Translate this page into:
Evaluating the structural and immune mechanism of Interleukin-6 for the investigation of goat milk peptides as potential treatments for COVID-19
⁎Corresponding authors. sbhavaniramya@gmail.com (Sundaresan Bhavaniramya), vaseeharanb@gmail.com (Baskaralingam Vaseeharan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
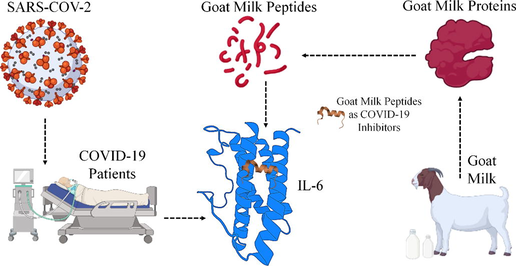
Abstract
The function of Immune control, haematopoiesis, and inflammation all depend on the cytokine Interleukin 6 (IL-6), and higher expression of IL-6 is seen in COVID-19 and other diseases. The immune protein IL-6 activation is dependent on binding interactions with IL-6Rα, mIL-6R, and sIL-6R for its cellular function. Termination of these reaction could benefit for controlling the over-expression in COVID-19 patients and that may arise as inhibitors for controlling COVID-19. Traditionally, the goat milk has been prescribed as medicine in ayurvedic practice and through this work, we have explored the benefits of peptides from goat milk as IL-6 inhibitors, and it have the potential of inhibiting the over expression of IL-6 and control the COVID-19 disease. Computational experiments have shown that goat peptides had strong interactions with IL-6, with higher scoring profiles and energy efficiency ranging from −6.00 kcal/mol to −9.00 kcal/mol in docking score and −39.00 kcal/mol in binding energy. Especially the YLGYLEQLLR, VLVLDTDYK and AMKPWIQPK peptides from goat milk holds better scoring and shows strong interactions were identified as the most potential IL-6 inhibitor candidates in this study. Peptides from Goat proteins, which are capable of binding to the IL-6 receptor with strong binding conformations, have no negative effects on other immune system proteins.
Keywords
Interleukin 6
Interleukin 6 receptor
Goat
Goat Milk
Peptide docking
MD simulation
COVID-19
1 Introduction
SARS-CoV-2, a new strain of coronavirus that was first detected in China's Hubei Province in late December 2019. On December 12, 2019, the first formal hospitalization was reported, and it was quickly labelled a pandemic with a rapid transmission rate, constituting a severe global public health hazard (Cascella et al., 2021). The initial name is said to WH Human coronavirus 1 (WHCV), then named as 2019-nCoV and finally, it has been called as SARS-CoV-2 and this virus has the high similarity with previous coronavirus and bat virus (Wu et al., 2020). As of right now, the death toll has risen to 5.32 million people, a number that is steadily rising. Fatigue, fever, headache, runny nose, and dry cough are the most common symptoms, but some instances are found to be asymptomatic (Belvis, 2020). So far, no effective antiviral drugs have been reported in the literature, with vaccination being the most effective means against the spread and genetic mutation of the virus (Bhavaniramya et al., 2021; Kaur and Gupta, 2020). But still there is a lack of effective compounds or drugs which can readily treat the serious illness caused by SARS-CoV-2 (Dong et al., 2020). The medical doctors are overcoming the problem by providing the medications based on suppressing the symptoms, and in addition, the SARS-CoV-2 hijacking of human immune mechanism is the major concern in dealing COVID-19 patients (Selvaraj et al., 2021b; Shah and Woo, 2021). SARS-CoV-2 has been shown to inhibit the lysosome's ability to combat illness in humans and based on this, other immune system processes may be affected if the lysosome is deacidified. Through this, we can better understand the immune system abnormalities associated with COVID-19 patients (Gorshkov et al., 2021; Pratap Reddy Gajulapalli, 2017). SARS-CoV-2 hijacks the human immune system, causing the entire immune system to be disrupted, as the human immune system relies significantly on white blood cells. Studies have found a link between COVID-19 severity and the cytokine Interleukin-6 (IL-6).
This IL-6 is an immune protein, initiate in response to any infections and tissue level injuries, contributes majorly to host defence mechanism. The IL-6 is coming under the IL cytokine family, and the signalling mechanism of IL-6 is triggered by IL-6Rα and gp130 (Liu et al., 2020). There are few reports that has discussed the involvement of IL-6 and IL-6R towards the COVID-19 infections with the additional term called cytokine storm. Severe sickness produced by SARS-CoV-2 in COVID-19 patients is associated with increased levels of the IL-6, which has been linked to respiratory failure, shock, and organ dysfunction (Coomes and Haghbayan, 2020). For COVID-19 patients, controlling IL-6 levels or its consequences is a critical treatment goal, and for that IL-6 can be treated as drug target for treating COVID-19 (Choy et al., 2020). The mechanism of IL-6 depends on IL-6 receptor (IL-6Rα) and gp130 for the activation and this ternary protein complex biologically forms the hexamer chain with all the three proteins. Initially the IL-6 and IL-6Rα forms the binary complex and it will show the interactions with domain II and domain III of gp130, and thus forms the IL-6|IL-6Rα|gp130 tertiary signalling complex (Foldvari-Nagy et al., 2021). By understanding this mechanism, there are few monoclonal antibodies reported in clinical development phase, which functionally block the IL-6 signalling/trans-signalling pathways (Tianyu et al., 2021). Patients with COVID-19 are currently being treated with the tocilizumab and Siltuximab to supress the IL-6 or cytokine storm, although orally consumed medicines are currently unavailable (Salama et al., 2021). There are few reports shows the Goat milk protein to having the tendency to protect against COVID-19 (Cakir et al., 2021; Chu et al., 2020), and in this work, we have explored the Indian variety of goat milk-based peptides for the inhibition or controlling the IL-6 in COVID-19 patients as shown in the Fig. 1.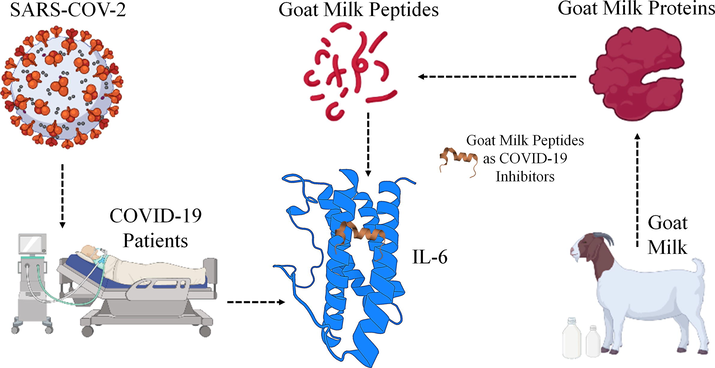
Graphical illustration of IL-6 as drug target for treating COVID-19 infected patients, and they can be treated by peptides obtained from the Goat Milk.
2 Materials and methods
2.1 Data source
The data information of peptides from various protein variants of goat milk from 10 native Indian goat breeds are reported in literature. An investigation into the post-translational changes of protein abundance in goat milk from genetically heterogeneous goat breeds in India has been published by Rout et al., 2021 (Rout and Verma, 2021). From the study, they have investigated ten Indian goat breeds for their genetic variation, proteome composition, and post translational alterations, and from that more than 50 goat milk peptides are reported and those peptides are source of this work. These peptide molecules are specifically extracted from the proteins of αS1-casein, αS2-casein, β-casein, κ-casein, β-lactoglobulin and α-lactalbumin (Verma et al., 2020).
2.2 Ligand and protein preparation
These peptide molecules are extracted and converted into 3D form using the chimera and prepared using the Ligprep with the conformations limited to 2 per peptide. The crystal structure of human Interleukin-6 (IL-6) is searched in PDB and the PDB ID: 1ALU is considered for the modelling studies (Somers et al., 1997). Here the protein is prepared for the in silico environmental setup by removing the unwanted crystallographic water molecules, ions and refinement of sidechains along with filling the missing residues and atoms, which modify the raw protein into the usable form (Yadav et al., 2021). Finally, the positional optimization of hydrogen bonds is processed and minimized until the final conformations of IL-6 reaches to 0.30 Å (Sastry et al., 2013).
2.3 Active site prediction
The region of active site where the potential goat milk peptides can bind efficiently are predicted by using the Active Site Prediction tool available in http://www.scfbio-iitd.res.in/dock/ActiveSite.jsp. The tool predicts the potential binding regions by computes the cavities in each protein that can be the position for the binding of goat milk peptides (Selvaraj et al., 2015; Singh et al., 2011).
2.4 Protein-Peptide docking
The obtained and prepared peptides are allowed to dock with the prepared IL-6 protein by using the HPepDock server and with the combination of Patchdock and firedock server (Zhou et al., 2018). The HpepDock server build based on the blind peptide-protein docking algorithm by rapid modelling of peptide conformations and global sampling of binding orientations that fit into the active sites. The same files given in the HpepDock server is given as input for the Patchdock sever, and here the Patchdock algorithm deals the protein-peptide Docking Algorithm Based on Shape Complementarity Principles, that can bind the goat peptides into the IL-6 shape complementary regions (Schneidman-Duhovny et al., 2005). The final conformations are subject to refinement using the firedock protocol. The pose matching in common between the HpepDock server and Patchdock server is subject to redocking with SP docking in Glide for the purpose of scoring and correctness of bonding/binding information’s (Siriwong et al., 2021; Tubert-Brohman et al., 2013).
2.5 Molecular dynamics simulations
The prepared crystal structure of IL-6 and along with final re-docked peptides from SP docking methods are given as inputs for the MD simulations in Gromacs (Van Der Spoel et al., 2005). Both apo form and complex peptide forms are allowed to simulate for the 10 ns in the dynamic aqueous environment (Selvaraj et al., 2018). The intense of performing the MD simulations is to check the biological conformation of the bioactive goat peptide to be stable inside the binding site of IL-6 (Gupta et al., 2021). For this, both apo and peptide bound IL-6 complex is subject to MD simulation filled with SPC water model in the orthorhombic box (Muralidharan et al., 2015). Neutralization of the system is done by adding suitable ion molecules (Na+ and Cl- ions) and the neutralized system is subject to energy minimization, followed by simulation for the timescale period of 10 ns. The trajectories are carefully evaluated for the analysis of RMSD, and conformations obtained from the various timeframes (Selvaraj et al., 2014).
2.6 ADME profiling and allergy prediction
Even though, the goat milk is the edible form, the peptides are better to check for the allergic conditions and thus the ADME and allergy tendency of the lead peptides are predicted by using the AllerCatPro and this server predicts the allergenicity potential from the amino acid sequence, tool available in https://allercatpro.bii.a-star.edu.sg/ along with Allergic ability of the peptides are also predicted through AllerTop 1.0 (Dimitrov et al., 2013; Maurer-Stroh et al., 2019). The best peptides based on docking, binding, and dynamics are acceded to in silico toxicity prediction through TOPKAT (Selvaraj et al., 2021a).
3 Results
3.1 Structure and function of IL-6
This research work aims to explore the peptide molecules extracted from the goat milk as drug candidates for the troubles initiated by IL-in COVID-19 patients, by blocking the binding site of the IL-6. This work will be highly valuable, especially for severe COVID-19 with serious illness patients. For checking the inhibitory profiles of goat milk peptides with IL-6 enzyme, the structure and function of the IL-6 is explored. The structure of IL-6 shows the 157 amino acid long protein structure shown in the Fig. 2a shows the 3D structure of IL-6, and here in Fig. 2b shows the site 1, site 2 and site 3 which are functionally playing the role in protein–protein binding of IL-6 with IL-6R and Il-6 with gp130. Fig. 2c shows the secondary structure of topology arrangement of IL-6 with the 6 helices, 7 helix-helix interacs, 3 beta turns and 2 disulphides to form the protein structure.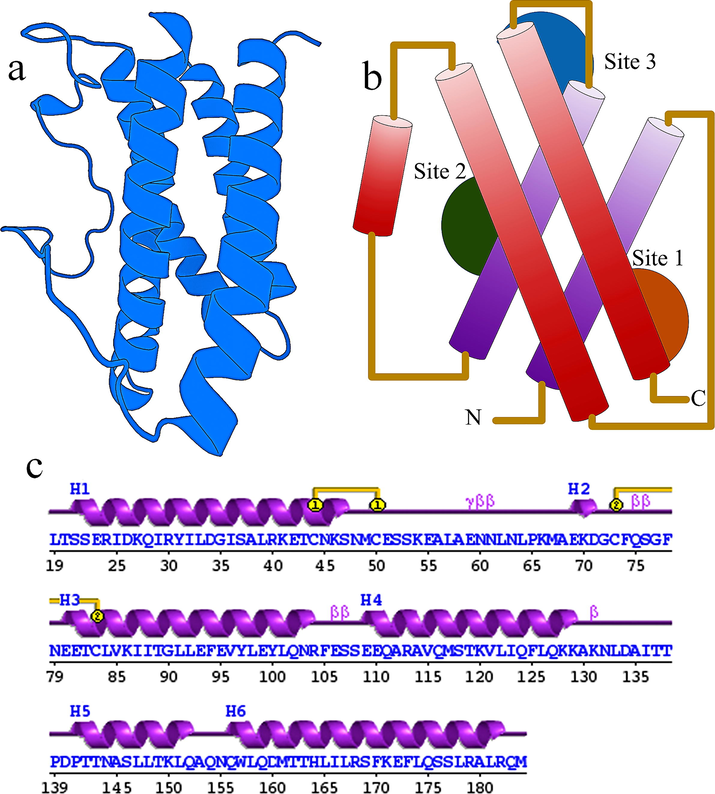
(a) Cartoon structure of IL-6 with the PDB ID: 1ALU (b) The Schematic representation of the domain structure of human IL-6 showing the site 1, site 2 and site 3 for the binding of IL-6R binding and gp130 binding (c) secondary structure of IL-6.
3.2 Active site information’s
The binding site information of IL-6 is checked for the amino acids, which can take responsibility of connecting the inhibitory peptides from goat milk. Tertiary Structure of IL-6 is comprised of four a-helices secondary structure connected via unstructured loops and the sites I, II and III represent the IL-6R and gp130 protein binding region. The Fig. 3a shows the predicted binding region of the IL-6, and here the red colour regions show the allocation of new peptide or new lead molecules can adopt itself in this predicted location. In addition, the position of the active site also explored and represented in Fig. 3b. The Fig. 3b shows the predicted active site is located in between the site2 and site3, which clarifies the peptides or ligands which bind in the predicted location will interrupt the binding of gp130 and disturb the protein–protein interactions.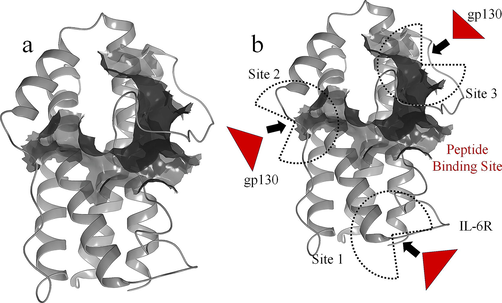
(a) Predicted binding region for peptide binding (red) in the IL-6 protein (Black). (b) Showing the peptide binding site in between the Site 2 and site 3, where the gp130 will be forming the protein–protein interactions.
3.3 Peptide docking and interactions
Thus, to understand the blocking of IL-6 and gp130, we have taken the prepared goat peptide molecules and allowed to interact with both docking servers and much correlated docking position is allowed to re-docked using the glide SP docking. The scoring of best scoring peptides in the SP docking is provided in the Table 1. The scoring values shows that, the goat milk peptides are able to form strong interactions with IL-6 by showing lower energy scores and better hydrogen bonds formations. The goat milk peptides are showing ∼ < −6 kcal/mol docking score and ∼ < −38 kcal/mol which shows the peptides exhibit lower energy profiles for interacting with IL-6. *Units Kcal/mol.
S. No
Peptide Sequence
Length
Docking Score
Binding Energy
H-Bonds
a
ALKALPMHIR
10
−6.39
−48.22
4
b
ALPMHIR
7
−7.73
−42.93
5
c
AMKPWIQPK
9
−8.59
−58.82
5
d
EMPFPK
6
−6.77
−43.10
4
e
HKEMPFPK
8
−5.96
−38.99
4
f
KETMVPK
7
−7.01
−53.03
4
g
TPEVDDEALEK
11
−8.38
−78.49
7
h
VLVLDTDYK
9
−8.89
−67.22
6
i
YIPIQYVLSR
10
−7.30
−60.24
6
j
YLGYLEQLLR
10
−8.91
−68.43
7
Scoring profiles are great for the goat milk peptides and shows strong interactions between the protein and peptide molecules. Best scoring poses of peptide molecules are showed along with protein conformations as shown in the Fig. 4, shows the center position with all the peptide conformation bound in between the site 2 and site 3 showing the tendency to disturb the gp130 binding. The dissection of each peptide poses, showing its binding orientation is provided in Fig. 4a–j and here the alphabets represent the (a) ALKALPMHIR, (b) ALPMHIR (c) AMKPWIQPK, (d) EMPFPK, (e) HKEMPFPK, (f) KETMVPK, (g) TPEVDDEALEK, (h) VLVLDTDYK, (i) YIPIQYVLSR and (j) YLGYLEQLLR are the best goat milk peptides. The goat milk peptides are positioned in between the site2 and site3, and clarifies that these peptides are having the ability to bind with IL-6 and can also block the interactions between the IL-6 and gp130. This can also say as, these peptide molecules are having the tendency to suppress the activation of IL-6 by blocking the gp130 binding with IL-6.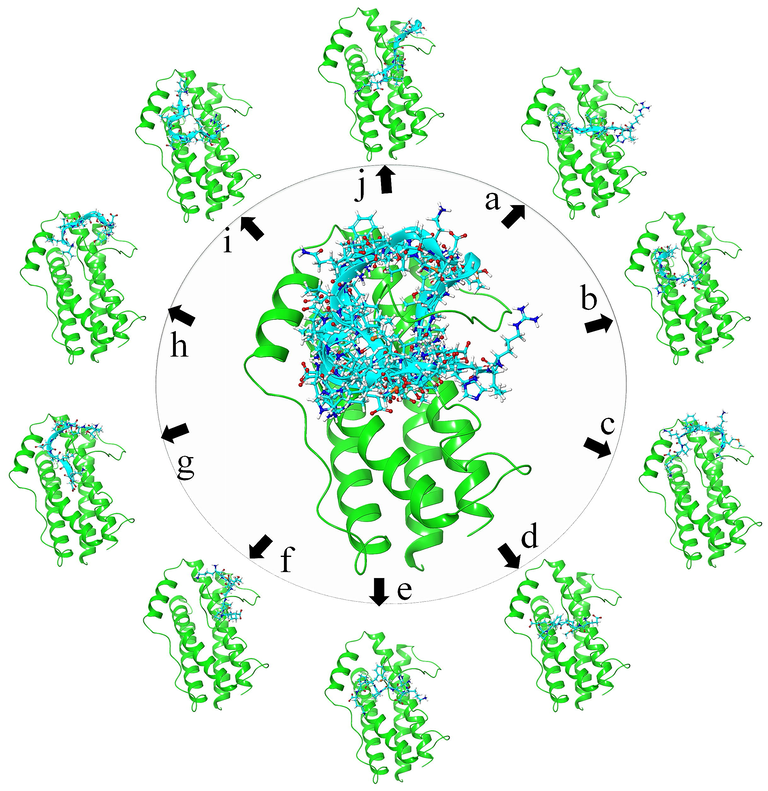
Visualization of protein-polymer interactions showing the binding mode of Goat peptides placed in between the site 2 and site 3. Here the common binding peptides are in center and round by each peptide pose represented with alphabets showing (a) ALKALPMHIR, (b) ALPMHIR (c) AMKPWIQPK, (d) EMPFPK, (e) HKEMPFPK, (f) KETMVPK, (g) TPEVDDEALEK, (h) VLVLDTDYK, (i) YIPIQYVLSR and (j) YLGYLEQLLR.
3.4 Molecular dynamics simulation
Firstly, the MD trajectories of IL-6 and its receptor either in the form of apoprotein or complex with 10 peptides were investigated during a simulation time of 10 ns to analyze the stability of these proteins, peptide and their interactions. For analysis of MD simulations, the RMSD of apo and complex MD simulation is averaged and those values are plotted in Fig. 5a shows the apo IL-6 is showing the average RMSD of ∼0.23 nm and all the goat milk peptides are surpassing the values by showing above the apo protein with the value of ∼0.28 nm and above. The minimum RMSD of goat milk is ∼0.28 nm and range maximum up to ∼0.65 nm. This may be due to the binding of goat peptides generate conformations on the dynamic equilibrium and possess strong binding. Those bindings are verified by using the H-bond validation and the average H-bonds between the protein and peptide are plotted in Fig. 5b. The average h-bonds between the protein and peptide shows the minimum h-bonds of 2.1 hydrogen bonds and maximum of 3.8 hydrogen bonds on average interactions in the 10 ns of MD simulations.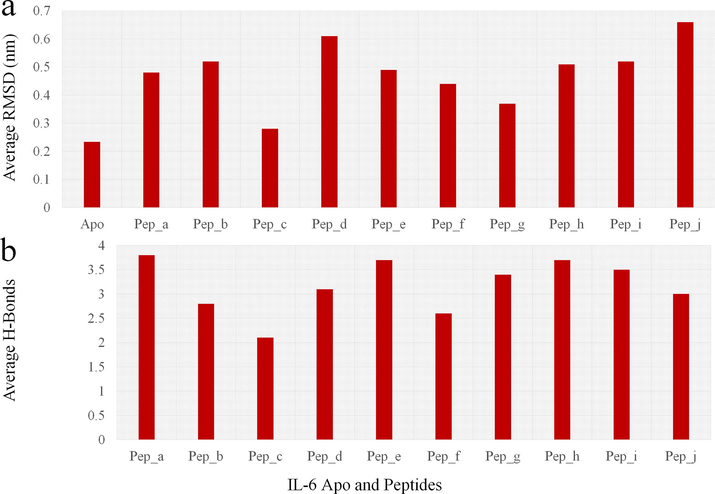
Molecular dynamics graphs for the timescale of 10 ns showing the (a) RMSD variations average between the apo and peptide bound complex. (b) Average number of hydrogen bonds formed between the IL-6 and goat peptides in the timescale of 10 ns.
3.5 Allergic and toxicity prediction
Final best docked peptides on scoring are allowed to extract the bioactive conformation and the peptides are clearly shows to be non-carcinogen and non-allergic in nature. Thus, these peptides are consumable in nature due to these properties. This is proven that, the goat milk is edible to consume by humans for multiple medicinal uses, and the prediction methods also confirms the peptides extracted from goat milk are also able to consume with non-carcinogenic and non-allergic property (Table 2).
S. No
Peptide Sequence
Carcinogen prediction
Allergic Prediction
a
ALKALPMHIR
Non-Carcinogen
Non-allergen
b
ALPMHIR
Non-Carcinogen
Non-allergen
c
AMKPWIQPK
Non-Carcinogen
Non-allergen
d
EMPFPK
Non-Carcinogen
Non-allergen
e
HKEMPFPK
Non-Carcinogen
Non-allergen
f
KETMVPK
Non-Carcinogen
Non-allergen
g
TPEVDDEALEK
Non-Carcinogen
Non-allergen
h
VLVLDTDYK
Non-Carcinogen
Non-allergen
i
YIPIQYVLSR
Non-Carcinogen
Non-allergen
j
YLGYLEQLLR
Non-Carcinogen
Non-allergen
4 Discussion
Through this work, the IL-6 interactions with gp130 are mechanistically broken with the goat milk peptides by allowing the peptides to interact in site 2 and site 3 region. As the IL-6 involved immune mechanism is primary on severe illness and rapid disease progression is a cytokine storm. Patient’s data on COVID-19 shows the increase effect on IL-6 confirms and leads to respiratory failure and other dysfunction. Thus, considering these, the IL-6 can be a potential drug target to study for the controlling the SARS-CoV-2. Here, the IL-6 is a human immunological protein and it cannot be completely inhibited and so, the immunomodulatory goat milk peptides are the best to apply as the IL-6 inhibitors. Thus, the goat milk peptides can able to modulate the levels of IL-6 or suppressing the higher active levels is the core point for curing the COVID-19 and thus, it can be a potential therapeutic drug target for dealing severe COVID-19 infections. For this, the structure and function of IL-6 protein is explored using the molecular modelling methods, which shows the protein topology is adopting the IL-6R in the site 1 and gp130 in site2 and site 3. Interestingly, our predicted active site for the binding of goat milk peptides is showing in between the site2 and site 3. Thus, the goat peptides on binding to these predicted regions, can able to interrupt the binding of gp130 with IL-6 and that can lead to suppress the levels of IL-6 in COVID-19 patients. For this, the bioactive peptides are prepared and allowed to interact with IL-6 using protein-peptide docking method using the HPepDock server and Patchdock server for getting more reliable conformations, and the best reported conformations are allowed to re-dock using the SP docking method for getting the scoring profiles of the protein-peptide docking. The scoring profiles shows all the best peptide bound pose of IL-6 have the docking score approximately lower than −6 kcal/mol and binding energy of approximately lower than −39 kcal/mol. This scoring profiles clarifies the strong binding is seen in between the goat milk peptides and IL-6 and that are also seen in the number of hydrogen bonds formed in between them. The minimum of four to maximum eight hydrogen bonds are seen in between the goat milk peptides and IL-6 shows the strong bonding interactions leads to peptides attaches into the IL-6 in tight forms. In addition, the binding conformation of pose interactions are also seen for each peptide shows the peptides are interact in between the site 2 and site 3, which confirms the peptide tendency of blocking the protein–protein interactions. For validation of binding poses, the goat milk bound peptide complex are simulated for 10 ns of MD simulations to verify the apo and peptide stability along with average interactions formed between the goat milk peptides and IL-6. RMSD graph shows the apo form of IL-6 is moderately flexible and shows ∼0.23 nm, but all the peptide bound IL-6 shows above these values. This is due to the presence of rotatable bonds in the goat milk peptides, which allow the bioactive conformation to generate more conformational changes inside the IL-6 peptide binding pocket in the dynamic environment. Final prediction of ADME property for checking the allergic and carcinogenic property shows all the peptides are free from allergic and carcinogenic property and edible in nature.
5 Conclusion
In humans, the Immune modulation, hemopoiesis, and inflammation are all aided by the cytokine interleukin-6 (IL-6). This protein IL-6 is linked to the disease like septic shock, acute respiratory distress syndrome (ARDS), and COVID-19 are all due to the elevated over expression of IL-6 levels. For IL-6 to exert its biological effects, it must first interact with IL-6R and gp130, which is found on both the membrane and the soluble levels (sIL-6R). IL-6-related disorders could benefit from a medication that prevents the interaction between these two proteins. IL-6-related disorders could benefit from a medication that prevents the interaction between IL and 6 with IL-6R and gp130 proteins. This work come up with strong bioactive peptides that able to bind with site2 and site 3, which says those goat milk peptides are having the strong inhibitory profiles to interrupt the binding of gp130 with IL-6. Thus, the goat milk peptides can able to suppress the over expressed levels of IL-6 and can be the best inhibitors for the serious illness of COVID-19 patients. In addition, these peptides are shown to have a non-carcinogenic and non-allergic property, and thus shows the edibleness of the goat milk peptides. Overall, through this work, we are providing 10 goat milk peptides, that strongly binds with IL-6 and interrupt the gp130 binding that leads to suppress the over expression of IL-6 levels. Further experiments on these goat milk peptides may support to evidence the potential of goat milk peptides against the COVID-19 and also to other diseases that happens due to the elevated levels of IL-6.
Acknowledgement
SB thankfully acknowledges UGC Kothari post-doctoral fellowship award (F.4-2/2006(BSRYBL/20-21/0014) and Alagappa University for providing the necessary facilities to carry out this work. The author would like to thank Deanship of Scientific Research at Majmaah University for supporting this work under project number R-2022-69.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Headaches during COVID-19: my clinical case and review of the literature. Headache. 2020;60(7):1422-1426.
- [Google Scholar]
- Comprehensive analysis of SARS-COV-2 drug targets and pharmacological aspects in treating the COVID-19. CMP. 2021;14
- [CrossRef] [Google Scholar]
- Investigation of beta-lactoglobulin derived bioactive peptides against SARS-CoV-2 (COVID-19): in silico analysis. Eur. J. Pharmacol.. 2021;891:173781.
- [CrossRef] [Google Scholar]
- Features, Evaluation, and Treatment of Coronavirus (COVID-19). Treasure Island (FL): StatPearls; 2021.
- Translating IL-6 biology into effective treatments. Nat. Rev. Rheumatol.. 2020;16(6):335-345.
- [Google Scholar]
- Design, synthesis, and biological evaluation of N-arylpiperazine derivatives as interferon inducers. Bioorg. Med. Chem. Lett.. 2020;30(24):127613.
- [CrossRef] [Google Scholar]
- Interleukin-6 in Covid-19: a systematic review and meta-analysis. Rev. Med. Virol.. 2020;30(6):1-9.
- [Google Scholar]
- AllerTOP–a server for in silico prediction of allergens. BMC Bioinformatics. 2013;14(Suppl 6):S4.
- [Google Scholar]
- A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target Ther.. 2020;5(1):237.
- [Google Scholar]
- On the role of bacterial metalloproteases in COVID-19 associated cytokine storm. Cell Commun. Signal. 2021;19(1):7.
- [Google Scholar]
- The SARS-CoV-2 cytopathic effect is blocked by lysosome alkalizing small molecules. ACS Infect. Dis.. 2021;7(6):1389-1408.
- [Google Scholar]
- Molecular dynamics analysis of the binding of human interleukin-6 with interleukin-6 alpha-receptor. Proteins. 2021;89(2):163-173.
- [Google Scholar]
- COVID-19 Vaccine: a comprehensive status report. Virus Res.. 2020;288:198114.
- [CrossRef] [Google Scholar]
- Can we use interleukin-6 (IL-6) blockade for coronavirus disease 2019 (COVID-19)-induced cytokine release syndrome (CRS)? J. Autoimmun.. 2020;111:102452.
- [CrossRef] [Google Scholar]
- Maurer-Stroh, S., Krutz, N.L., Kern, P.S., Gunalan, V., Nguyen, M.N., Limviphuvadh, V., Eisenhaber, F., Gerberick, G.F., 2019. AllerCatPro-prediction of protein allergenicity potential from the protein sequence. Bioinformatics 35(17), 3020-3027.
- Structure-based virtual screening and biological evaluation of a Calpain inhibitor for prevention of selenite-induced cataractogenesis in an in vitro system. J. Chem. Inf. Model. 2015;55(8):1686-1697.
- [Google Scholar]
- Organocatalytic Asymmetric Synthesis of 2, 3′-Connected Bis-Indolinones by Mannich Reactions of N-Acetylindolin-3-ones with Isatin N-Boc Ketimines. Synthesis. 2017;49(22):4986-4995.
- [Google Scholar]
- Post translational modifications of milk proteins in geographically diverse goat breeds. Sci. Rep.. 2021;11(1):5619.
- [Google Scholar]
- Tocilizumab in patients hospitalized with Covid-19 pneumonia. N. Engl. J. Med.. 2021;384(1):20-30.
- [Google Scholar]
- Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des.. 2013;27(3):221-234.
- [Google Scholar]
- PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res.. 2005;33(Web Server issue):W363-367.
- [Google Scholar]
- Artificial intelligence and machine learning approaches for drug design: challenges and opportunities for the pharmaceutical industries. Mol. Divers 2021
- [Google Scholar]
- Selvaraj, C., Dinesh, D.C., Krafcikova, P., Boura, E., Aarthy, M., Pravin, M.A., Singh, S.K., 2021b. Structural understanding of SARS-CoV-2 drug targets, active site contour map analysis and COVID-19 therapeutics. Curr. Mol. Pharmacol.
- Molecular insights of protein contour recognition with ligand pharmacophoric sites through combinatorial library design and MD simulation in validating HTLV-1 PR inhibitors. Mol. Biosyst.. 2015;11(1):178-189.
- [Google Scholar]
- Molecular dynamics simulations and applications in computational toxicology and nanotoxicology. Food Chem. Toxicol.. 2018;112:495-506.
- [Google Scholar]
- Investigations on the interactions of lambdaphage-derived peptides against the SrtA mechanism in Bacillus anthracis. Appl. Biochem. Biotechnol.. 2014;172(4):1790-1806.
- [Google Scholar]
- Molecular perspectives of SARS-CoV-2: pathology, immune evasion, and therapeutic interventions. Mol. Cells. 2021;44(6):408-421.
- [Google Scholar]
- AADS–an automated active site identification, docking, and scoring protocol for protein targets based on physicochemical descriptors. J. Chem. Inf. Model. 2011;51(10):2515-2527.
- [Google Scholar]
- Identification of a chitooligosaccharide mechanism against bacterial leaf blight on rice by in vitro and in silico studies. Int. J. Mol. Sci.. 2021;22(15)
- [Google Scholar]
- 1.9 A crystal structure of interleukin 6: implications for a novel mode of receptor dimerization and signaling. EMBO J.. 1997;16(5):989-997.
- [Google Scholar]
- New tale on LianHuaQingWen: IL6R/IL6/IL6ST complex is a potential target for COVID-19 treatment. Aging (Albany NY). 2021;13(21):23913-23935.
- [Google Scholar]
- Improved docking of polypeptides with Glide. J. Chem. Inf. Model. 2013;53(7):1689-1699.
- [Google Scholar]
- Functional milk proteome analysis of genetically diverse goats from different agro climatic regions. J. Proteomics. 2020;227:103916
- [Google Scholar]
- A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265-269.
- [Google Scholar]
- Investigating into the molecular interactions of flavonoids targeting NS2B-NS3 protease from ZIKA virus through in-silico approaches. J. Biomol. Struct. Dyn.. 2021;39(1):272-284.
- [Google Scholar]
- HPEPDOCK: a web server for blind peptide-protein docking based on a hierarchical algorithm. Nucleic Acids Res.. 2018;46(W1):W443-W450.
- [Google Scholar]







