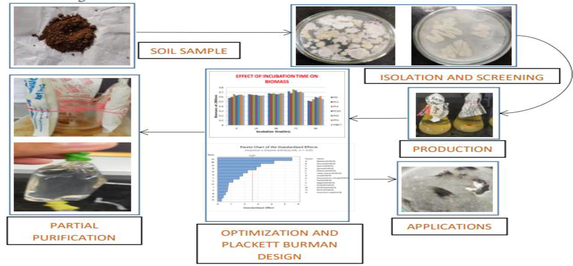
Translate this page into:
Extracellular Protease Production, Optimization, and Partial Purification from Bacillus nakamurai PL4 and its Applications
⁎Corresponding authors at: Department of General Science, Ibn Sina National College for Medical Studies, Al Mahajar Street: 31906, Jeddah 21418, Saudi Arabia (S.M. Shakeel Iqubal). muddapur@kletech.ac.in (Uday M. Muddapur), shakeeliqubal@gmail.com (S.M. Shakeel Iqubal)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
The major goal of the study was to isolate bacteria synthesizing protease enzyme from a soil sample taken in Dandeli, Karnataka, India. Furthermore, screening, production, and optimization of medium components for maximum protease activity, partial purification of crude enzymes, and application of protease produced by Bacillus nakamurai were carried out. At 72 hrs and pH 6, the optimum incubation time, pH, and temperature were evaluated (acidic and thermophilic). As a substrate, casein was employed. Plackett-Burman screening was performed, and KH2PO4, xylose, MnCl2, and peptone were discovered to be essential components in protease production media. Following the production and optimization processes, partial purification was performed using ammonium sulfate precipitation, with the maximum protease activity at 60% ammonium sulfate, and further dialysis was performed using precipitated enzyme, yielding enzyme activity of 0.747 U/mL. The protease enzyme proved effective at removing egg yolk stains as well as degrading (dehairing) chicken feathers and hairs from goat skin. Bacillus nakamurai PL4 can be utilized for the industrial-scale production of proteases to fulfill current demands. Thus, such optimized parameters can optimize protease production and their application across various industries.
Keywords
Extracellular Protease
Enzyme activity
Optimization
Bacillus nakamurai PL4
Protease applications
Microbial enzyme
1 Introduction
Enzymes are considered the essential molecules that include many metabolic activities in the body and are useful for survival, enzyme-based products are considered safe compared to other chemical-based products (Gupta et al., 2002). For producing the enzymes microbes are used, where the product may be extracellular or may be intracellular, microbes are preferred in many industrial applications for their wide advantages (Kirk et al., 2002). Proteases are a significant class of enzymes with numerous industrial uses in different sectors. Proteases, proteinases, peptidases, and proteolytic enzymes cleave the peptide bonds in proteins, but the most commonly used name is protease (Rao et al., 1998a). The main purpose of the protease enzyme is to hydrolyze/catalyze the peptide bond present in the proteins (Mótyán et al., 2013).
Exo-peptidases and endo-peptidases are different types of proteases. Depending on whether they function at the N or C site, exopeptidases are classified as either aminopeptidases or carboxypeptidases and have the ability to cleave the peptide bonds at the end of an amino acid chain. Enzymes called endopeptidases break peptide bonds within amino acid chains. Serine proteases, aspartic proteases, cysteine proteases, and metalloproteases are the four groups of endo-peptidases based on the functional group located at the active site (Dalal, 2015). Based on pH, proteases can be categorised as alkaline, acidic, or neutral (Mótyán et al., 2013; Dalal, 2015). Plants, mammals, and microorganisms may all generate protease enzymes; however, microbes are typically favoured due to their low cost and high efficiency (Doddapaneni et al., 2007).
In many industrial applications, such as waste treatment, the bioremediation process, the detergent industry, the textile business, and the leather industry, protease is given great priority as the most significant enzyme. Proteases are commonly employed in the baking industry for a variety of tasks, including the creation of crackers, waffles, baked goods, and bread. These enzymes are employed to speed up mixing, increase texture and flavour, control bread's gluten strength, and increase uniformity and consistency of the dough (Kumar et al., 2002).
The majority of the commercially significant alkaline proteases are produced by Bacillus spp., and the alkaline protease produced by Bacillus spp. is stable in a wide range of pH, temperature, and substrate (Adinarayana et al., 2004). Protease enzymes produced by bacteria can be easily modified through genetic manipulation. Due to their great growth and stability, the microbial community is chosen above others for the large-scale manufacture of protease (Oberoi et al., 2001). The focus of the current study is on the partial purification and optimization of Bacillus nakamurai PL4 alkaline protease and its uses in dairy, poultry and detergent industries.
2 Materials and methods
2.1 Isolation
The soil samples were taken from a lake in Dandeli, Karnataka, India. Using a saline solution, 1 g of soil was weighed and serially diluted (0.9 percent NaCl). The diluted samples were applied to nutrient agar plates with pH 7 agar, 0.1 percent beef extract, 0.2 percent yeast extract, 0.5 percent peptone, and 0.1 percent sodium chloride. Colonies were morphologically evaluated, added to nutrient broth (1.3 %), plated on nutrient agar slants, and stored in glycerol stocks (50 %) for further use after plates were incubated at 37 °C for 24 hrs (Dubey et al., 2007; Beg et al., 2003; Uyar et al., 2011).
2.1.1 Screening for proteolytic activity
Screening of protease activity is done by streaking colonies onto skim milk agar containing casein (0.5 %), yeast (0.25 %), skim milk (0.1 %), glucose (0.1 %), agar (1.05 %), and incubate for 24–48 hrs to observe the clear zones, once we observe the clear zone one can measure the zone of clearance by scale to identify the protease activity. Skim milk agar contains casein which acts as the protein source, when the bacterial colony is streaked onto skim milk agar if the bacterial colony is capable of producing protease, this protease acts on casein and breaks the peptide bond present in casein hence the clear zone is observed (Rathakrishnan et al., 2012).
2.2 Identification of bacteria
Beginning with Gram staining, endospore staining, and the motility test, bacteria were identified and compared with the reference Bergey's manual of determinative bacteriology (Uyar et al., 2011) before being further characterised using 16 s rRNA gene sequencing. Phylogeny used blast sequences to do phylogenetic analysis. Using the Tree View application, a phylogenetic tree was created using the neighbor-joining technique. A chi2-based parameter was used to verify the branching pattern's repeatability.
2.3 Production of protease-producing bacteria
2.3.1 Media composition and inoculum development for submerged fermentation
The inoculum was prepared by subculturing the Bacillus nakamurai strain in a 250 mL conical flask in the production media containing components that play an important role including carbon source, nitrogen source, metal ions, minerals, and salts. A pH 8.0 optimised production medium containing 1 percent galactose, 0.5 percent casein, 0.55 percent peptone, 0.2 percent KH2PO4, 1 percent Na2CO3, and 0.2 percent MgSO47H2O was added to the production medium and incubated with continuous shaking to perform the shake flask fermentation. 72 hrs of shaker fermentation at 37 °C with controlled agitation at 150–200 rpm were completed. The entire culture broth was centrifuged at 6000 rpm for 15 min at the conclusion of the fermentation period to remove debris, and the supernatant was then collected and used as a crude enzyme in subsequent experiments (Rathakrishnan et al., 2012).
2.4 Optimization of fermentation conditions
For one factor at a time (OFAT) experiments, parameters such pH (4,6,8,10 & 12), incubation duration (at intervals of 24 hrs), and substrate (casein, gelatin, azoacasein & albumin) were used to explore the impact of various parameters on the protease activity and the biomass (Rathakrishnan et al., 2012; Alkarkhi et al., 2021).
2.4.1 Effect of incubation time
The isolate Bacillus nakamurai was infused into the optimal production medium with pH 8 to examine the impact of incubation duration on protease synthesis. Additionally, the organism was shaker-incubated at 37 °C and 120 rpm to ensure uniform development. The protease activity and biomass were monitored at regular time intervals of 24, 48, 72, and 96 hrs duration. At the regular interval of 24 hrs, cell growth by wet method and proteolytic activity was estimated by centrifuging the fermentation media at 6000 rpm for 15 mins to remove the cell debris (Rathakrishnan et al., 2012; Rao et al., 1998b).
2.4.2 Effect of pH
By incubating the isolates in an optimized production medium for 72 hrs at 120 rpm and 37 °C with various pH values in the range of 4–12 while using the necessary concentrations of 1 N NaOH and 1 N HCl, it was possible to determine the impact of pH on protease production by the isolate Bacillus nakamurai. Centrifuging the fermentation media at 6000 rpm for 15 min to remove the cell debris allowed researchers to determine biomass using the wet technique and the amount of proteolytic activity (Rao et al., 1998b).
2.4.3 Effect of substrate
The bacterium Bacillus nakamurai was cultured in optimum production media using a variety of substrates (Casein, Azocasein, Albumin, and Gelatin), with the pH and period of incubation remaining constant at 8 and 72 hrs at 37 °C and 120 rpm, respectively. The biomass was measured using the wet method, and the proteolytic activity was evaluated by centrifuging the fermentation media at 6000 rpm for 15 mins to remove the cell debris (Beg et al., 2003).
2.5 Designing statistically-based experiments
For screening tests, Plackett-Burman (PB) designs are used to evaluate the relative significant components in the media for the synthesis of protease from Bacillus nakamurai strain. The variables were tested at two levels of high (+) and low (-) concentrations (Table. 1), and considering the midpoint, 14 variables were tested at 2 levels and total experimental runs were 20. The matrix can be designed for fourteen factors in twenty trials. All trials were done in 250 mL Erlenmeyer flasks containing 100 mL of the medium. The selected media components were carbon source (Maltose, Glucose, Starch, Xylose), Organic nitrogen source (Peptone, Yeast extract), Inorganic nitrogen (Urea, Ammonium nitrate), minerals (NaCl, MgSO4, ZnSO4, KH2PO4, MnCl2) and inoculum size to remove debris for each of the trials/runs. The fermentation medium was optimized based on OFAT results (pH 6, 72 hrs of incubation, and casein as the substrate) (Olajuyigbe, 2013; Yang et al., 2000). The significant factors were screened by pareto chart, main effect plots, regression equation and variance, PB design was performed using minitab software version 20 (Table 2). A = Maltose, B = Glucose, C = Starch, D = Xylose, E = Peptone, F = Yeast extract, G = Urea, H = Ammonium nitrate, J = Sodium Hydroxide, K = Magnesium sulphate, L = Zinc sulphate, M = Potassium dihydrogen phosphate, N = Zinc chloride, O = Magnesium (II) chloride.
Factor
Low (-1)
Mid point
High (+1)
Carbon source
0.5 %
1 %
1.5 %
Nitrogen source
0.3 %
0.55 %
0.8 %
Minerals
0.1 %
0.2 %
0.3 %
Inoculum size
2.0 %
4.0 %
6.0 %
Run
Blk
A
B
C
D
E
F
G
H
J
K
L
M
N
O
1
1
+
–
+
+
–
–
–
–
+
–
+
–
+
+
2
1
+
+
–
+
+
–
–
–
–
+
–
+
–
+
3
1
–
+
+
–
+
+
–
–
–
–
+
–
+
–
4
1
–
–
+
+
–
+
+
–
–
–
–
+
–
+
5
1
+
–
–
+
+
–
+
+
–
–
–
–
+
–
6
1
+
+
–
–
+
+
–
+
+
–
–
–
–
+
7
1
+
+
+
–
–
+
+
–
+
+
–
–
–
–
8
1
+
+
+
+
–
–
+
+
–
+
+
–
–
–
9
1
–
+
+
+
+
–
–
+
+
–
+
+
–
–
10
1
+
–
+
+
+
+
–
–
+
+
–
+
+
–
11
1
–
+
–
+
+
+
+
–
–
+
+
–
+
+
12
1
+
–
+
–
+
+
+
+
–
–
+
+
–
+
13
1
–
+
–
+
–
+
+
+
+
–
–
+
+
–
14
1
–
–
+
–
+
–
+
+
+
+
–
–
+
+
15
1
–
–
–
+
–
+
–
+
+
+
+
–
–
+
16
1
–
–
–
–
+
–
+
–
+
+
+
+
–
–
17
1
+
–
–
–
–
+
–
+
–
+
+
+
+
–
18
1
+
+
–
–
–
–
+
–
+
–
+
+
+
+
19
1
–
+
+
–
–
–
–
+
–
+
–
+
+
+
20
1
–
–
–
–
–
–
–
–
–
–
–
–
–
–
2.6 Analytical methods
The cell biomass of the fermentation media is obtained from the wet method and protease activity was estimated by lowry’s modified method by using a tyrosine standard graph for converting optical density to enzyme activity (Beg et al., 2003).
2.6.1 Assay for protein concentration
Six test tubes were used to prepare tyrosine for various dilutions for the setup. The following amounts in millilitres (mL) of 1.1 mM tyrosine standard stock solutions are added to the six test tubes: 0.05, 0.10, 0.20, 0.40, and 0.50. With the blank removed, the volume is increased to 2 mL, and then 5 mL of sodium carbonate (0.5 M) and 5 mL of Folin phenol (0.5 M) reagent are added. At 280 nm, the absorbance was measured [20].
2.6.2 Assay for protease activity
For the assay, suitable vials were considered, at the beginning 5 mL of casein (0.65 %) is added and let to incubate at 37 °C for 5 mins for uniform mixing, after the incubation 0.5 mL of supernatant (enzyme) is added to each test tube and then the solution is incubated at 37 °C for 10 mins, after the incubation 5 mL of trichloro acetic acid (110 mM) is added, again 0.5 mL of the enzyme is added and then incubated at 37 °C for 30 mins, the solution is filtered and 2 mL of test filtrate is considered for further process, to this filtrate 5.0 mL of sodium carbonate (0.5 M) is added and the reaction is stopped by adding 1.0 mL of FC reagent (0.5 M) and incubated at 37 °C for 30 mins, blank is prepared by adding all the components excluding enzyme (Keay and Wildi, 1970; Sellami-Kamoun et al., 2008).
The absorbance is measured by UV visible spectrophotometer at 660 nm, once the optical density is obtained, the OD values are converted to Enzyme activity (U/mL) by using tyrosine standard graph (Sellami-Kamoun et al., 2008; assar et al., 2015).
2.6.3 Cell growth analysis
Cell Biomass is estimated by adding 2 mL of fermentation broth to the cuvette and analyzing the absorbance at 600 nm under UV visible spectrophotometer (Sellami-Kamoun et al., 2008; Nassar et al., 2015).
2.7 Partial purification
Partial purification of protease activity is performed to remove unwanted substances from crude enzymes and to have the high enzyme activity, specific activity, and possible recovery of the enzyme (Uyar and Baysal, 2004).
2.7.1 Partial purification by ammonium sulpate precipitation
Ammonium sulphate precipitation is an initial step in enzyme purification, the technique is performed based on the solubility difference and is classified into two processes”salting in” and “salting out”, when the concentration of salt has increased the solubility of protein decreases, ionic strength increases and the protein precipitates out termed as “salting out”, ammonium (NH4+) and sulphate (SO4-2) ions are within the aqueous phase they are attracted to the opposite poles and the enzyme gets purified (Uyar and Baysal, 2004; Debnath et al., 2021).
The purification of an enzyme is performed by considering the different concentrations of ammonium sulphate (20–80 %) at every concentration enzyme activity is analyzed. The crude enzyme is taken in the beaker and a magnetic stirrer is placed into it, then ammonium sulphate is added slowly to bring the final concentration to 0–20 %, this setup is kept at 40 °C for 3 hrs, and the solution is centrifuged at 3000 rpm for 15 mins and pellets are stored in the tris-HCl buffer for further analyses of enzyme activity, specific activity and fold purification. The process is continued for the different concentrations of ammonium sulphate till there is high enzyme activity in the pellets compared to the supernatant (Debnath et al., 2021).
2.7.2 Partial purification by dialysis
After the precipitation of crude enzyme dialysis is performed, dialysis is the separation technique that is used to separate small molecules from large molecules based on size, the unwanted substances easily pass through the dialysis bag based on the principle of diffusion and the partially purified enzyme retain in the dialysis bag (Devi et al., 2008).
The dialysis bag was cut to the proper length and was soaked in phosphate buffer, the dialysis bag is then sealed with a metal clip or a rubber band with crude enzyme into it and stirred by using a magnetic stirrer for 3 hrs, then stored at 40 °C and centrifuged at 6000 rpm for 20 mins and the supernatant is stored at −20 °C (Devi et al., 2008).
2.8 Application of protease enzyme
2.8.1 Detergent industry
The application of protease in the removal of egg yolk stain was observed, the clean cloths of the size (3*3 cm^2) were soaked in egg yolk for 3 hrs and dried in a hot air oven at 80 °C for 10 mins, then the dried clothes are subjected to the crude enzyme, crude enzyme + detergent, only detergent, and only crude enzyme, to estimate the effectiveness of crude enzyme in the removal of egg yolk stain. Egg yolk is protein, when the protease enzyme acts on the egg yolk there is breakage of peptide bonds, due to this stain can be removed from the cloth (Najafi et al., 2005).
2.8.2 Feather degradation
Feathers from the chicken were collected and cleaned with sterile water to remove blood stains, further it was cut into proper length and soaked in the crude enzyme for 24 hrs to estimate the effectiveness of crude enzyme in feather degradation (Jo et al., 2008).
2.8.3 Degradation of hairs from goat skin
The goat skin with hairs was collected and subjected to the crude enzyme, the goat skin was cut into uniform length and was placed in Petri plates and then crude enzyme was added and incubated for 24 hrs, then the results were analyzed (Kumar et al., 2008).
3 Results and discussion
3.1 Isolation of microorganisms producing proteases
A total of 10 bacterial strains were identified and purified in the current investigation through repeated subculturing and streaking. The use of skim milk agar was reported by other researchers (Beg et al., 2002; Suganthi et al., 2013; D Odia et al., 2006). The 10 bacterial strains were screened for proteolytic activity on skim milk agar plates, and 4 bacterial strains displayed clear zones around their colonies ranging from small to large clear zones indicating the activity. Among these isolates, the strain PL4 showed the maximum proteolytic activity, indicating the zone of clearance of 26 mm (Figs. 1, 2) and was selected for the further study.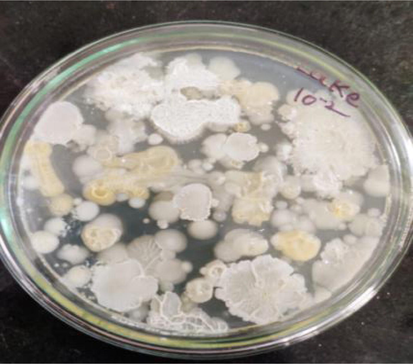
Isolation of bacterial colonies from lake soil.
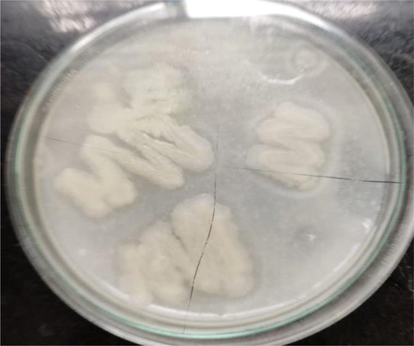
Screening for proteolytic acitivity by skim milk agar.
3.2 Identification of bacteria
Gram staining and endospore staining, respectively, revealed that the strain PL4 was rod-shaped and spore-forming (Fig. 3), and the Bacillus nakamurai strain PL4 displayed the most homology with Bacillus sp (Accession number-ON817261-ON817263) provided by the National Collection of Industrial Microorganisms of the Council of Scientific and Industrial Research (CSIR-National Chemical Laboratory) (NCL Pune). The bacterium Bacillus nakamurai strain PL4 is closely related to the strains Bacillus velezensis strain CMB 205 and Bacillus amyloliquefaciens strain MPA 1034, according to the phylogenetic tree created by neighbor-joining (Fig. 4). The findings concurred with those of (Ali et al., 2016; Sfg et al., 2008; Banerjee et al., 1999; Pastor et al., 2001), who discovered many bacterial strains connected to bacillus genera. Additionally, it is well known that bacteria, notably Bacillus species, produce the majority of the commercially available protease (Sfg et al., 2008). Indian lake soil was sought after by many researchers as a rich source of microorganisms that create protease enzymes (Banerjee et al., 1999; Pastor et al., 2001).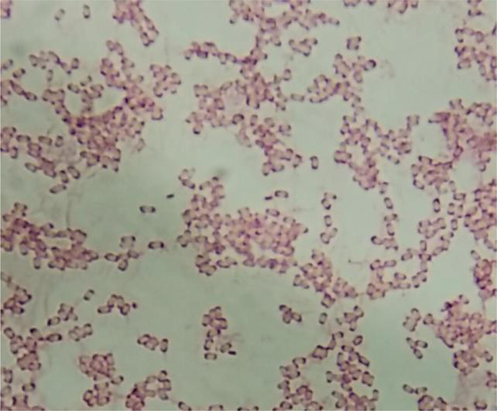
Gram positive Bacilli (spores).
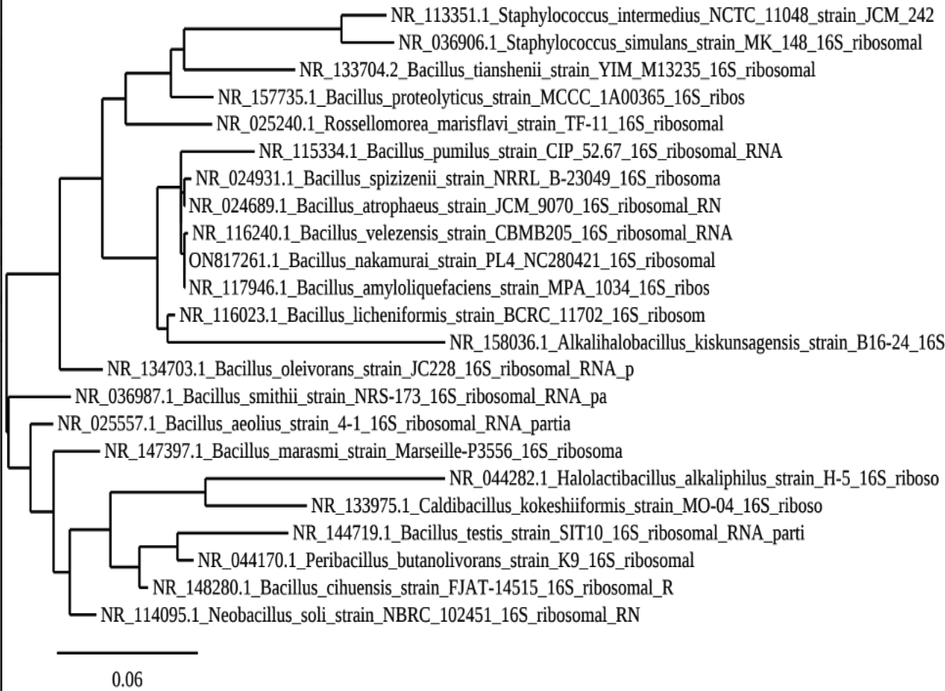
The 16 s rRNA from strain PL4 was used to create a phylogenetic tree illustrating the taxonomic position of the strain. The tree was created using the neighbor-joining method.
3.3 Production of protease-producing bacteria
The production media with the inoculum was incubated at 37 °C and at different time intervals showed the variation in growth of the Bacillus nakamurai PL4 and protease production, At 72 hrs of incubation the cell growth was observed to be high (OD at 600 nm: 0.7612) and there was a high protease activity (0.92 U/mL) determined by tyrosine standard graph (Fig. 5) and there was low cell growth (OD: 0.66) and protease production (0.8715 U/mL) at 24 hrs of incubation (Fig. 6). These results showed a direct connection between cell growth and protease synthesis, the reason could be that as there is the consumption of carbon sources, the microbes can produce protease in the high amount (Rahman et al., 1994). These results (Ferrero et al., 1996) recorded Bacillus licheniformis MIR 29′s maximal growth and protease production at 37 °C. And (Aruna et al., 2014), noted that Bacillus tequilensis CSGAB0139 displayed its peak proteolytic activity at 37 °C.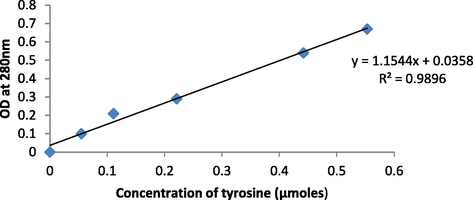
Tyrosine standard graph.
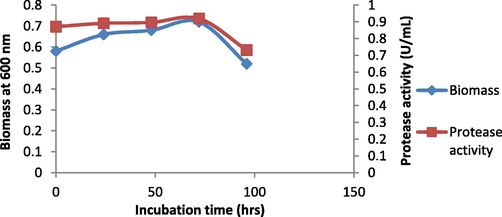
Effect of incubation time on cell growth and protease activity by B. Nakamurai PL4.
3.4 Optimization of fermentation conditions
3.4.1 Effect of pH on biomass and protease production.
Different pH values showed a different effect on protease yield and growth of B. nakamurai PL4 strain, where pH 4, 6, 8,10 & 12 were considered and the isolate showed high biomass (OD: 0.56) and protease yield (0.8105 U/mL) at pH 8 (Fig. 7) which indicated that the enzyme is alkaline in nature and low protease activity at pH 12 (0.613 U/mL), the alkaline pH of the media should be maintained for maximum protease activity, (Sharmin et al., 2005; Yossan et al., 2006; Nisha and Divakaran, 2014; Adinarayana et al., 2003; Razzaq et al., 2019), who reported high protease activity in alkaline pH from Bacillus amovivorus, Bacillus megaterium, Bacillus subtilis NS and Bacillus subtilis PE-1.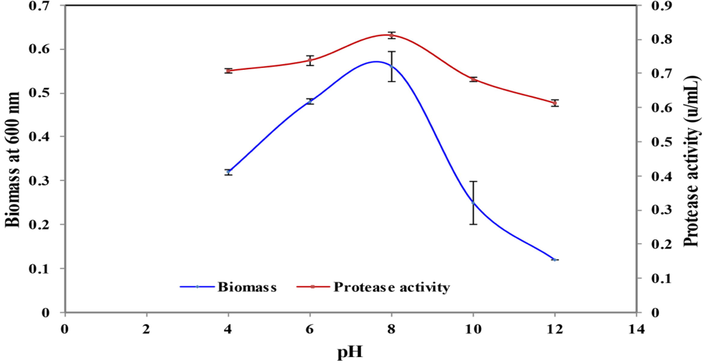
Effect of pH on cell growth and protease activity by B. Nakamurai PL4.
3.4.2 Effect of substrate on biomass and protease production.
Different substrates were considered for determining variation in protease and biomass yield, the substrates such as casein, azocasein, albumin, and gelatin were considered, in this casein was reported as the best substrate through which there was high biomass (OD:0.548) and protease yield (0.4436 U/mL) (Fig. 8) which indicated that when casein was used as substrate the microbes consumed it and there was high yield. Other researchers (Bhaskar et al., 2007; Haulon et al., 1982; Kole et al., 2007) also reported casein as the best substrate for Bacillus proteolyticus-CFR3001, Bacillus subtilis, and Bacillus licheniformis.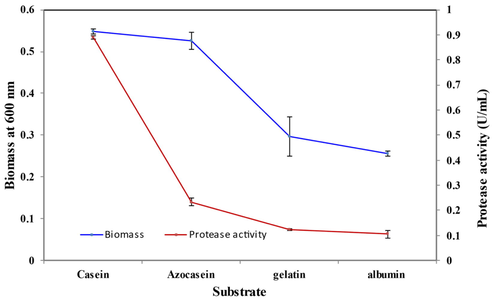
Effect of substrate on cell growth and protease activity by B. Nakamurai PL4.
3.5 Statistical experimental design
For the rapid evaluation of the effects of various media components, PB design is one of the statistical tools (Beg et al., 2003; Bhagwat et al., 2015). The first optimization step identified the significant factors for protease production from B. Nakamurai PL4 using 14 variables and 20 runs with two levels of low and high (Table 1), Table 3 represents the protease activity (U/mL) for each runs, the effect of each variable along with mean square, F-values, and p-values, the p-values < 0.05 were considered as the significant variables. In our study Xylose (p-value: 0.025), peptone (0.035), KH2PO4 (0.009) and MnCl2 (0.003) were considered as significant effect on protease production (Espoui et al., 2022; Javaid, 2022) from p-values in Table 4 (ANOVA table). Whereas other variables do not play an important/significant role in protease production. The observed values (Table 5) is identical to R-sq (94.19 %) (Table 6). Regression equation was analyzed to report the relationship between the set of data (From regression equation in uncoded units) and all other variables were neglected. Analysis of the impact of protease production was done by Pareto chart (Fig. 9) at a 95 % confidence level which indicated that MnCl2, KH2PO4, xylose, and peptone have a significant effect. Where MnCl2 plays a major role in protease production in 14 variables, from the main effect plots (Fig. 10) the negative and positive effects were analyzed for enzyme activity, where MnCl2 has the positive effect and xylose, peptone and KH2PO4 have negative effects.
Run
Blank
A
B
C
D
E
F
G
H
J
K
L
M
N
O
EA (U/mL)
1
1
+
–
+
+
–
–
–
–
+
–
+
–
+
+
0.25
2
1
+
+
–
+
+
–
–
–
–
+
–
+
–
+
0.12
3
1
–
+
+
–
+
+
–
–
–
–
+
–
+
–
0.25
4
1
–
–
+
+
–
+
+
–
–
–
–
+
–
+
0.125
5
1
+
–
–
+
+
–
+
+
–
–
–
–
+
–
0.23
6
1
+
+
–
–
+
+
–
+
+
–
–
–
–
+
0.15
7
1
+
+
+
–
–
+
+
–
+
+
–
–
–
–
0.25
8
1
+
+
+
+
–
–
+
+
–
+
+
–
–
–
0.23
9
1
–
+
+
+
+
–
–
+
+
–
+
+
–
–
0.11
10
1
+
–
+
+
+
+
–
–
+
+
–
+
+
–
0.12
11
1
–
+
–
+
+
+
+
–
–
+
+
–
+
+
0.28
12
1
+
–
+
–
+
+
+
+
–
–
+
+
–
+
0.14
13
1
–
+
–
+
–
+
+
+
+
–
–
+
+
–
0.23
14
1
–
–
+
–
+
–
+
+
+
+
–
–
+
+
0.3
15
1
–
–
–
+
–
+
–
+
+
+
+
–
–
+
0.18
16
1
–
–
–
–
+
–
+
–
+
+
+
+
–
–
0.189
17
1
+
–
–
–
–
+
–
+
–
+
+
+
+
–
0.3
18
1
+
+
–
–
–
–
+
–
+
–
+
+
+
+
0.25
19
1
–
+
+
–
–
–
–
+
–
+
–
+
+
+
0.23
20
1
–
–
–
–
–
–
–
–
–
–
–
–
–
–
0.22
Source
DF
Adj SS
Adj MS
F-value
P-value
Remarks
Model
14
0.066859
0.004776
5.79
0.032
–
Linear
14
0.066859
0.004776
5.79
0.032
–
Maltose (%W/V)
1
0.000154
0.000154
0.19
0.684
Non-significant
Glucose (%W/V)
1
0.000285
0.000285
0.35
0.582
Non-significant
Starch (%W/V)
1
0.001073
0.001073
1.30
0.306
Non-significant
Xylose (%W/V)
1
0.008344
0.008344
10.11
0.025
Significant
Peptone (%W/V)
1
0.006790
0.006790
8.23
0.035
Significant
Yeast extract
(%W/V)1
0.000456
0.000456
0.55
0.491
Non-significant
Urea(%W/V)
1
0.004104
0.004104
4.97
0.076
Non-significant
Ammonium nitrate(%W/V)
1
0.000078
0.000078
0.09
0.771
Non-significant
NaCl(%W/V)
1
0.000505
0.000505
0.61
0.470
Non-significant
MgSO4(%W/V)
1
0.002892
0.002892
3.50
0.120
Non-significant
ZnSO4(%W/V)
1
0.002153
0.002153
2.61
0.167
Non-significant
KH2PO4(%W/V)
1
0.014392
0.014392
7.44
0.009
significant
MnCl2(%W/V)
1
0.025099
0.025099
30.41
0.003
significant
Incoulum size
(%V/V)1
0.00536
0.000536
0.65
0.457
Non-significant
Error
4
0.004127
0.000825
–
–
–
Total
19
0.070986
–
–
–
–
Term
Effect
Coef
T-Value
Remarks
Constant
0.2097
32.84
Maltose (%W/V)
−0.00555
−0.00278
−0.43
Negative effect
Glucose (%W/V)
0.00755
0.00378
0.59
Positive effect
Starch (%W/V)
−0.01465
−0.00733
−1.14
Negative effect
Xylose (%W/V)
−0.04085
−0.02042
−3.18
Negative effect
Peptone (%W/V)
-0.0.03685
−0.01843
−2.87
Negative effect
Yeast extract (%W/V)
−0.00955
−0.00478
−0.74
Negative effect
Urea (%W/V)
0.02865
0.01432
2.23
Positive effect
Ammonium nitrate (%W/V)
0.00395
0.00197
0.31
Positive effect
NaCl (%W/V)
−0.01005
−0.00502
−0.78
Negative effect
MgSO4 (%W/V)
0.02405
0.01203
1.87
Positive effect
ZnSO4(%W/V)
0.02075
0.01037
1.61
Positive effect
KH2PO4(%W/V)
−0.05365
−0.02683
−4.18
Negative effect
MnCl2(%W/V)
0.07085
0.03542
5.51
Positive effect
Incoulum size
(%V/V)−0.01035
−0.00518
−0.81
Negative effect
MODEL SUMMARY
S
R-sq
R-sq(adj)
R-sq(pred)
0.0287300
94.19 %
77.91 %
6.98 %
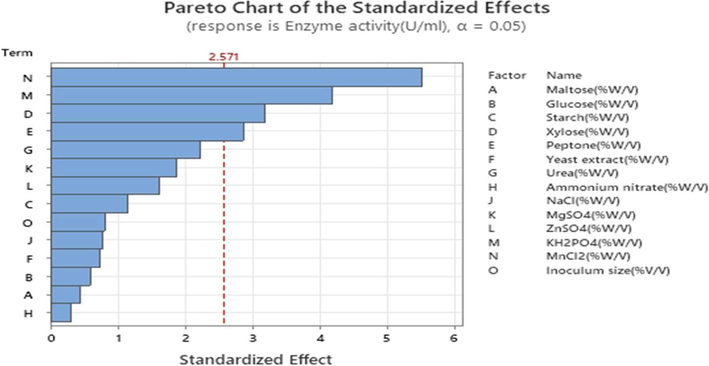
Pareto chart in response of enzyme activity (U/mL).
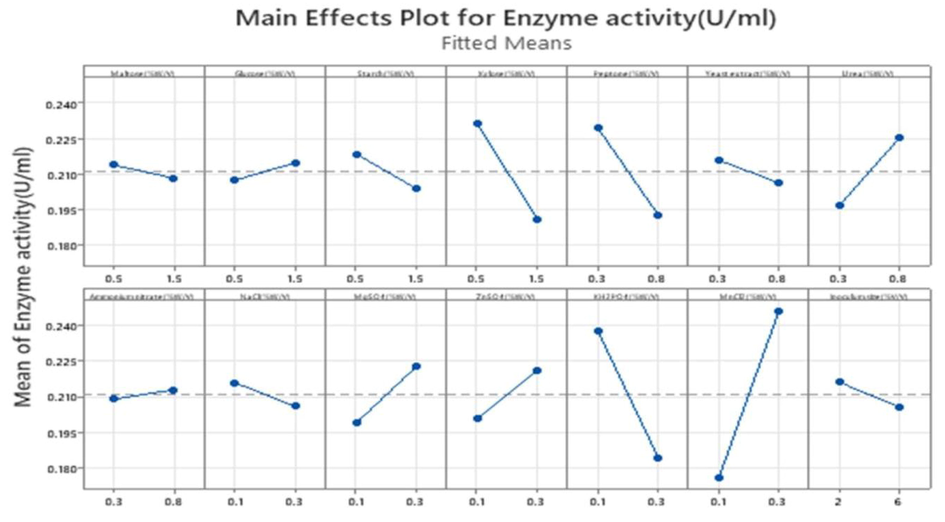
Main effects plot for enzyme activity.
From the above Table 5, Maltose, starch, xylose, peptone, yeast extract, NaCl, KH2PO4, and inoculum size have negative effects, and Urea, ZnSO4, MnCl2, ammonium nitrate, MgSO4, and Glucose have positive effects, this optimization technique helps to reduce the cost of the certain process by optimizing the parameters so that non-significant parameters can be removed from the design. In our study xylose plays a major role as a carbon source, MnCl2, and KH2PO4 as the metal ions, and peptone as the nitrogen source.
3.6 Regression equation
Enzyme activity (U/mL) = 0.2381–0.0056 Maltose (%W/V) − 0.0076 Glucose (%W/V) − 0.0146 Starch (%W/V) − 0.0408 Xylose (%W/V) − 0.0737 Peptone (%W/V) − 0.0191 Yeast extract (%W/V) + 0.0573 Urea (%W/V) + 0.0079 Ammonium nitrate (%W/V)- 0.0502 NaCl (%W/V) + 0.1202 MgSO4 (%W/V) + 0.1038 ZnSO4 (%W/V)-0.2682 KH2PO4 (%W/V) + 0.3543 MnCl2 (%W/V) −0.00259 Inoculum size (%W/V).
3.7 Partial purification by ammonium sulphate precipitation and dialysis
Ammonium sulphate precipitation (Table 7) was performed at the different concentration of ammonium sulphate to determine the protease activity at each stage, the enzyme purity was observed to be increased by increasing the ammonium sulphate concentration of crude enzyme and there was a decrease in enzyme activity of supernatant and increase in enzyme activity of pellets with increase in concentration of ammonium sulphate precipitation (40–60 %) Fig. 11, high protease activity was observed when 60 % of ammonium sulphate concentration was added to crude enzyme and it was observed that purity was increased.
Purification step
Protease activity (U/mL)
Crude enzyme
0.3
Ammonium sulphate precipitation
0.72
Dialysis
0.83
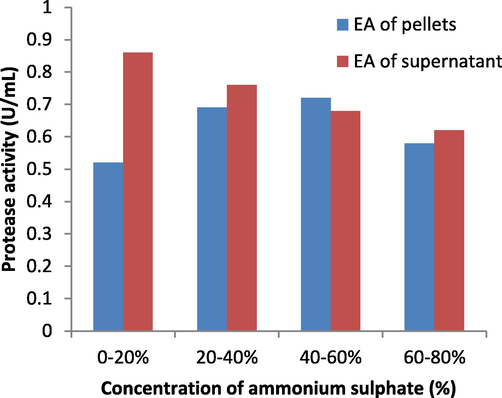
Enzyme activity of supernatant and pellets.
Further purification was done by dialysis to get high purity enzyme, this was ensured by calculating protease activity, it was observed that protease activity was increased by dialysis.
3.8 Applications of protease production by B. Nakamurai PL4 in detergent, feather and goat hair degradation
Crude protease enzyme was examined for stain removal activity, the egg yolk-stained cloth was soaked with crude enzyme, detergent, and water for about 3 hrs, a desirable change was noticed and was effective in removing stain from cloth, Fig. 12. Which concluded that crude enzyme in combination with water and detergent gives good results. This application has a major contribution to the formulation of detergent additives, also other researchers (Biosci et al., 2016; Rao et al., 2009; Al-Ghanayem and Joseph 2020) examined the ability of protease enzyme in stain removal from Bacillus circulans.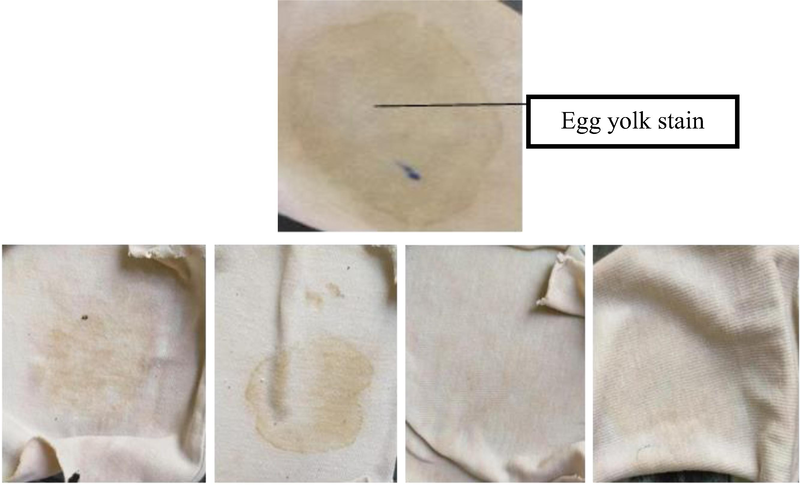
Removal of egg yolk stain by crude protease enzyme., Only water (b) Water and protease (c)Water and detergent (d)Water detergent and protease.
The chicken feathers were considered for the experiment, the main aim was to check the ability of crude protease enzyme is degradation of the feathers, the crude enzyme was successful in degrading the feathers when kept at the incubation for 24 hrs, Fig. 13, which acts as keratin applications. Other researchers (Zaghloul, 1998; Mohamedin, 1999) also reported the ability of feather degradation in Bacillus subtilis and Streptomyces strain.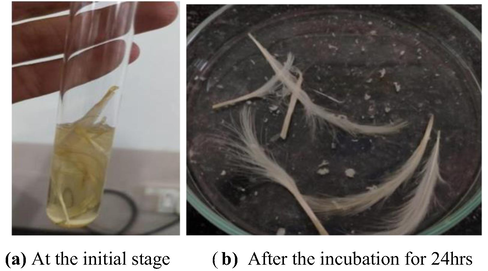
Degradation of chicken feathers from crude protease enzyme.
Goat skin was considered for the experiment, the main aim was to check the proteolytic activity on goat skin for the dehairing activity, Fig. 14, the crude enzyme was effective in removing the hairs after the incubation for 24 hrs, this has a huge role in leather industry and degumming (Grbavcic et al., 2011; Prakash et al., 2005; Hammami et al., 2018). Other researchers (Dayanandan et al., 2003; Annapurana et al., 1996) also examined the protease enzyme from Bacillus species on dehairing activity.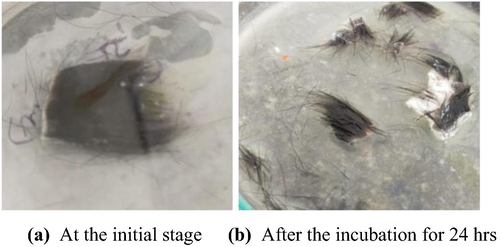
Dehairing by crude protease enzyme.
4 Conclusions
The present study revealed the production of alkaline protease from B. Nakamurai PL4 and its homology with other strain through phylogenetic analysis, the optimization studies helped to understand the nature of cell growth and protease production, where protease production was high at 72 hrs of incubation, at pH 8 and casein was considered best suitable substrate, further statistical optimization was done by using tool named Plackett-Burman design, in which the significant factors were peptone, MnCl2, KH2PO4 and xylose from 14 variables and 20 runs, However further optimization is required, partial purification was done by ammonium sulphate precipitation and dialysis to increase the proteolytic activity from crude enzyme. The protease activity was increased after ammonium sulphate precipitation to 0.72 U/mL and 0.83 U/mL after dailysis. Furthermore, the crude enzyme confirmed its importance in certain industrial applications like detergent additives, feather degradation and dehairing activity. Thus, Bacillus nakamuria PL4 can be utilized for the industrial-scale production of proteases to fulfill current demands. Furthermore, such improved parameters can optimize protease production and their application across various industries.
Funding
The authors are grateful to the Deanship of Scientific Research, Najran University, Najran, Saudi Arabia, for funding this research through grant research code NU/RG/MRC/11/9; this work was supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R199), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia; this work was supported by AlMaarefa University researchers supporting program (TUMA-2021–36), AlMaarefa University, Riyadh, Saudi Arabia.
Acknowledgments
The authors are thankful to Najran University, Najran, KSA; KLE Technological University, BVB Campus, Hubballi-580031, Karnataka, India. The authors are grateful to the Deanship of Scientific Research, Najran University, Najran, Saudi Arabia, for funding this research through grant research code NU/RG/MRC/11/9; this work was supported by Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R199), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia; this work was supported by AlMaarefa University researchers supporting program (TUMA-2021–36), AlMaarefa University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Purification and partial characterization of thermostable serine alkaline protease from a newly isolated Bacillus subtilis PE-11. AAPS PharmSciTech. 2003;4:E56.
- [CrossRef] [Google Scholar]
- Investigation on alkaline protease production with B. subtilisPE-11 immobilized in calcium alginate gel beads. Process Biochem.. 2004;39:1331-1339.
- [Google Scholar]
- Current prospective in using cold-active enzymes as eco- friendly detergent additive. Appl. Microbiol. Biotechnol 2020:2871-2882.
- [Google Scholar]
- Molecular characterization and growth optimization of halo-tolerant protease producing Bacillus Subtilis Strain BLK-1.5 isolated from salt mines of Karak, Pakistan. Extremophiles. 2016;20:395-402.
- [CrossRef] [Google Scholar]
- Evaluation and mechanism of glucose production through acid hydrolysis process: Statistical approach. Biocatal. Agric. Biotechnol.. 2021;36:102157
- [CrossRef] [Google Scholar]
- Eco-friendly enzymatic dehairing using extracellular protease from Bacillus species isolate. J. Am. Leath Chem. Assoc.. 1996;91:115-119.
- [Google Scholar]
- Production and partial characterization of alkaline protease from Bacillus tequilensis strains CSGAB0139 isolated from spoilt cottage cheese. Int. J. Appl. Biol. Pharma. Tech.. 2014;5:201-221.
- [Google Scholar]
- Thermostable alkaline protease from Bacillus brevis and its characterization as a laundry detergent additive. Process Biochem.. 1999;35:213-219.
- [CrossRef] [Google Scholar]
- Kinetic constants determination for an alkaline protease from Bacillus mojavensis using response surface methodology. Biotechnol. Bioeng.. 2002;78:289-295.
- [CrossRef] [Google Scholar]
- Statistical media optimization and alkaline protease production from Bacillus mojavensis in a bioreactor. Process Biochem.. 2003;39:203-209.
- [CrossRef] [Google Scholar]
- Statistical medium optimization for the production of collagenolytic protease by Pseudomonas sp. SUK using response surface methodology. Microbiology. 2015;84:520-530.
- [CrossRef] [Google Scholar]
- Bhaskar, N., Sudeepa, E.S., Rashmi, H.N., Selvi, A.T., 2007. Partial purification and characterization of protease of Bacillus proteolyticus-CFR3001 isolated from fishprocessing wasteand its antibacterial activities.
- Production, purification and characterization of detergent-stable, halotolerant alkaline protease for ecofriendly application in detergents’ industry. Int. J. Biosci.. 2016;8:47-65.
- [Google Scholar]
- Screening and isolation of protease producing bacteria from soil collected from different areas of Burhanpur Region (MP) India. Int. J. Curr. Microbiol. Appl. Sci.. 2015;4:597-606.
- [Google Scholar]
- Application of an alkaline protease in leather processing: an ecofriendly approach. J. Clean. Prod.. 2003;11:533-536.
- [CrossRef] [Google Scholar]
- Partial purification and characterization of a thermophilic and alkali-stable laccase of Phoma herbarum isolate KU4 with dye-decolorization efficiency. Prep. Biochem. Biotechnol.. 2021;51:901-918.
- [CrossRef] [Google Scholar]
- Purification, characterization of alkaline protease enzyme from native isolate Aspergillus niger and its compatibility with commercial detergents. Indian J. Sci. Technol.. 2008;1:1-6.
- [CrossRef] [Google Scholar]
- Purification and characterization of a solvent and detergent stable novel protease from Bacillus cereu”s. Bacillus cereu”s. Micro. Res. 2007;159:135-140.
- [Google Scholar]
- Papain-like proteases: applications of their inhibitors. Afr. J. Biotechnol.. 2007;6:1077-1086.
- [Google Scholar]
- Optimization of protease production process using bran waste using Bacillus licheniformis. Korean J. Chem. Eng.. 2022;39:674-683.
- [CrossRef] [Google Scholar]
- Thermo stable alkaline protease of Bacillus licheniformis MIR 29: isolation, production and characterization. Appl. Microbiol. Biotechnol. 1996;45:327-332.
- [Google Scholar]
- Production of lipase and protease from an indigenous Pseudomonas aeruginosa strain and their evaluation as detergent additives: compatibility study with detergent ingredients and washing performance. Bioresour. Technol.. 2011;102:11226-11233.
- [CrossRef] [Google Scholar]
- Bacterial alkaline proteases: molecular approaches and industrial applications. Appl. Microbiol. Biotechnol.. 2002;59:15-32.
- [CrossRef] [Google Scholar]
- Low-cost culture medium for the production of proteases by Bacillus mojavensis SA and their potential use for the preparation of antioxidant protein hydrolysate from meat sausage by-products. Ann. Microbiol.. 2018;68:473-484.
- [CrossRef] [Google Scholar]
- The influence of glucose, ammonium and magnesium availability on the production of protease and bacitracin by Bacillus licheniformis. J. Gen. Microbiol. 1982;128:845-851.
- [Google Scholar]
- Enhanced phytase production from indigenous Bacillus subtilis KT004404 by response surface methodology to be used as poultry feed supplement. Pak. J. Zool. 2022
- [CrossRef] [Google Scholar]
- Screening of protease-producing Serratia marcescens FS-3 and its application to deproteinization of crab shell waste for chitin extraction. Carbohydr. Polym.. 2008;74:504-508.
- [CrossRef] [Google Scholar]
- Protease from genus iBacillus. I: neutral proteases. Biotechnol. Biotechnol. Bioeng. 1970;12:179-212.
- [Google Scholar]
- Industrial enzyme applications. Curr. Opin. Biotechnol.. 2002;13:345-351.
- [CrossRef] [Google Scholar]
- Production of protease by bacillus subtilis using simultaneous control of glucose and ammonium concentrations. J. Chem. Technol. Biotechnol.. 2007;41:197-206.
- [CrossRef] [Google Scholar]
- Optimization of conditions for production of neutral and alkaline protease from species of Bacillus and Pseudomonas”. Ind. J. Micro. 2002;42:233-236.
- [Google Scholar]
- Characterization of an alkaline active-thiol forming extracellular serine keratinase by the newly isolated Bacillus pumilus. J. Appl. Microbiol.. 2008;104:411-419.
- [CrossRef] [Google Scholar]
- Isolation, identification and some cultural conditions of a protease-producing thermophilic Streptomyces strain grown on chicken feather as substrate. Int. Biodeterior. Biodegrad.. 1999;43:13-21.
- [Google Scholar]
- Research applications of proteolytic enzymes in molecular biology. Biomolecules. 2013;3:923-942.
- [CrossRef] [Google Scholar]
- Potential application of protease isolated from Pseudomonas aeruginosa PD100. Electron. J. Biotechnol.. 2005;8:197-203.
- [CrossRef] [Google Scholar]
- Proteases production by a bacterial isolate Bacillus amyloliquefaciens 35s obtained from soil of the Nile delta of Egypt. Br. Microbiol. Res. J.. 2015;6:315-330.
- [CrossRef] [Google Scholar]
- Optimization of alkaline protease production from Bacillus subtilis NS isolated from sea water. Afr. J. Biotechnol.. 2014;13:1707-1713.
- [CrossRef] [Google Scholar]
- Characterization and wash performance analysis of an SDS-resistant alkaline protease from a Bacillus sp. World J. Microbiol. Biotechnol. 2001;17:493-497.
- [Google Scholar]
- Characterization and stability of extracellular alkaline proteases from halophilic and alkaliphilic bacteria isolated from saline habitat of coastal Gujarat. India. Brazil. J. Microbiol. 2006;37:276-282.
- [Google Scholar]
- Optimized production and properties of thermostable alkaline protease from Bacillus subtilis SHS-04 grown on groundnut (Arachishypogaea) meal. Adv. Enzyme Res.. 2013;1:112-120.
- [Google Scholar]
- Proteases production using Bacillus subtilis- 3411 and amaranth seed meal medium at different aeration rate. Brazil. J. Microbiol. 2001;32:6-9.
- [Google Scholar]
- Purification and characterization of Bacillus cereus protease suitable for detergent industry. Appl. Biochem. Biotechnol.. 2005;127:143-155.
- [CrossRef] [Google Scholar]
- Purification and characterization of a heat stable alkaline protease from Bacillus stearothermophilus F1. Appl Microbiol Biotechnol. 1994;40:822-827.
- [Google Scholar]
- Characterization of thermo- and detergent stable serine protease from isolated Bacillus circulans and evaluation of eco-friendly applications. Process Biochem. 2009
- [Google Scholar]
- Molecular and biotechnological aspects of microbial proteases. Microbiol. Mol. Biol. Rev.. 1998;62:597-635.
- [CrossRef] [Google Scholar]
- Molecular and biotechnological aspects of microbial protease. Microbiol. Mol. Biol. 1998;62:597-635.
- [Google Scholar]
- Optimization of process parameters using a statistical approach for protease production by Bacillus subtilis using cassava waste. Int. J. Chem. Tech. Res. 2012;4:749-760.
- [Google Scholar]
- Microbial proteases applications. Front. Bioeng. Biotechnol.. 2019;7:110.
- [CrossRef] [Google Scholar]
- Stability of thermostable alkaline protease from Bacillus licheniformis RP1 in commercial solid laundry detergent formulations. Microbiol. Res.. 2008;163:299-306.
- [CrossRef] [Google Scholar]
- Response surface optimization of medium composition for alkaline protease production by Bacillus clausii. Biochem. Eng. J. 2008;39:37-42.
- [Google Scholar]
- Isolation and characterization of a protease producing bacteria Bacillus amovivorus and optimization of some factors of culture conditions for protease production. J. Biol. Sci. (Faisalabad). 2005;5:358-362.
- [CrossRef] [Google Scholar]
- Screening and optimization of protease production from a halotolerant Bacillus licheniformis isolated from saltern sediments. J. Genet. Eng. Biotechnol.. 2013;11:47-52.
- [CrossRef] [Google Scholar]
- Production and optimization of process parameters for alkaline protease production by a newly isolated Bacillus sp. under solid state fermentation. Process Biochem.. 2004;39:1893-1898.
- [CrossRef] [Google Scholar]
- Optimal conditions for production of extracellular protease from newly isolated Bacillus cereus strain CA15. Eurasian J. Biosci.. 2011;1–9
- [CrossRef] [Google Scholar]
- Production and purification of protease from a Bacillus subtilis that can deproteinize crustacean wastes. Enzyme Microb. Technol.. 2000;26:406-413.
- [CrossRef] [Google Scholar]
- Sci. Asia.. 2006;32:377.
- [CrossRef]
- Cloned Bacillus subtilis alkaline protease (aprA) gene showing high level of keratinolytic activity. Appl. Biochem. Biotechnol.. 1998;70–72:199-205.
- [CrossRef] [Google Scholar]







