasi, Picariello, and Ferranti, 2009; George et al., 2010), toxicometabolomics (Bouhifd et al., 2013; Lindon et al., 2005), AOPs modelling (Vinken, 2013; Ankley et al., 2010) and the exposomes (Wild, 2012; Rappaport, 2011; Miller and Jones, 2014) amongst others. One such current advancement is the emergence of synthetic biology (Mc Auley et al., 2015; Haynes, 2016; Osório, 2015). The purpose of synthetic biology is to design and engineer novel biological entities from genes (Aitken and Akman, 2013) and the generation of virtual organs that can be used to improve health (Polizzi, 2013). The toxicological aspects of synthetic biology are known as synthetic toxicology (Schmidt and Pei, 2011). Synthetically engineered biological gene circuits are designed to assess the effect of input has towards the output. These gene circuits will allow the impact of a stressor on transcription to be evaluated. An example of such circuit is the synthetic Deg-On system (a system consisting of two plasmids) that converts proteasomal degradation of TetR (transcriptional regulator) into a fluorescent signal, thereby linking ubiquitin–proteasome system (UPS) activity to an easily detectable signal (Zhao et al., 2014). This bioproduct will be able to screen for UPS activating molecules and will aid in selecting mammalian cells with different levels of proteasome activity. One such application is in toxicant studies as UPS activation can be employed as a mean of detecting toxicant exposure such as that of arsenic trioxide that deactivates the UPS system (Chiu et al., 2015). Synthetic biology in general and synthetic toxicology specifically offers the capacity to study cellular regulation and the pathways by re-designing bio parts, devices, and systems. In terms of challenges, it will generate hitherto unknown and unnatural molecules that need to be tested for their toxicity to natural and future synthetic biological systems (Behrendorff and Gillam, 2017; Polizzi, 2013; Aitken and Akman, 2013; Mc Auley et al., 2015).
Though the advancement in systems toxicology is heading in the right direction, it is equally important to address and acknowledge the existing challenges facing systems toxicology. Among the pertinent issues that need to be addressed are need of validation and scientific credibility, understanding the limitations of new and alternative methods, practicality and cost of methods, standardize and systematic test systems and improved computational models and modelling approaches, the need of robust, repeatable data-driven multicellular systems biology, linking network perturbations to phenotypes, uncertainties is systems toxicology computational models that allows for reliable predictive power, safe and secure sharing of high quality, multiscale datasets through curated public data libraries. Understanding and addressing these challenges in itself requires development of new algorithm and data integration methods. The success of this will ensure systems toxicology move toward regulatory acceptance, provided that systematic and coherent international standards are in place.
6 Conclusion
A key player in toxicological research is the need to predict the adverse outcomes of chemicals. The ability to fully delineate the biological phenotype describing an adverse outcome is a pre-requisite in understanding which chemical is responsible for eliciting a toxic response including the mechanism by which this is achieved. Perceptibly, the more accurate and robust downstream prediction is likely to be provided when more detailed descriptions are included in the analysis. However, the setback is with coming up with a rational explanation to relate data and to identify useful information from to the vast amount of available data. There now exist a paradigm shift in toxicological research that deviates from the –omics era towards that of the systems’ era (i.e. from generating data to understanding datasets). It is important to note that these deviations are not completely void from drug discovery/development and safety assessment paradigms but rather building upon massive experimental knowledgebase and integrating it into a more user-friendly and easily accessible format that will give rise to digital cells, organs and ultimately organisms. This will increase our ability to better understand the progression of diseases and to develop safer and more efficacious network-based drugs. Such a system would be a huge asset and advantage and would directly influence the way toxicity studies are conducted in the near future.
Acknowledgements
Special thanks to Nick Plant for providing the foundation for this review (Plant, 2015).
Funding
This review is a preliminary work from a research that was financially supported by the Fundamental Research Grant Scheme (FRGS/2/2013/SKK10/UPM/02/3; 2013), Ministry of Higher Education, Malaysia. F. A. is a recipient of Universiti Putra Malaysia’s graduate research fellowship.
Author contributions
Conceptualization, F. A. and M. S. C.; writing—original draft preparation, F. A. and M. S. C.; writing—review and editing, D. A. I. A., T. C. L., N, F. M. H. and M. S. C. The manuscript is submitted with the agreement and approval of all the authors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Systems Toxicology: Systematic Approach to Predict Toxicity. CPD. 2016;22(46):6911-6917.
- [CrossRef] [Google Scholar]
- Toxicology and genetic toxicology in the new era of “toxicogenomics”: impact of “-omics” technologies. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2002;499(1):13-25.
- [CrossRef] [Google Scholar]
- The Evolution of Bioinformatics in Toxicology: Advancing Toxicogenomics. Toxicol. Sci.. 2011;120(Supplement 1):S225-S237.
- [CrossRef] [Google Scholar]
- The use of systems biology in chemical risk assessment. Current Opinion in Toxicology. 2019;15:48-54.
- [CrossRef] [Google Scholar]
- Nested sampling for parameter inference in systems biology: application to an exemplar circadian model. BMC Syst. Biol.. 2013;7(1):72.
- [CrossRef] [Google Scholar]
- Proteome and proteomics: New technologies, new concepts, and new words. Electrophoresis. 1998;19(11):1853-1861.
- [CrossRef] [Google Scholar]
- Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem.. 2010;29(3):730-741.
- [CrossRef] [Google Scholar]
- Battista, Christina, Kyunghee Yang, Simone H Stahl, Jerome T Mettetal, Paul B Watkins, Scott Q Siler, and Brett A Howell, 2018. Using quantitative systems toxicology to investigate observed species differences in CKA-mediated hepatotoxicity. Toxicological Sci. 166 (1):123-130. DOI: 10.1093/toxsci/kfy191
- Prospects for Applying Synthetic Biology to Toxicology: Future Opportunities and Current Limitations for the Repurposing of Cytochrome P450 Systems. Chem. Res. Toxicol.. 2017;30(1):453-468.
- [CrossRef] [Google Scholar]
- Proteomics: quantitative and physical mapping of cellular proteins. Trends Biotechnol.. 1999;17(3):121-127.
- [CrossRef] [Google Scholar]
- Toxicology Strategies for Drug Discovery: Present and Future. Chem. Res. Toxicol.. 2016;29(4):473-504.
- [CrossRef] [Google Scholar]
- Arsenic trioxide induces programmed cell death through stimulation of ER stress and inhibition of the ubiquitin–proteasome system in human sarcoma cells. Cancer Lett.. 2015;356(2):762-772.
- [CrossRef] [Google Scholar]
- The role of early in vivo toxicity testing in drug discovery toxicology. Expert Opinion on Drug Safety. 2008;7(2):107-110.
- [CrossRef] [Google Scholar]
- Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol.. 2011;7(1):501.
- [CrossRef] [Google Scholar]
- Toxicoproteomics: New paradigms in toxicology research. Toxicol. Mech. Methods. 2010;20(7):415-423.
- [CrossRef] [Google Scholar]
- Hartung, Thomas, George Daston, 2009. “Are in vitro tests suitable for regulatory use?” Toxicological Sci. 111 (2):233-237. DOI: 10.1093/toxsci/kfp149
- Systems Toxicology: Real World Applications and Opportunities. Chem. Res. Toxicol.. 2017;30(4):870-882.
- [CrossRef] [Google Scholar]
- Hayes, A Wallace, Roman Li, Julia Hoeng, Anita Iskandar, Manuel C Peistch, and Michael L Dourson, 2019. “New approaches to risk assessment of chemical mixtures.” Toxicology Research and Application 3:2397847318820768. DOI: 10.1177/2397847318820768
- Hodgson, Ernest, 2004. A textbook of modern toxicology: John Wiley & Sons. ISBN: 0-471-26508-X
- Network pharmacology: the next paradigm in drug discovery. Nat. Chem. Biol.. 2008;4(11):682-690.
- [CrossRef] [Google Scholar]
- Protein identification in the post-genome era: the rapid rise of proteomics. Quart. Rev. Biophys.. 1997;30(4):279-331.
- [CrossRef] [Google Scholar]
- Kitano, Hiroaki, 2002. Systems biology: a brief overview. Science 295 (5560):1662-1664. DOI: 10.1126/science.1069492
- Klaassen, Curtis D, Mary O Amdur, 2013. Casarett and Doull's toxicology: the basic science of poisons. Vol. 1236: McGraw-Hill New York. ISBN: 0071347216
- Applicability of computational systems biology in toxicology. Basic Clin. Pharmacol. Toxicol.. 2014;115(1):45-49.
- [CrossRef] [Google Scholar]
- Systems toxicology approach for the assessment of zebrafish cardiotoxicity. Toxicol. Lett.. 2018;295:S102.
- [CrossRef] [Google Scholar]
- The Consortium for Metabonomic Toxicology (COMET): aims, activities and achievements. Pharmacogenomics. 2005;6(7):691-699.
- [CrossRef] [Google Scholar]
- Quantitative systems toxicology analysis of in vitro mechanistic assays reveals importance of bile acid accumulation and mitochondrial dysfunction in TAK-875-induced liver injury. Toxicol. Sci.. 2019;167(2):458-467.
- [CrossRef] [Google Scholar]
- Systems Toxicology Approach to Unravel Early Indicators of Squamous Cell Carcinoma Rate in Rat Nasal Epithelium Induced by Formaldehyde Exposure. International Conference on Practical Applications of Computational Biology & Bioinformatics 2019
- [CrossRef] [Google Scholar]
- A systems toxicology paracetamol overdose framework: accounting for high-risk individuals. Comput. Toxicol.. 2019;12:100103
- [CrossRef] [Google Scholar]
- Systems biology and synthetic biology: A new epoch for toxicology research. Adv. Toxicol.. 2015;2015
- [CrossRef] [Google Scholar]
- The nature of nurture: refining the definition of the exposome. Toxicol. Sci.. 2014;137(1):1-2.
- [CrossRef] [Google Scholar]
- Proteomic approaches to study structure, functions and toxicity of legume seeds lectins. Perspectives for the assessment of food quality and safety. J. Proteomics. 2009;72(3):527-538.
- [CrossRef] [Google Scholar]
- Nelson, J.H., 2003. Nuclear Magnetic Resonance Spectroscopy. 2003. Upper Saddle River: Pearson Education, Inc. ISBN: 0130334510
- Metabonomics: a platform for studying drug toxicity and gene function. Nat. Rev. Drug Discovery. 2002;1(2):153-161.
- [CrossRef] [Google Scholar]
- 'Metabonomics': understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29(11):1181-1189.
- [CrossRef] [Google Scholar]
- Microarrays and toxicology: the advent of toxicogenomics. Molecular Carcinogenesis: Published in cooperation with the University of Texas MD Anderson Cancer Center. 1999;24(3):153-159.
- [CrossRef] [Google Scholar]
- Synthetic biology: Genetic kill switches—a matter of life or death. Nat. Rev. Genet.. 2015;17(2):67.
- [CrossRef] [Google Scholar]
- In Silico Toxicology Data Resources to Support Read-Across and (Q) SAR. Front. Pharmacol.. 2019;10:561.
- [CrossRef] [Google Scholar]
- Insights into mechanisms and severity of drug-induced liver injury via computational systems toxicology approach. Toxicol. Lett.. 2019;312:22-33.
- [CrossRef] [Google Scholar]
- The principles and practice of toxicogenomics: applications and opportunities. Toxicol. Sci.. 2000;54(2):277-283.
- [CrossRef] [Google Scholar]
- Testing Times in Toxicology–In Vitro vs In Vivo Testing. Proceedings of Animal Alternatives in Teaching, Toxicity Testing and Medicine. 2011;2:53-59.
- [Google Scholar]
- A six-month systems toxicology inhalation/cessation study in ApoE−/− mice to investigate cardiovascular and respiratory exposure effects of modified risk tobacco products, CHTP 1.2 and THS 2.2, compared with conventional cigarettes. Food Chem. Toxicol.. 2019;126:113-141.
- [CrossRef] [Google Scholar]
- Genomics, proteomics and metabonomics in toxicology: Hopefully not ‘fashionomics’. Pharmacogenomics. 2004;5(7):879-893.
- [CrossRef] [Google Scholar]
- A Systems Toxicology Approach for the Prediction of Kidney Toxicity and Its Mechanisms In Vitro. Toxicol. Sci.. 2019;169(1):54-69.
- [CrossRef] [Google Scholar]
- Implications of the exposome for exposure science. J. Eposure Sci. Environ. Epidemiol.. 2011;21(1):5-9.
- [CrossRef] [Google Scholar]
- Synthetic toxicology: where engineering meets biology and toxicology. Toxicol. Sci.. 2011;120(suppl_1):S204-S224.
- [CrossRef] [Google Scholar]
- Slikker Jr, William, Merle G Paule, Linnzi KM Wright, Tucker A Patterson, and Cheng Wang, 2007. Systems biology approaches for toxicology. J. Appl. Toxicol.: An Int. J. 27 (3):201–217. DOI: 10.1002/jat.1207.
- High-throughput concentration–response analysis for omics datasets. Environ. Toxicol. Chem.. 2015;34(9):2167-2180.
- [CrossRef] [Google Scholar]
- Evaluation of the Tobacco Heating System 2.2. Part 1: Description of the system and the scientific assessment program. Regul. Toxicol. Pharm.. 2016;81:S17-S26.
- [CrossRef] [Google Scholar]
- Systems toxicology: from basic research to risk assessment. Chem. Res. Toxicol.. 2014;27(3):314-329.
- [CrossRef] [Google Scholar]
- Te, Jerez A, Mohamed Diwan M AbdulHameed, and Anders Wallqvist, 2016. Systems toxicology of chemically induced liver and kidney injuries: histopathology‐associated gene co‐expression modules. J. Appl. Toxicol. 36 (9):1137-1149. DOI: 10.1002/jat.3278
- Vermeeren, Veronique, Luc Michiels, 2011. Evolution Towards the Implementation of Point-Of-Care Biosensors. Biosensors for Health, Environment and Biosecurity:127. DOI: 10.5772/19432
- Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol. Sci.. 2014;142(2):312-320.
- [CrossRef] [Google Scholar]
- Adverse outcome pathway development II: best practices. Toxicol. Sci.. 2014;142(2):321-330.
- [CrossRef] [Google Scholar]
- The adverse outcome pathway concept: a pragmatic tool in toxicology. Toxicology. 2013;312:158-165.
- [CrossRef] [Google Scholar]
- Toward a comprehensive molecular design framework for reduced hazard. Chem. Rev.. 2010;110(10):5845-5882.
- [CrossRef] [Google Scholar]
- Toxicogenomics and systems toxicology: aims and prospects. Nat. Rev. Genet.. 2004;5(12):936-948.
- [CrossRef] [Google Scholar]
- The exposome: from concept to utility. Int. J. Epidemiol.. 2012;41(1):24-32.
- [CrossRef] [Google Scholar]
- From proteins to proteomes: large scale protein identification by two-dimensional electrophoresis and arnino acid analysis. Bio/Technology. 1996;14(1):61-65.
- [CrossRef] [Google Scholar]
- Analyzing the mechanisms behind macrolide antibiotic-induced liver injury using quantitative systems toxicology modeling. Pharm. Res.. 2019;36(3):48.
- [CrossRef] [Google Scholar]
- Protein microarrays for systems biology. Acta Biochim Biophys Sin. 2011;43(3):161-171.
- [CrossRef] [Google Scholar]
- Sensitive detection of proteasomal activation using the Deg-On mammalian synthetic gene circuit. Nat. Commun.. 2014;5(1):1-12.
- [CrossRef] [Google Scholar]







