Translate this page into:
Differentiation of ‘Candidatus Liberibacter asiaticus’ in Saudi Arabia based on tandem repeat variability in genomic locus
⁎Corresponding author. yasereid@ksu.edu.sa (Yasser E. Ibrahim)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Citrus greening, or huanglongbing, is a destructive disease threatening many citrus worldwide, and drastically altering the global dynamics of the citrus industry. The disease is caused by one of several unculturable bacterial species belonging to ‘Candidatus Liberibacter’. The recent availability of complete genome sequences of ‘Candidatus Liberibacter asiaticus’ (CLas) has facilitated comprehensive assessments of genomic variability using a range of approaches, including short tandem repeat analysis. The objective of this study was to evaluate the genetic diversity of CLas populations in Saudi Arabia based on tandem repeat number (TRN) within the CLIBASIA_01645 locus, predicted to encode the bacteriophage C1 repressor protein. Results indicated that the genotype richness of the Saudi Arabian CLas isolates was conserved by 27% based on the TRN locus. Four different genotypes TRN2, TRN3, TRN4, and TRN5 were identified. However, the TRN2 and TRN5 were the most dominant genotypes. All four of the TRN genotypes were associated with CLas-positive mandarin (Citrus reticulata) or sweet orange (C. sinensis) citrus trees. The diversity (H = 0.69) and evenness (H'=0.914) were overall relatively high, with the northern region of Saudi Arabia harboring the highest diversity (0.7) and evenness score (0.9–1.0). Phylogenetic analysis of the CLas-bacteriophage C1 repressor protein of the Saudi Arabian isolates indicated CLas was more closely related to ‘Candidatus Liberibacter africanus’ than to ‘Candidatus Liberibacter americanus’.
Keywords
Bacteriophage C1 repressor protein
Citrus
Genetic diversity
Huanglongbing
Minisatellite
1 Introduction
Citrus greening (CG), or huanglongbing, is the most important disease of cultivated citrus trees worldwide (Albrecht and Bowman, 2012). Citrus greening has been detected in at least 40 countries in Africa, Asia, Oceanian, and North and South American countries, with approximately 100 million infected trees (Bové, 2006; FAO, 2013). The disease is known as a yellow dragon in China (Reinking, 1919), yellow shoot in South Africa (Oberholzer et al., 1965), dieback in India (Capoor, 1963), and phloem necrosis and vein phloem degeneration in Indonesia (Tirtawidjaja et al., 1965). The causal agent of CG is obligate phloem-restricted gram-negative bacteria belonging to alpha Proteobacteria, family Rhizobiaceae. The bacteria were characterized in 1994 based on their 16S rDNA sequence and shown to be a genus named Candidatus Liberibacter (Jagoeuiex et al., 1994). Three different species of Candidatus (‘Ca’) Liberabacter that infect the rutaceous family were identified, including ‘Ca Liberibacter asiaticus’ (CLas), ‘Ca Liberibacter africanus’ (CLaf), and ‘Ca Liberibacter americanus’ (CLam). ‘Candidatus Liberibacter asiaticus’ is endemic to Asia and is also known to occur in Brazil (Sao Paulo), the United States (Florida) (Bové, 2006), Africa (Saponari et al., 2010), and Arabian Peninsula (Bové, 2006). ‘Candidatus Liberibacter africanus’ was found in African countries (Oberholzer et al., 1965). ‘Candidatus Liberibacter americanus’ was found in Brazil and South America (Texeira et al., 2005). ‘Candidatus Liberibacter asiaticus’ and ‘Ca Liberibacter americanus’ are transmitted by the Asian citrus psyllid (ACP) Diaphorina citri, (Kuwayama, 1908) and CLaf by the two-spotted citrus psyllid Trioza erytreae (Del Gurecio), respectively C. This citrus greening disease was first reported in Saudi Arabia in 1980 based on characteristic symptoms and electron microscope. Infected trees were reported to occur in the Makkah and Asir regions (Bové and Garnier, 1984). In 2022 the disease was reported in Najran, Al Baha, Hail, Al Madinah, Al Jawf, Asir, Riyadh, Makkah, and Tabuk regions (Ibrahim et al., unpublished data). Saudi Arabia occupied almost four to fifths of the Arab Peninsula, with a total area of around 2,000,000 square kilometers (https://www.stats.gov.sa/en/page/259). Citrus in Saudi Arabia has been recognized as one of the important crops after dates and grapes, with total local production reaching 100,000 tons (Fiaz et al., 2018). In the era of genomics, molecular data such as genotyping is required for discriminating between CLas variants within the same species. The ability to differentiate species and/or variants of the same species provides valuable epidemiological data for tracking sources of infection, determining the genetic diversity of, and developing effective management practices (Al Obaidi et al., 2018; Singh et al., 2019). ‘Liberibacter asiaticus’ has traditionally been classified based on the most conserved genomic locus (16S rDNA). Although the 16S rDNA genomic region exhibits minimal genetic diversity of CLas (Nelson, 2012), somehow, it has been highly useful for the detection of Liberibacter species and strains or variants using PCR primers designed to amplify conserved regions (Coletta-Filho et al., 2005; Roberts et al., 2015). Other genomic regions, such as beta operon of ribosomal protein, and outer membrane protein genes have been somewhat useful for detecting CLas strains, but have not been useful for genetic differentiation studies (Bastianel et al., 2005; Deng et al., 2008; Hocquellet et al., 2009). Variable numbers of tandem repeats (VNTR) have been frequently used to distinguish other bacterial species (Katoh et al., 2011; Lin et al., 2005; Ma et al., 2014). For CLas, Katoh (2011) reported that VNTRs might discriminate CLas isolates more precisely than single nucleotide polymorphisms. In addition, two related and hypervariable genes (hyvI and hyvII) were identified in the prophage region of CLas strain Psy62 may be used to differentiate CLas strains not only from samples from various geographical locations origin and even from a single CLas-infected sample (Putammuk et al., 2014, Zhou et al., 2011). Locus CLIBASIA 01645 encoding bacteriophage repressor proteins which regulate phage/prophage activity (Duan et al., 2009), employed as a sensitive indicator in CLas strain distinction. Environmental adaptations and pathogenicity may be affected by variations in this chromosomal region (Liu et al., 2011). To now, there is no data regarding intraspecies variation in a population of Saudi Arabian CLas strains based on tandem repeat number (TRN). This study aimed to assess the genetic diversity in the CLas population prevalent in Saudi Arabia based on the TRN in the CLIBASIA_01645 locus.
2 Materials and methods
2.1 Bacterial strains
DNA samples of CG-infected citrus trees from the previous study (Ibrahim et al., unpublished data) were used for the genetic diversity assessment of Saudi Arabian CLas strains. Detailed information on the samples is presented in Table 1.
Region
Latitude
Longitude
Altitude
(MASL)*Host
Makkah
21°25′21″N
39°49′24″E
232–269
Citrus reticulata
Najran
17°29′30″N
44°7′56″E
1238–1244
C. limon, C. reticulata C. sinensis
Al Baha
20°00′45″N
41°27′55″E
1608–1913
C. reticulata, C. sinensis
Tabuk
28°23′50″N
36°34′44″E
774–802
C. reticulata, C. sinensis
Riyadh
24°38′N
46°43′E
599–666
C. reticulate
Hail
27°31′N
41°41′E
841–852
C. aurantofolia, C. reticulata, C. sinensis
Al Madinah (Al Ula)
24°28′N
39°36′E
767–827
C. sinensis
2.2 Amplification of CLIBASIA_01645 genomic locus
PCR amplification targeting CLIBASIA_01645 was carried out using specific primer pairs LapGP-1f (5′-GACATTTCAACGGTATCGAC-′3) and LapGP-1r (5′-GCGACATAATCTCACTCCTT-′3) (Chen et al., 2010). A 25 µl mixture containing 12.5 µl 2x green PCR ready mix (Promega), 0.4 mM forward/reverse primers, and 100 ng of template DNA was used for the reaction. PCR amplification was performed with an initial denaturation at 95˚C for 4 min followed by 35 cycles of 95˚C for 30 s, 58˚C for 45 s, 72˚C for 60 s, and the final extension at 72˚C for 10 min on Nexus gradient master cycler. The PCR products were separated into 1.5 % (w/v) agarose gels and stained with acridine orange.
2.3 Nucleotide sequencing, similarity, TRN Analysis, genetic diversity, and phylogeny analysis
The expected PCR products of CLIBASIA 01645 locus were cleaned and followed bidirectional sequencing on Macrogen Inc, Seoul, South Korea. Bioedit version 7. was used to clean, align and used for in silico translation of the nucleotide sequences. Cleaned sequences were analyzed further for their similarity to other deposited sequences on the gene bank using online BLAST tools on National Center for Biotechnology Information (NCBI) at https://blast.ncbi.nlm.nih.gov/Blast.cgi. Tandem repeat analysis was conducted using online Tandem Repeats Finder online software, Boston University (https://tandem.bu.edu/trf/trf.html) (Benson, 1999). The tandem repeats obtained in the software were further verified manually. The genetic diversity among different populations of CLas was calculated using Nei’s H value as H = 1 - ∑pi2, where ‘pi’ referred to the frequency of the allele ‘i’ at the locus (Nei, 1973). The evenness index was calculated as follows: H relative (H'); H'=H/Hmax where H Max = 1–1/Z; Z = number of groups of allel. The value of H' was between 0 and 1, where H'=1 when there was evenness. Statistical analysis was conducted on the frequency of different TRN using the F test employing analysis of variance test (Gomez and Gomez, 1984). Phylogeny of Candidatus Liberibacter based on bacteriophage repressor protein C1 was constructed using the MEGA version 11. The bootstrap consensus tree inferred from 1000 replicates is taken to represent the evolutionary history.
3 Results
3.1 Detection and analysis of locus CLIBASIA 01645 from citrus samples
The locus CLIBASIA 01645 was detected in field samples, and 50 samples from various citrus growing regions were positive with a single PCR amplicon group. Positive samples were distributed in 22 citrus farms and nurseries throughout seven distinct citrus growing regions (Table 2). Positive samples were found in the leaves, midrib, and/or petiole, as well as the placenta within the fruit. Positive samples were predominantly found in the petiole, accounting for 36 % of the total. The loci had varying copy numbers of the motif repeat sequence AGACACA, ranging from 2, 3, 4, and 5 copies. The number of copies of the theme in positive samples varied, with 34 % having two copies, 8 % having three copies, 40 % having four copies, and 18 % having five copies (Table 3). *MASL: meter above sea level. **Midrib. ***Petiole. ****Fruit. *The highest percentage among CLas TRN genotypes in Saudi Arabia.
Region
Altitude
(MASL)*Host
Source
TRN
Accession Number
Makkah
269
Citrus reticulata
M**
5
OK362253
Makkah
232
C. reticulata
M
4
OK362257
Makkah
232
C. reticulata
M
4
OK362261
Makkah
232
C. reticulata
M
4
OK362264
Najran
1225
C. sinensis
P***
4
OK362281
Najran
1225
C. sinensis
P
4
OK362282
Najran
1244
C. sinensis
M
4
OK362266
Najran
1244
C. sinensis
M
4
OK362270
Najran
1239
C. limon
P
5
OK362255
Najran
1239
C. limon
P
4
OK362259
Najran
1238
C. reticulata
P
5
OK362283
Najran
1238
C. reticulata
P
2
OK362256
Najran
1238
C. reticulata
P
4
OK362260
Najran
1238
C. reticulata
P
4
OK362284
Najran
1238
C. reticulata
P
2
OK362263
Najran
1238
C. reticulata
P
4
OK362276
Najran
1238
C. reticulata
P
2
OK362271
Najran
1238
C. reticulata
P
5
OK362267
Najran
1238
C. reticulata
P
4
OK362279
Al Baha
1913
C. sinensis
M
5
OK362262
Al Baha
1913
C. sinensis
F****
5
OK362265
Al Baha
1608
C. reticulata
F
2
OK362254
Al Baha
1608
C. reticulata
F
2
OK362268
Al Baha
1608
C. reticulata
M
2
OK362274
Al Baha
1608
C. reticulata
F
2
OK362273
Al Baha
1608
C. reticulata
F
2
OK362277
Al Baha
1608
C. reticulata
F
3
OK362258
Tabuk
795
C. sinensis
M
5
OK362269
Tabuk
774
C. reticulata
M
3
OK362272
Tabuk
774
C. sinensis
F
2
OK362275
Tabuk
802
C. sinensis
F
4
OK362280
Riyadh
666
C. reticulata
M
4
OK362278
Riyadh
666
C. reticulata
M + P
2
OK362250
Riyadh
666
C. reticulata
M + P
2
OK362252
Riyadh
599
C. reticulata
M + P
2
OK362251
Hail
852
C.aurantofolia
P
5
OK362235
Hail
852
C.aurantofolia
P
2
OK362246
Hail
852
C.aurantofolia
P
5
OK362236
Hail
852
C. reticulata
P
2
OK362247
Hail
852
C. reticulata
M + P
4
OK362237
Hail
841
C. sinensis
M + P
3
OK362243
Hail
841
C.reticulate
M + P
4
OK362238
Hail
841
C. reticulata
P
4
OK362239
Al Madinah (Al Ula)
827
C. sinensis
P
4
OK362240
Al Madinah (Al Ula)
827
C. sinensis
P
2
OK362248
Al Madinah (Al Ula)
778
C. sinensis
P
4
OK362241
Al Madinah (Al Ula)
781
C. sinensis
P
2
OK362249
Al Madinah (Al Ula)
767
C. sinensis
P
4
OK362242
Al Madinah (Al Ula)
767
C. sinensis
P
2
OK362245
Al Madinah (Al Ula)
767
C. sinensis
P
3
OK362244
Region
Number of repeat motif
Index
5
4
3
2
Diversity
Evenness
Tabuk
2
2
2
2
0.75
1.0
Hail
4
6
2
4
0.72
0.958
Al Madinah (Al Ula)
6
2
6
0.61
0.816
Najran
6
18*
6
0.56
0.747
Al Baha
4
2
10
0.53
0.708
Makkah
2
6
0.38
0.5
Riyadh
2
6
0.38
0.5
Total
18
40
8
34
0.69
0.914
3.2 Mapping of the CLas population in Saudi Arabia based on TRN profiling in locus CLIBASIA 01645
TRN profiling revealed that the CLas population in Saudi Arabia had four genotypes: TRN2, TRN3, TRN4, and TRN5. TRN4 genotype was the most common, followed by TRN2, TRN5, and TRN3, with distribution frequencies of 40 %, 34 %, 18 %, and 8 %, respectively. TRN4 was found in six out of seven citrus-growing regions. The TRN4 genotype was found in Najran (18 %), Makkah, Hail, Al Madinah (Al-Ula) (6 %), Tabuk, and Riyadh (2 %). The TRN 5 genotype was found in Najran (6 %), Al Baha and Hail (4 %), Makkah, and Tabuk (2 %). TRN2 genotype was found in six regions, including Al Baha (10 %), Najran, Riyadh, Al Madinah (Al-Ula) (6 %), Hail (4 %), and Tabuk (4 %). TRN3 genotype was found in four regions: Al Baha, Tabuk, Hail, and Al Madinah (Al-Ula), with a percentage frequency of 3 % in each. The regions of Tabuk and Hail were the wealthiest regions for CLIBASIA 01645 in terms of genotype detection in citrus growing regions, followed by Al Madinah (Al-Ula), Najran, Al Baha, Makkah, and Riyadh. Tabuk and Hail shared four genotypes in common. Three genotypes were found in Al Baha, Najran, and Al Ula, whereas two genotypes were found in Makkah and Riyadh. With a variety rating, the diversity index (H) equal to 0.75, and the evenness index (H') equal to 1, Tabuk was the most diverse region. Hail had a high level of variety (H = 0.72) and an evenness index of 0.958. Al Madinah (Al-Ula) was the third most diverse (0.61) and evenly distributed region (0.816). Najran and Al Baha had the same diversity with index values of 0.56 and 0.53, respectively. Makkah and Riyadh were the lowest regions for diversity, with an index value of 0.38 and the evenness value was 0.5. CLas genotype distribution was different between citrus hosts, according to TRN CLIBASIA 01645. In cultivars C. reticulata and C. sinensis, all TRN genotypes were identified. Each host, including C. auranntifolia and C. limon, had two TRN. TRN 5 and TRN 2 genotypes were found in C. aurantifolia, while TRN 5 and TRN 4 genotypes were found in C. limon. C. reticulata conserved the majority of genotypes, accounting for 56 % of the total, followed by C. sinensis (34 %), C. aurantifolia (6 %), and C. limon (4 %) (Fig. 1). A comparison of the CLas population of the Saudi Arabian isolates with other CLas populations throughout the world revealed that the CLas genotype of Saudi Arabia was relatively diversified. We noted 17 distinct CLas genotypes from different regions (countries) worldwide. The CLas population variation in TRN ranged from 2 to 21 copies of the motif AGACACA. Saudi Arabia retained four of the seventeen CLas genotypes, making the country a diverse genotype compared to other countries. Saudi Arabia had more CLas diversity (H = 0.69) than China (H = 0.66), Iran (H = 0.32), and the United States (H = 0.4) (Table 4). Among the four genotypes in the Saudi Arabia population, only genotypes with TRN2 and TRN5 were in the frame, with frequencies of 34 % and 18 % in Saudi Arabia, respectively. Both TRN had been found in Tabuk, Hail, and Al Baha. The TRN 2 genotype had 651 bp and encoded 216 amino acids. The TRN 2 genotype was deficient in the amino acid RHKTQDT. Eight percent of the population of CLas genotype was detected from the low altitude regions with composition 2 % of TRN5 genotype and 6 % of TRN4 genotype. Most of the Clas genotype was distributed in high altitude regions with a composition of 34 % TRN 2 and TRN 4, and 16 % were TRN5. Cluster analysis based on CLas diversity, region, and altitude data showed that the CLas population in Saudi Arabia could be clustered in three different main regions, i.e., north, middle, and south regions (Fig. 2). Numbers of the total sample and diversity index (H’/Nei value) are highlighted in bold. NC: not calculated because the total number of samples is<30.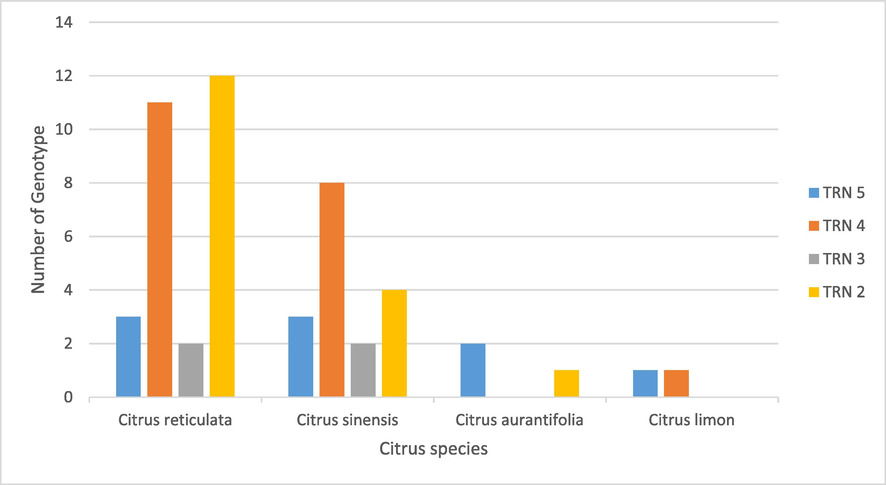
Graphical illustration of ‘Candidatus Liberibacter asiaticus’ genotypes according to tandem repeat numbers CLIBASIA 01645 distribution in different citrus species in Saudi Arabia.
Region (country)
TRN
Total
H’
Ref.
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
21
Iran
4
26
1
1
32
0.32
Saberi et al., 2018
China
1
9
19
31
3
2
65
0.66
Chen et al., 2010
USA
2
6
97
4
1
11
1
3
1
126
0.40
Chen et al., 2010
Matos et al., 2013
India
2
2
14
27
21
23
13
12
2
3
2
6
1
1
1
130
0.87
Ghosh et al., 2015
Katoh et al., 2012
Japan
8
19
8
3
8
4
15
9
4
2
1
81
0.86
Katoh et al., 2011
Taiwan
1
1
1
1
4
NC
Katoh et al., 2011
Timor leste
2
1
3
NC
Katoh et al., 2012
Papua New Guinea
1
1
NC
Katoh et al., 2012
Indonesia
2
1
2
3
3
1
12
NC
Katoh et al., 2011
Saudi Arabia
17
4
20
9
50
0.69
This study
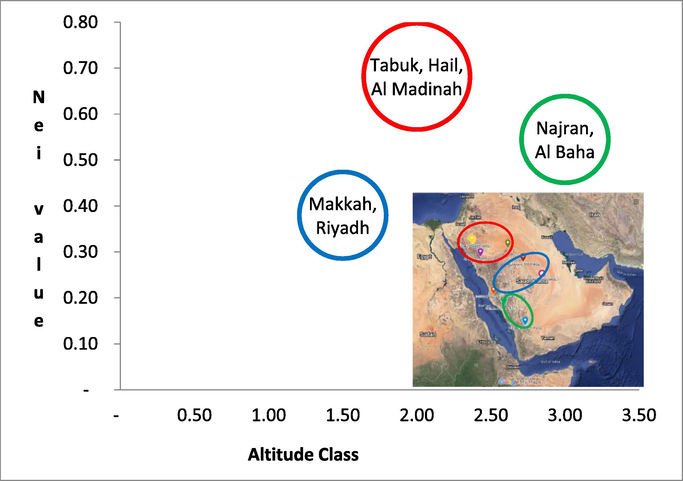
Cluster Analysis of ‘Candidatus Liberibacter asiaticus’ genotypes in Saudi Arabia based on tandem repeat numbers diversity.
4 Discussion
The genetic variability of the CLas population connected to the CG disease of citrus in Saudi Arabia was explored based on the diversity of different citrus species using TRN in variable CLIBASIA 01645 genomic regions. Our findings revealed the heterogeneity of the Saudi Arabia CLas isolates, which conserved 27 % of the total genotype CLas reported worldwide (i.e., 17 genotypes). Our data also revealed that Saudi Arabia’s population was not only heterogeneous but also diversified when compared to other populations worldwide, such as those in Iran and the United States. This finding could be explained since richness (i.e., the total number of genotypes found in a given location) and evenness (i.e., distribution frequency of genotypes among each other, i.e., evenness) will influence the high diversity value (Pyron, 2010). Although the number of genotypes found in Saudi Arabia was lower than in the United States (9 genotypes) and Iran (4 genotypes), the frequency of genotypes was even, and the dominance genotype was quite low. As a result, Saudi Arabia had a high level of diversity. The dominating genotype TRN 4, had the highest frequency in Saudi Arabia (40 %) across the country, while TRN 5 had the highest frequency in the United States (77 %) and Iran (81 %), respectively. Interestingly, the Saudi Arabian dominance genotype differed from that of other countries. Saudi Arabia possesses TRN 4 and 2 as dominant genotypes; however, TRN 5 and 7 were the predominant genotype in many countries. ‘Ca Liberibacter asiaticus’ population was significantly different between citrus hosts (P < 0.05). ‘Ca Liberibacter asiaticus’ populations were abundant in C. reticulata (Mandarin) and C. sinensis (Sweet Orange). All of the Saudi Arabian genotypes were likewise conserved in those two citrus species. On the other hand, C. aurantifolia (Mexican lime) and C. limon (Lemon) had only two genotypes. Citrus reticulata and C. sinensis were favorable for CG; however, C. aurantifolia and C. limon were not. Folimonova et al. (2009) divided Citrus genotypes into four categories based on their reaction to CLas: sensitive, moderately tolerant, tolerant, and resistant. Mandarin and sweet orange were classified as sensitive, while lime and lemon were classified as moderately tolerant or tolerant. In Saudi Arabia, the CLas genotype was rather evenly distributed. The CLas genotype was abundant in Tabuk and Hail, followed by Al Madinah (Al Ula), Najran, and Al Baha. However, Makkah and Riyadh were the lowest regions in richness and diversity. This finding could imply that the CLas had already established themselves in those three regions, i.e., Najran, Makkah, and Riyadh, much earlier. The altitude of the region may also influence the distribution of the CLas genotype. In this study, we found that the distribution of the CLas genotype was significantly different (P < 0.05) between low-altitude regions (<500 m above sea level) and high-altitude regions (>500 m above sea level), and our data were cooperative with Sigh et al. (2019) and Wang et al. (2012). Interestingly, our finding showed that TRN 2 was prevalent in high altitudes, in contrast reported before by Singh et al. (2019), who reported that TRN2 was prevalent in low altitudes. This may suggest that TRN2 was adaptive to different altitudes. Based on CLas diversity, region, and altitude data, cluster analysis revealed that the CLas population in Saudi Arabia could be divided into three primary regions: north, middle, and south. This finding leads to the conclusion that the north region of Saudi Arabia was most likely the source of CLas genetic variation and maybe a significant region in the origin or speciation of CLas, which is still unknown. This notion was reinforced by the previously given historical evidence on citrus farming in Al Madinah (Al Ula) and the theory proposed by Beattie et al. (2005 and 2008) that CLas originated in Africa. Beattie et al. (2005 and 2008) proposed that the origin of CLas was not from China or even India but Africa. CLas was predicted to come from Africa and transmitted from Vepris lanceolata (family Rutaceae); the original host of CLas; to orange or mandarin trees by T. erytreae in one of the European colonies on the southeast coast of Africa and then taken to the Indian subcontinent in infected plants or budwood some 300–500 years ago. It was then acquired and spread by D. citri. The CLIBASIA 01645 locus was annotated as a bacteriophage repressor protein C1 gene (Duan et al., 2009). Only TRN2, TRN5, TRN8, TRN11, TRN14, TRN17, and TRN 20 genotypes were in the frame, according to in silico analysis of sequences on locus CLIBASIA 01645. The amino acid RHKTQDT was encoded by the majority of the in-frame genotypes. According to the TRN, the amino acids will be repeated numerous times. TRN 2, found in nearly every region of Saudi Arabia had lost these amino acid sequences. TRN 2 had a 216-amino-acid genotype code. TRN 2 amino acid sequences were compared to other amino acid sequences NCBI database and found to be 81.94 % and 81.59 % similar to helix turn helix transcriptional regulator and bacteriophage repressor protein C1 of CLaf, respectively. Meanwhile, a low similarity was found with bacteriophage repressor protein C1 of Clam with a value of 69.44 %. This may indicate relatively close evolutionary history between the CLas population and the CLaf population rather than Clam, which was also supported by phylogeny analysis (Fig. 3). TRN variation in locus CLIBASIA 01645 was formed due to environmental adaptation or pathogenicity in countries such as India, Iran, and the United States (Liu et al., 2011). A Tandem repeat number was formed in bacterial populations by DNA strand slippage (Bichara et al., 2006; Verstrepen et al., 2005) and had a role in pathogenic or environmental fitness (Boles et al., 2004). On the other hand, the TRN 4 genotype predominated in the CLas community in Saudi Arabia's southern region. This variation could be attributable to an environmental effect, as TRN 4 was only found in the Najran region and not in Al Baha. We also believe that the CLas population was influenced by a neighboring country, since Yemen host CLaf strain. A precise genetic characterization of greening strains in Yemen is required. Generally, these results support the use of CLIBASIA 01645 locus for analyzing intraspecies CLas population structure and genetic diversity, and the potential for the use of different loci for more detailed analysis of CLas population structure.
Phylogeny of Candidatus Liberibacter spp, the causal agent citrus greening based on bacteriophage repressor protein C1 showing relatively close evolutionary history of ‘Candidatus Liberibacter asiaticus’ (CLas) with ‘Candidatus Liberibacter africanus’ (CLaf) (AKK20211) rather than ‘Candidatus Liberibacter americanus’ (CLam) (EMS36728). CLas and CLaf are close to CLso ‘Candidatus Liberibacter solanacerum’ (CLso). The amino acid of Saudi Arabia isolates and some selected CLas were translated in-silico using BioEdit version 7.0.5.3 before phylogenetic tree construction. The evolutionary history was inferred using the Neighbor-Joining method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches.
Ethical statement
Authors declared that this manuscript has not published elsewhere. All authors read and approved the final version of this manuscript. The authors declare that the present work was developed without any potential conflict of interest, with no human or animal participants.
Acknowledgments
This Work was funded by the National Plan for Science, Technology, and Innovation (MAARIFAH), King Abdul-Aziz City for Science and Technology, Kingdom of Saudi Arabia, grant Number (14-BIO-627-02).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Tolerance of trifoliate citrus rootstock hybrids to Candidatus Liberibacter asiaticus. Sci. Hortic.. 2012;147:71-80.
- [Google Scholar]
- Al-Obaidi, M., Z. Suhaili, M. Desa, and M. Nasir. 2018. Genotyping Approaches for Identification and Characterization of Staphylococcus aureus Genotyping Approaches for Identification and Characterization of Staphylococcus aureus. 10.5772/intechopen.75969.
- Diversity of ‘Candidatus Liberibacter asiaticus’, based on the omp gene sequence. Appl. Environ. Microbiol.. 2005;71:6473-6478.
- [Google Scholar]
- Beattie GAC, Holford P, Mabberley DJ, Haigh AM, Broadbent P. 2008. Australia and huanglongbing. Proc. FFTC-PPRI-NIFTS Joint Workshop Manag. Citrus Green. Virus Dis. Rehabil. Citrus Ind. ASPAC, eds. TY Ku, THH Pham, pp. 75–100. Ha Noi, Viet Nam: Plant Prot. Res. Inst.
- Beattie, G.A.C., D.J. Mabberley, P. Holford, P. Broadbent, and P. De Barro. 2005. Huanglongbing: its possible origins, collaborative research in Southeast Asia, and developing incursion management plans for Australia. In: Proceedings of 2nd International Citrus Canker and Huanglongbing Research Workshop. Gottwald, T.R., W.N. Dixon, J.H. Graham, and P. Berger (eds.), November 7–11 Orlando 2005 Florida Pp 52.
- Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res.. 1999;27(2):573-580.
- [Google Scholar]
- Mechanisms of tandem repeat instability in bacteria. Mutat. Res.. 2006;598:144-163.
- [Google Scholar]
- Self-generated diversity produces “insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. U. S. A.. 2004;101:16630-16635.
- [Google Scholar]
- Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J Plant Pathol. 2006;88:7-37.
- [Google Scholar]
- Bové J.M., and Garnier, M. 1984. Citrus greening and psylla vectors of the disease in the Arabian Peninsula. Proceedings of 9th Conference IOCV, Riverside, 109-114.
- Guangdong and Florida populations of ‘Candidatus Liberibacter asiaticus’ distinguished by a genomic locus with short tandem repeats. Phytopathology. 2010;100:567-572.
- [Google Scholar]
- Analysis of 16S rDNA sequences from citrus huanglongbing bacteria reveal a different “Ca. Liberibacter” strain associated with citrus disease in São Paulo. Plant Dis. 2005;89:848-852.
- [Google Scholar]
- Identification and characterization of the Huanglongbing bacterium in pummelo from multiple locations in Guangdong, P.R. China. Plant Dis.. 2008;92:513-518.
- [Google Scholar]
- Complete genome sequence of citrus huanglongbing bacterium, “Candidatus Liberibacter asiaticus” obtained through metagenomics. Mol. Plant Microbe Interact.. 2009;22:1011-1020.
- [Google Scholar]
- Achieving food security in the Kingdom of Saudi Arabia through innovation: Potential role of agricultural extension. J. Saudi Soc. Agric. Sci.. 2016;17:365-375.
- [Google Scholar]
- Examination of the responses of different genotypes of citrus to huanglongbing (citrus greening) under different conditions. Phytopathology. 2009;99:1346-1354.
- [Google Scholar]
- Food and Agricultural Organization (FAO) Managing Huanglongbing/Citrus Greening Disease in the Caribbean Issue brief #4 2013 www.fao.org/3/a-ax739e.pdf.
- Ghosh D.K. S. Bhose M. Motghare A. WArghane, K. Mukherjee, D.K. Ghosh Sr., A.K. Sharma, M.S. LAdaniya, and S. Gowda. 2015. Genetic diversity of Indian Populations of ‘Candidatus Liberibacter asiaticus’ based on the tandem repeat variability in genomic locus Phytopathology 105 8 2015 1043 1049.
- Statistical procedures for agricultural research (2nd ed.). New York, NY, USA: John Wiley and Sons; 1984. p. :680.
- Detection and identification of the two ‘Candidatus Liberibacter species’ associated with citrus Huanglongbing by PCR amplification of ribosomal protein genes of the Î2 operon. Mol. Cell. Probes. 1999;13:373-379.
- [Google Scholar]
- The phloem-limited bacterium of greening disease of citrus is a member of the a subdivision of the Proteobacteria. Int. J. Syst. Bacteriol.. 1994;44:379-386.
- [Google Scholar]
- Differentiation of “Candidatus Liberibacter asiaticus” isolates by variable-number tandem-repeat analysis. Appl. Environ. Microbiol.. 2011;77:1910-1917.
- [Google Scholar]
- Differentiation of Indian, East Timorese, Papuan and Floridian‘Candidatus Liberibacter asiaticus’ isolates on the basis of simple sequence repeat and single nucleotide polymorphism profiles at 25 loci. Ann. Appl. Biol.. 2012;160:291-297.
- [Google Scholar]
- Multilocus simple sequence repeat markers for differentiating strains and evaluating genetic diversity of Xylella fastidiosa. Appl. Environ. Microbiol.. 2005;71:4888-4892.
- [Google Scholar]
- Analysis of a prophage gene frequency revealed population variation of ‘Candidatus Liberibacter asiaticus’ from two citrus-growing provinces in China. Plant Dis.. 2011;95:431-435.
- [Google Scholar]
- Population structures of ‘Candidatus Liberibacter asiaticus’ in southern China. Phytopathology. 2014;104:158-162.
- [Google Scholar]
- Validation of ‘Variable Number of Tandem Repeat’-Based Approach for Examination of ‘Candidatus Liberibacter asiaticus’ Diversity and Its Applications for the Analysis of the Pathogen Populations in the Areas of Recent Introduction. PLoS One. 2013;8(11):e78994.
- [Google Scholar]
- Reported single nucleotide polymorphisms on the 16S rRNA gene do not support haplotypes of “Candidatus Liberibacter asiaticus”. Citrus Res. Technol.. 2012;33(2):75-79.
- [Google Scholar]
- Oberholzer P.C.J., von Standen D.F.A., Basson W.J., 1965. Greening disease of sweet orange in South Africa. In: Proceedings of 3rd Conference IOCV, IOCV, University Florida Press, Gainesville 1965, 213- 219.
- Genetic Diversity of Candidatus Liberibacter asiaticus Based on Two Hypervariable Effector Genes in Thailand. PLoS One. 2014;9(12):e112968.
- [Google Scholar]
- Diseases of economic plants in southern China. Philippine Agricultural. 1919;8:109-135.
- [Google Scholar]
- Three novel lineages of ‘Candidatus Liberibacter africanus’ associated with native rutaceous hosts of Trioza erytreae in South Africa. Int J Syst Evol Microbiol. 2015;65(Pt 2):723-731.
- [CrossRef] [Google Scholar]
- Diversity and characterization of Candidatus Liberibacter asiaticus strains causing huanglongbing disease in Iran, based on two prophage loci. J. Phytopathol.. 2018;166:623-632.
- [CrossRef] [Google Scholar]
- First Report of ‘Candidatus Liberibacter asiaticus’ Associated with Huanglongbing in Sweet Orange in Ethiopia. Plant disease. 2010;94(4) 482–482
- [CrossRef] [Google Scholar]
- Genetic Variability Based on Tandem Repeat Numbers in a Genomic Locus of ‘Candidatus Liberibacter asiaticus’ Prevalent in North East India. Plant Pathol. J.. 2019;35(6):644-653.
- [Google Scholar]
- First report of a Huanglongbing-like disease of Citrus in São Paulo State, Brazil and association of a new Liberibacter species, “Candidatus Liberibacter americanus”, with the disease. Plant Dis.. 2005;89:107.
- [Google Scholar]
- Tirtawidjaja S., Hadewidjaja T., Lasheen A.M., 1965. Citrus vein phloem degeneration virus, a possible cause of citrus chlorosis in Java. Proceedings of the American Society of Horticultural Science 86, 235-243.
- Intragenic tandem repeats generate functional variability. Nat. Genet.. 2005;37:986-990.
- [Google Scholar]
- Diversity and plasticity of the intracellular plant pathogen and insect symbiont “Candidatus Liberibacter asiaticus” as revealed by hypervariable prophage genes with intragenic tandem repeats. Appl. Environ. Microbio.. 2011;77(18):6663-6673.
- [Google Scholar]







