Translate this page into:
Functional characterization of promiscuous tryptophan decarboxylase from indole alkaloids producing Rauvolfia tetraphylla L.
⁎Corresponding author. dramuthaswaminathan@gmail.com (Amutha Swaminathan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University
Abstract
The enzyme Tryptophan decarboxylase (TDC, EC 4.1.1.28) gene facilitates the conversion of tryptophan to tryptamine. A new gene encoding TDC was identified from the alkaloid producing plant Rauvolfia tetraphylla by transcriptome analysis, termed as RtTDC. It contains 1,500 base pair which encodes an open reading frame for 499-amino-acid polypeptide with molecular mass of 55729.29 kDa and isoelectric point of 5.37. Multiple sequence alignment and phylogenetic tree analysis showed the closest similarity (95.3 %) with the TDC from the Rauvolfia verticillata. This enzyme has property of recombinant tryptophan decarboxylase from R. tetraphylla was characterized. The potential activity of tryptophan decarboxylase specific to L-tryptophan may contribute to the biosynthesis of indole alkaloids in R. tetraphylla. The finding of tryptophan metabolites in R. tetraphylla plants is a novel report, lead to hypothesize the existence of TDC enzymatic activity, from which aromatic amino acid decarboxylases is formed. These results support the in-silico annotation of the examined protein sequences of R. tetraphylla as TDC and suggest the involvement of TDC enzymatic activity in this plant. Molecular modeling of the TDC gene evidencing the reliability, stability and the structural similarities of the R. tetraphylla TDC gene with R. verticillata TDC gene. The L-tryptophan used as ligand in docking analysis to verify the TDC gene enzymatic activity for synthesis of Indole alkaloids. High performance liquid chromatography data analyses of RtTDC catalyzed reaction mixture confirmed the catalytically decarboxylative activity of RtTDC.
Keywords
Rauvolfia tetraphylla
Tryptophan decarboxylase
Indole alkaloid
Gene Sequence
Molecular Docking
1 Introduction
The plant Rauvolfia tetraphylla L. belongs to the family Apocynaceae, commonly known as ‘Devil Pepper’ and ‘Be Still Tree’ has interest because of its capacity to produce compounds with medicinal properties. R. tetraphylla has been traditionally used for medicinal purposes in various regions of India and it is sometimes utilized as a replacement for R. serpentine (Mahalakshmi et al., 2019). The Malaraya tribes of Tamil Nadu use a decoction of the bark of R. tetraphylla as an external application to treat chronic cutaneous diseases and eliminate parasites. Moreover, an extract of this plant is blended with castor oil to form a liniment, which is recommended for specific types of chronic and refractory skin ailments (Vanjari et al., 2022).
A wide range of alkaloids with modern biological effects are still being discovered and studied. Currently, around 60 alkaloids from plants have been approved as drugs in different countries (Cordell et al., 2001). Indole alkaloids are a group of alkaloids that contain an indole structure. Numerous compounds were identified as alkaloids, which contain isoprene groups and known as terpene indole or secologanin tryptamine alkaloid. Indole alkaloids are a major class of alkaloids found in several important plant groups, particularly in the Catharanthus and Rauvolfia plants from the Apocynaceae family. These plants have been scientifically studied for their pharmacological properties, with some undergoing clinical trials and others already approved for medicinal usage (Murugesan & Kaliappan, 2023).
The biosynthesis of indole alkaloids, which are the secondary plant metabolites primarily accumulated in plants, is mainly regulated by the enzyme tryptophan decarboxylase (Jeevanandham et al., 2022). Rauvolfia species are capable of synthesizing secondary metabolites, known as monoterpenoid indole alkaloids (MIAs), which exhibit remarkable biological activities. Indole alkaloids are a diverse class of plant metabolites that have various biological activities, such as anti-cancer, anti-tumour, anti-microbial, anti-inflammatory, and anti-viral properties (Lorensen et al., 2023). Over the past few decades, biosynthesis pathways of terpenoid indole alkaloids (TIAs) have been partially elucidated in Catharanthus roseus (L.) G. Don, a medicinal plant that contains various alkaloids (Liu et al., 2021). These mechanisms are believed to be similar in Rauvolfia species (Liu et al., 2012). Recently, (Stander et al., 2023) also identified enzymes in R. tetraphylla responsible for yohimbane monoterpene indole alkaloid biosynthesis.
R. tetraphylla is known to produce secondary metabolites such as ajmaline, alstonine, aricine, lankanescine reserpine, reserpiline, deserpidine, isoreserpine, sarpagine, rauvotetraphyllines, serpentine, and yohimbine (Lorensen et al., 2023; Mahalakshmi et al., 2019). It remains a fascinating plant species due to its production of unique heterocyclic alkaloids with monoterpene indole skeletons. These compounds have generated significant interest from both biological and therapeutic perspectives. Reserpine is a significant indole alkaloid synthesized by R. tetraphylla. It is commonly used to treat hypertension and several psychiatric illnesses by serving as a tranquilizing agent. The biosynthesis of reserpine initiates from tryptophan, which serves as the starting material (Anitha & Ranjitha, 2006). In the quest for an effective strategy to source pharmaceutical indole alkaloids, exploring the molecular techniques of R. tetraphylla biosynthetic pathway for indole alkaloids offers a promising avenue to boost their production and meet pharmaceutical demands.
Tryptophan decarboxylase (TDC) is an enzyme involved in the biosynthesis of indole alkaloids (Goddijn et al., 1995). L-tryptophan is the substrate that converted into tryptamine by TDC through a decarboxylation reaction. This study was carried out with a primary objective to explore the potential of R. tetraphylla for alkaloid biosynthesis using TDC gene and docking analysis.
2 Materials and methods
2.1 Genomic DNA extraction and PCR amplification
Total genomic DNA of the R. tetraphylla was extracted from young leaf tissues by using DNeasy plant Mini Kit (Qiagen, Germany). Purified total DNA was quantified and its quality verified on 0.8 % agarose gel. The gene sequence of R. verticillata was retrieve from the NCBI (ABP96805.1) to design the primer for identifying the RtTDC. The target gene was amplified with specific Forward primer 5′-ATGGGCAGCATTGATTCAACAG-3′ and Reverse primer 5′-TCAAGCTTCCTTGAGCAAATCA-3′ using PCR amplification (Dharshini et al., 2020). All the PCR products obtained by TDC gene were resolved by electrophoresis on 0.8 % agarose gel in 1x TBE Buffer, stained with ethidium bromide and gel documentation system was used for further calibration.
2.2 DNA elution and ligation
The amplified fragment of TDC was extracted from the gel using QIAquick Gel Extraction Kit (Qiagen, Germany) and ligated into pTZ57R/T vector using InsTAclone PCR Cloning Kit (Thermo Fisher Scientific, USA). The isolated DNA fragment was cloned into the pTZ57R/T vector (Mirahmadi et al., 2015). Following this, the vector containing the inserted DNA was transformed into DH5α Escherichia coli strain (Froger & Hall, 2007). A single transformed colony from each ampicillin-containing plate was selected and transferred to separate PCR tubes. Colony PCR was performed and the desired DNA fragment was confirmed by electrophoresis using 1 % agarose gel run at 70 V for 30 min. The transformed bacteria were cultured and the plasmid containing the cloned DNA fragment was isolated by QIAprep Spin Miniprep kit (Qiagen, Germany), following established plasmid isolation procedures (Sambrook et al., 1989).
2.3 Gene sequencing
The isolated plasmid was sequenced using the Sanger sequencing method, which is a widely used DNA sequencing technique. Sequencing was performed using the BigDye Terminator kit v. 3.1 and cleaned up with BigDye XTerminator v. 3.1 (Applied Biosystems, Foster City, CA). The DNA fragment is sequenced by adding a mixture of dideoxy nucleotides (ddNTPs) to the reaction, which terminate the DNA synthesis at each nucleotide. The resulting fragments are separated by size and analysed to determine the sequence of the DNA fragment (Crossley et al., 2020). The M13 primer is a commonly used primer in Sanger sequencing that anneals to the pTZ57R/T vector and sequence the cloned DNA fragment. After obtaining the sequencing results, Snap gene viewer was used to analyse the sequence and obtained the complete sequence of TDC.
2.4 Computational analysis of the gene sequence
Sequence similarity analysis between RtTDC and other TDCs from plants was performed using the basic local alignment search tool (BLAST) provided by the National Centre for Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/Blast). Nucleotide and amino acid sequences were aligned using CLUSTALW2 (https://www.ebi.ac.uk/Tools/msa/clustalw2). To identify the coding sequence, the open reading frame (ORF) finder graphical analysis by NCBI (https://www.ncbi.nlm.nih.gov/projects/gorf) and ExPASy Bioinformatics resource portal of the Swiss Institute of Bioinformatics (https://www.expasy.org/proteomics/) are used.
2.5 Protein sequence and structure prediction
The protein sequence of the TDC gene was determined using the Translate tool. The homology-based 3D model for the RtTDC protein was generated through the SWISS-MODEL (https://swissmodel.expasy.org), utilizing the 6eew.1.A structure as a template. The conserved regions and multiple sequence were identify using MUSCLE (Sievers & Higgins, 2018). The phylogenetic tree was built using Mega11 software (Kumar et al., 2018). The Neighbour-Joining (NJ) method with 1000 bootstrap pseudo replicates was used to carry out the phylogenetic analysis. The physiochemical properties were assessed using Expasy's ProtParam server (https://us.expasy.org/tools/protparam.html). Secondary structure of TDC were analysed by HNN (https://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_hnn.html) (Hierarchical Neural Network) (Kashani-Amin et al., 2019). Using the ProtScale program (https://web.expasy.org/protscale/), a Kyte and Doolittle hydropathy plot was generated (Kyte & Doolittle, 1982).
2.6 Ligand-receptor interactions through molecular docking and visualization tools
To investigate the interaction between a gene’s protein and a ligand, it is essential to obtain the receptor (TDC) through a 3D model and the ligand (L-Tryptophan) from the pubchem database (https://pubchem.ncbi.nlm.nih.gov/). Docking has been carried out by Autodock vina following the protocol described by Morris et al., (Morris et al., 2008). Visualization of the docking results is achieved using PyMOL software, allowing them to explore the complex 3D structures and interactions between the ligand and receptor (Rauf et al., 2015). To evaluate the interaction between the substrate Trp and the enzyme binding site, docking simulations were conducted using Ligplot + with the best 3D model of TDC (Laskowski & Swindells, 2011).
2.7 TDC enzymatic assay
The catalytic activity of TDC from R. tetraphylla was evaluated using the established procedures (Islas et al., 1994; Jadaun et al., 2017). Frozen leaves (2 g) were mechanically pulverized in a cold mortar and pestle, followed by homogenized with 1.25 mL of 0.1 M HEPES buffer at pH 7.5. The homogenate was centrifugated at 12,000 rpm for 30 min and the supernatant was immediately utilized for the enzyme source. The TDC assay mixture (100 μL) comprised enzyme extract (25 μL), 4 μM Pyridoxal 5-phosphate (PLP) (25 μL), 5 mM L-tryptophan (25 μL) and 50 mM HEPES buffer (25 μL). The reaction mixtures were incubated at 37 ℃ for 30 min. Then the reaction was stopped by adding 100 μL of prechilled methanol to precipitate proteins. Followed by centrifugation at 12,000 rpm for 10 min, the supernatant was used for further High-Performance Liquid Chromatography (HPLC) analysis to confirm the tryptophan converted into tryptamine in the presence of TDC enzyme. The HPLC system employed a C18 column and the mobile phase, composed of solvent A (Acetonitrile) and solvent B (H2O containing phosphoric acid) at 280 nm. All enzymatic reactions were analysed in triplicate and Tryptamine served as the standard for comparative analysis.
3 Results and discussion
3.1 Isolation of TDC gene
TDC is a key enzyme in the biosynthesis of monoterpenoid indole alkaloids (MIAs) (Liu et al., 2012) and plays a crucial role in the medicinal properties of R. tetraphylla. TDC genes have been identified in several plant species, including R. verticillata (ABP96805.1). In our study, TDC sequence from R. verticillata was used to design primers for PCR amplification of TDC from R. tetraphylla. PCR amplification was performed using specific primers designed to target the TDC gene (Fig. 1). PCR product was sequenced and confirmed as the TDC gene as well as submitted to NCBI (Accession No: OR105870). It showed a high degree of sequence similarity 95.39 % between the TDC sequences of R. verticillata and R. tetraphylla, suggesting that the TDC gene is conserved across different species within the genus Rauvolfia.
Electrophoresis of PCR products from genomic DNA on 1 % agarose gel of known molecular weight. Lane 1: DNA molecular weight marker (DNA ladder mix – 1 kb ladder, Gene Ruler); Lane 2&3: A total of 20 µL of PCR product amplification of TDC gene stained with ethidium bromide; Lane 4–7: Colony PCR for the confirmation of transformation; Lane 8&9: Isolation of Plasmid after plasmid extraction process by QIAprep Spin Miniprep kit.
TDC are a class of PLP-dependent enzymes responsible for catalysing the conversion of tryptophan into tryptamine (O'Connor & Maresh, 2006). The TDC-encoding gene was first isolated from Catharanthus roseus, a plant known for producing the antitumor monoterpene indole alkaloid vincristine (De Luca et al., 1989). Several TDC genes have been cloned from various plants, such as Capsicum annuum, Ophiorrhiza pumila, Oryza sativa, and R. verticillata, using a homology cloning strategy (Park et al., 2009).
TDC-encoding gene from R. verticillata was cloned and exhibit catalytic activity (Liu et al., 2012). Pepper fruits (Capsicum annuum) were found to contain TDC-encoding genes, namely CanTDC1 and CanTDC2. CanTDC1 gene showed three times higher specific catalytic activity compared to CanTDC2 (Park et al., 2009). The genome of Oryza sativa holds a sum of seven TDC genes, with only the genes OsTDCAK31 and OsTDCAK53 exhibited effective expression in Escherichia coli. It was confirmed that these genes exhibited catalytic decarboxylation activities towards L-tryptophan, leads to one or more TDC-encoding genes in a specific plant species (Kang et al., 2007).
Camptotheca acuminata Decne. is known for producing the renowned antitumor compound camptothecin, has been found to possess TDC genes such as CaTDC1 and CaTDC2. The decarboxylation activity of CaTDC1 and CaTDC2 towards L-tryptophan has been substantiated, resulting in the integration of the decarboxylation product camptothecin (López‐Meyer & Nessler, 1997). CaTDC3 from C. acuminata was cloned through heterologous overexpression and functional characterization, it was confirmed that CaTDC3 exhibits catalytic decarboxylation activity toward L-tryptophan (Qiao et al., 2022).
3.2 Model building and evaluation
The unknown 3D structure of R. tetraphylla TDC monomer was modelled using comparative protein modelling, with the crystallographically solved structure of C. roseus as the template. It is valuable when experimental structural data for the target protein is unavailable, providing valuable insights into protein structure and function in the field of structural biology (Facchiano, 2017).
The amino acid sequences of TDC from R. tetraphylla that currently lack structural data in the RCSB Protein Databank (PDB) were obtained using SWISS-MODEL. The best template was selected based on high score, lower e-value, and maximum sequence identity, and utilized as a reference structure to construct a 3D model (Waterhouse et al., 2018). The target protein under consideration is Aromatic-L-amino-acid decarboxylase, with a Uniport ID of P17770 and consists of 500 amino acids. For structural modelling purposes, the crystal structure of C. roseus TDC is being utilized as a template and is represented by the PDB ID 6EEW.
The Global Model Quality Estimate (GMQE) for the template stands at an impressive 0.97, indicating high confidence in its overall quality. The Template-Target Sequence-Overlap Quality (QSQE) score is calculated to be 0.88, suggesting significant structural similarities between the target protein and the template. Furthermore, when comparing the sequence of the target protein to that of the template, there is a substantial sequence identity of 88.96 % and indicates a strong relationship between the target and template.
The quality of the homology models was evaluated using the Saves server, with a focus on the Ramachandran plot (Fig. 2) and is a valuable tool for determining the stereochemical quality of protein structures by analysing the distribution of phi (ϕ) and psi (ψ) angles of the protein backbone (Sheik et al., 2002). It provides insights into the conformational stability and overall reliability of the modelled protein structures (LASKOWSKI et al., 2013). The Ramachandran plot demonstrated that over 90 % of the residues in the modelled proteins occupied the “favoured” and “allowed” regions of the plot, indicating their reliability and structural accuracy. To further validate the stability of the modelled proteins, the Deep View Swiss PDB viewer (Guex et al., 2009) was analysed for steric hindrances between residues in the models. The absence of steric hindrances suggests that the modelled protein structures are free from clashes representing their stability and suitability.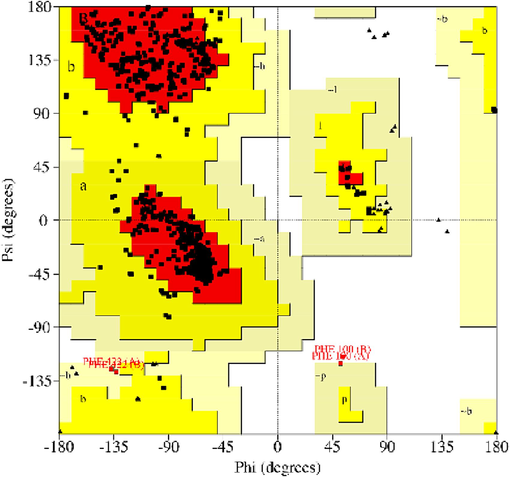
Ramachandran Plot analysis for structural 3D model protein of TDC gene.
3.3 Multiple sequence alignment of RT TDC were performed with RvTDC and CrTDC
The cloned gene of RtTDC encompasses 1,500 bp and encodes a TDC containing 499 amino acid residues with high similarity to those of RvTDC. In the multiple sequence alignment of TDC sequences from R. verticillata (Rv), R. tetraphylla (Rt), and C. roseus (Cr), a functional and highly conserved region was identified (Fig. 3a). Conserved regions in the alignment are areas where the amino acid residues are highly similar across the different sequences, suggesting that these regions have important functional or structural roles that are conserved throughout evolution. The identification of such a conserved region in the TDC sequences is significant because it indicates that this region plays a critical role in the function of TDC. The conserved region is involved in the catalytic function of the enzyme, which is the conversion of L-tryptophan to tryptamine is a key step in the biosynthesis alkaloids.
Multiple amino acid residue sequences alignment of biochemically characterized plant TDCs. (a) TDCs are from Catharanthus roseus, Rauvolfia verticillata, and Rauvolfia tetraphylla. (b) Phylogenetic tree showed an occurrence and evolutionary relationship between TDC from different plant species. The tree was built using Mega11 software. Bootstrap values are all above 95%. COFCA- Coffea canephora, CAPAN - Capsicum annuum, SOLLC - Solanum lycopersicum, SOLTU- Solanum tuberosum, ORYSJ- Oryza sativa, CATRO–Catharanthus roseus, RAUVE- Rauvolfia verticilla, TDC RT – Rauvolfia tetraphylla, 9ROSI - Castanea mollissima, CANSA - Cannabis sativa, CAMAC - Camptotheca acuminata.

Multiple amino acid residue sequences alignment of biochemically characterized plant TDCs. (a) TDCs are from Catharanthus roseus, Rauvolfia verticillata, and Rauvolfia tetraphylla. (b) Phylogenetic tree showed an occurrence and evolutionary relationship between TDC from different plant species. The tree was built using Mega11 software. Bootstrap values are all above 95%. COFCA- Coffea canephora, CAPAN - Capsicum annuum, SOLLC - Solanum lycopersicum, SOLTU- Solanum tuberosum, ORYSJ- Oryza sativa, CATRO–Catharanthus roseus, RAUVE- Rauvolfia verticilla, TDC RT – Rauvolfia tetraphylla, 9ROSI - Castanea mollissima, CANSA - Cannabis sativa, CAMAC - Camptotheca acuminata.
The similar pursuit unveiled that RtTDC show resemblances in amino acid residues with plant TDCs. Among the enzymatic mechanisms of TDCs from plant (Liu et al., 2012; López‐Meyer & Nessler, 1997), it is predictable that Lys319 plays a key role in RtTDC as the catalytic amino acid. RtTDC is grouped in the same clade as functionally characterized TDCs from C. roseus (CrTDC) (De Luca et al., 1989), Ophiorrhiza pumila (OpTDC) (Yamazaki et al., 2003), R. verticillata (RvTDC) (Liu et al., 2012) and Gelsemium sempervirens (GsTDC) (Franke et al., 2019). These TDCs have been known to participate in the biosynthesis of indole alkaloids. It suggests that RtTDC likely shares a similar catalytic function with these characterized TDCs.
3.4 Phylogenetic tree with related sequences
Phylogenetic tree showed three clades (Fig. 3b), in which one of the branches show common ancestors for R. tetraphylla (RtTDC), C. roseus (TDC CATRO), R. verticillata (RAUVE) and Oryza sativa (ORYSJ). Among them RtTDC exhibited close relationship with RAUVE. The results from the neighbour-joining phylogenetic analysis of 12 demonstrative species within flowering plants suggest that R. tetraphylla is closely related to R. verticillata. C. roseus, R. verticillata (Liu et al., 2012), Oryza sativa evaluated the TDC catalyses to produce Indole alkaloids.
3.5 Physicochemical characterization of TDC
The physicochemical properties of TDC were predicted using Expasy's Prot-param program (Table 1). The protein sequence length is 499 amino acids and molecular weight is 55,729.29 Daltons. The theoretical isoelectric point (pI) is 5.37, indicating that protein may be acidic. TDC contains a total of 61 negatively charged residues and 48 positively charged residues. The balance between positively and negatively charged residues contributes to the overall charge of the protein under specific conditions. The extinction coefficient of the protein is determined to be 70,860 M−1 cm−1, a value often utilized for quantifying the protein concentration in UV absorption measurements. The instability index is 47.92 indicates a moderate propensity for protein instability, suggesting potential sensitivity to environmental factors.
Protein
Tryptophan decarboxylase
Sequence Length
499 amino acids
Molecular weight
55729.29 kDa
Theoretical Pi
5.37
Total number of negatively charged residues (Asp + Glu)
61
Total number of positively charged residues (Arg + Lys)
48
Extinction Coefficient
70860 M−1cm−1
Instability Index
47.92
Aliphatic Index
94.19
Grand average of hydropathicity (GRAVY)
0.029
The aliphatic index is computed to be 94.19, characterizes the protein thermostability, as it represents the relative volume of aliphatic side chains. The Grand Average of Hydropathicity (GRAVY) is 0.029, reflecting the overall hydrophobicity. The Kyte and Doolittle hydrophobicity plot of RtTDC protein showed a pattern of high hydrophobicity with the GRAVY value and hydrophobic residues (Fig. 4). These parameters provide valuable information about the protein biophysical properties, stability, and solubility.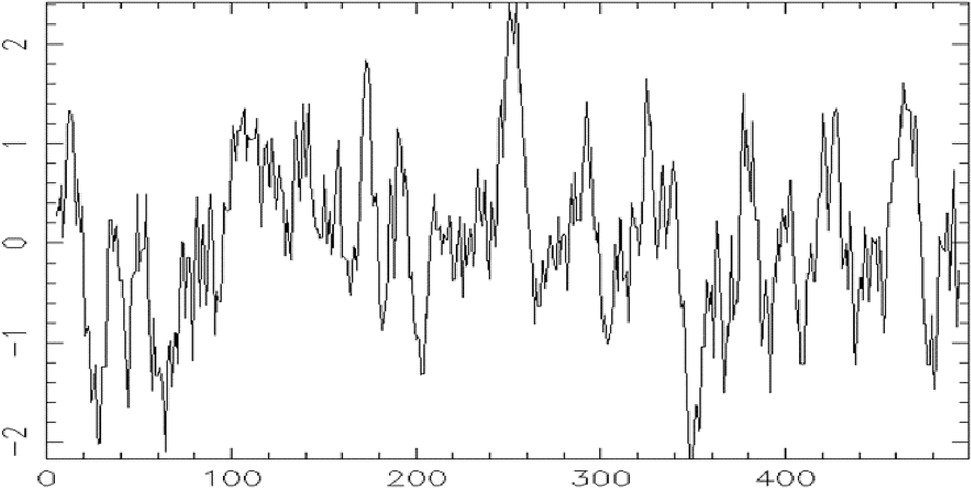
Hydropathy plot of residues 1 to 499 of sequence Emboss.001 using Kyte and Doolittle hydropathy parameter.
The amino acid composition of the TDC protein is presented in Table 2. It provides valuable insights into the protein primary structure, shedding light on the relative abundance of each amino acid residue present. Leucine (L), alanine (A), serine (S), valine (V), glutamic acid (E), threonine (T), glycine (G), aspartic acid (D), proline (P), isoleucine (I), lysine (K), and methionine (M), phenylalanine (F), arginine (R), asparagine (N), tyrosine (Y), cysteine (C), glutamine (Q), histidine (H), and tryptophan (W) are the 20 amino acids, among them leucine is the most abundant, with a total count of 57 residues constituting 11.4 % of the protein.
S.No
Composition of amino acids
Percentage
1
Ala (A) 40
8.0
2
Arg (R) 23
4.6
3
Asn (N) 16
3.2
4
Asp (D) 26
5.2
5
Cys (C) 9
1.8
6
Gln (Q) 9
1.8
7
Glu (E) 35
7.0
8
Gly (G) 25
5.0
9
His (H) 10
2.0
10
Ile (I) 25
5.0
11
Leu (L) 57
11.4
12
Lys (K) 25
5.0
13
Met (M) 17
3.4
14
Phe (F) 23
4.6
15
Pro (P) 25
5.0
16
Ser (S) 41
8.2
17
Thr (T) 32
6.4
18
Trp (W) 9
1.8
19
Tyr (Y) 14
2.8
20
Val (V) 38
7.6
The hydropathy profiles of C. roseus TDC and D. melanogaster DDC1 proteins showed 39 % structural similarity. The deduced amino acid sequence of C. roseus TDC revealed an unexpected similarity to animal DDC in the secondary structure of conserved domains (De Luca et al., 1989; Goddijn et al., 1994). The comparison of Campotheca TDC1 and Catharanthus TDC revealed a significant 69 % identity between the two sequences. This high level of sequence conservation highlights the potential functional importance of this region in the catalytic activity and stability of TDC enzymes (López‐Meyer & Nessler, 1997). It showed that amino acid composition provides information for understanding its chemical nature and potential implications for its function and structure.
3.6 Secondary structure prediction of TDC
The secondary structure of TDC gene R. tetraphylla protein sequences were calculated using HNN (Fig. 5 a and b) and reveals the distribution of various structural elements. The most predominant secondary structure is alpha helix (Hh), which comprises 253 residues, accounting for 50.70 % of the protein. The protein contains 61 residues in the extended strand (Ee) conformation representing 12.22 %. The majority of the protein, consisting of 185 residues (37.07 %), is categorized as random coil (Cc).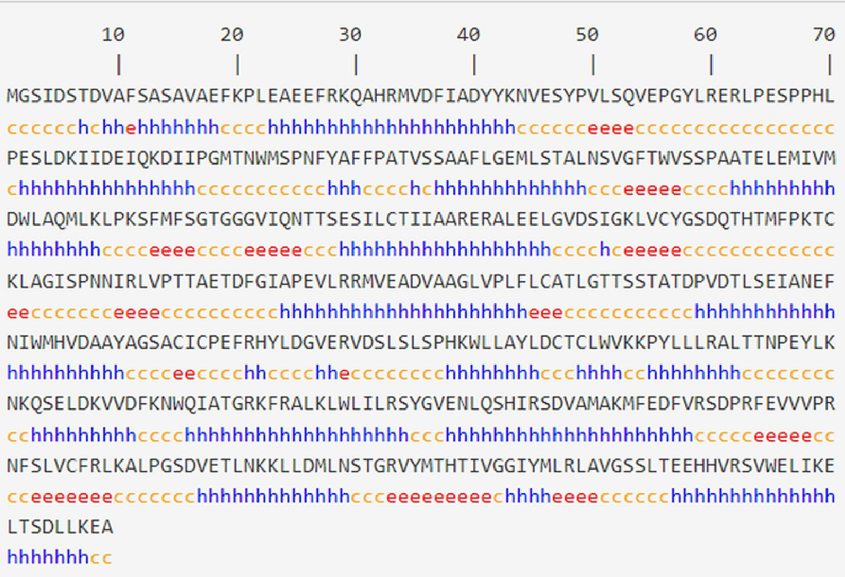
The HNN software was used to predict the secondary structure of the TDC gene. (a) Structural codes in blue (h-Helix), orange (c-coil), red (e-sheet). (b) Secondary Structure prediction of TDC gene sequence of 499 amino acids in Rauvolfiate traphylla. Lines in different colours represent different secondary structures. Blue line indicates α helix; red line indicates extended strand and purple line indicates random coil. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
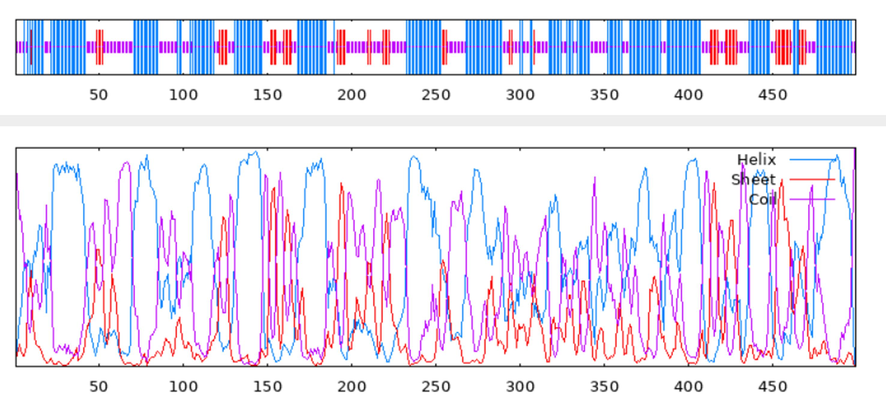
The HNN software was used to predict the secondary structure of the TDC gene. (a) Structural codes in blue (h-Helix), orange (c-coil), red (e-sheet). (b) Secondary Structure prediction of TDC gene sequence of 499 amino acids in Rauvolfiate traphylla. Lines in different colours represent different secondary structures. Blue line indicates α helix; red line indicates extended strand and purple line indicates random coil. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In this study, isolated and characterized a full-length cDNA encoding TDC from R. tetraphylla, marking the first time this gene has been studied in this species. The deduced amino acid sequence of RtTDC exhibited significant similarity to TDC counterparts found in other species. Prior research on RvTDC (Liu et al., 2012) has shown that the Lys319 residue directly participates in binding to PLP-binding site. It is conserved in other decarboxylases, including 3,4-dihydroxyphenylalanine (DOPA) decarboxylases from C. acuminata (López‐Meyer & Nessler, 1997), D. melanogaster DDC (Clark et al., 1978), feline glutamate decarboxylase (Kobayashi et al., 1987) and periwinkle TDC (De Luca et al., 1989), strongly suggests that lysine-319 of TDC is responsible for binding to PLP. The conservation of lysine-319 in the PLP binding site emphasizes the importance of conserved domains in facilitating subunit assembly, catalytic function, and substrate specificity of TDC enzymes.
3.7 Ligand binding to TDCs from R. tetraphylla by molecular docking
Molecular docking was analysed by utilizing the predicted three-dimensional structure of RtTDC and L-tryptophan as the substrate. The results of the highest scoring ligand are summarized in Table 3. The table contains rankings and properties of substructures based on Binding Energy, Cluster RMSD (Root Mean Square Deviation), and Reference RMSD. The top-ranked substructure is listed with Sub-Rank 1, Run-Rank 14, a Binding Energy of −4.69, and Cluster RMSD and Reference RMSD values of 0.00, 28.56, respectively. Pharmacophore is a critical concept in docking studies as it refers to the set of molecular features that are essential for a ligand to bind to a receptor (Fig. 6a). Understanding the pharmacophore enabled to design and modify ligand to improve binding affinity and selectivity for the target receptor, leading to more effective and safer drugs.
Rank
Sub- Rank
Run
Binding Energy
Cluster RMSD
Reference RMSD
Grep Pattern
1
1
14
−4.69
0.00
28.56
RANKING
2
1
27
−4.58
0.00
61.50
RANKING
3
1
4
−4.49
0.00
40.28
RANKING
3
2
19
−4.25
0.35
40.19
RANKING
3
3
47
−3.58
1.78
40.42
RANKING
4
1
16
−4.49
0.00
54.44
RANKING
5
1
39
−4.48
0.00
60.37
RANKING
5
2
8
−4.39
1.39
59.42
RANKING
6
1
45
−4.47
0.00
49.99
RANKING
7
1
2
−4.40
0.00
49.03
RANKING
8
1
18
−4.15
0.00
48.81
RANKING
9
1
40
−4.11
0.00
61.04
RANKING
10
1
23
−4.09
0.00
36.98
RANKING

Pharmacophore model of TDC gene derived from 3D model and its mapping with L- tryptophan ligand. (a) The active amino acids were mapped to the best pharmacophore model. The pharmacophore model was illustrated using Catalyst notation, which encompassed of hydrogen bond acceptor, hydrogen bond donor and hydrophobic region. (b) Hydrogen Bond Interaction of RtTDC protein (Receptor) and L-tryptophan (Ligand); hydrogen bond is showed in yellow dotted line; Hydrogen bonds are observed in Lys319, Phe101, His458 and Thr204 amino acids residues. (c) Ligplot diagram using the model RtTDC and highlights the interaction with Trp Ligand. The ligand is depicted with bold (blue-coloured) bonds, while the hydrogen-bonded residues from the protein are represented with thin (brown-coloured) bonds. Dashed lines illustrate the hydrogen bonds formed between the ligand and the protein. Additionally, hydrophobic contacts made with the protein are indicated by spoked arcs pointing towards the ligand, and corresponding spokes on the ligand atoms reveal which atoms are engaged in these contacts. Likewise, spokes pointing towards the contact atoms mark the atoms in the hydrogen-bonded groups engaged in hydrophobic contacts. The residue names are annotated with letters in parentheses, indicating the corresponding chain identifiers. The diagram showcases the catalytic triad comprising Lys319, Phe101, His458, and Thr204, along with the ligand's TRP residue snuggled within the enzyme's highly hydrophobic specificity pocket in the active site. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)

Pharmacophore model of TDC gene derived from 3D model and its mapping with L- tryptophan ligand. (a) The active amino acids were mapped to the best pharmacophore model. The pharmacophore model was illustrated using Catalyst notation, which encompassed of hydrogen bond acceptor, hydrogen bond donor and hydrophobic region. (b) Hydrogen Bond Interaction of RtTDC protein (Receptor) and L-tryptophan (Ligand); hydrogen bond is showed in yellow dotted line; Hydrogen bonds are observed in Lys319, Phe101, His458 and Thr204 amino acids residues. (c) Ligplot diagram using the model RtTDC and highlights the interaction with Trp Ligand. The ligand is depicted with bold (blue-coloured) bonds, while the hydrogen-bonded residues from the protein are represented with thin (brown-coloured) bonds. Dashed lines illustrate the hydrogen bonds formed between the ligand and the protein. Additionally, hydrophobic contacts made with the protein are indicated by spoked arcs pointing towards the ligand, and corresponding spokes on the ligand atoms reveal which atoms are engaged in these contacts. Likewise, spokes pointing towards the contact atoms mark the atoms in the hydrogen-bonded groups engaged in hydrophobic contacts. The residue names are annotated with letters in parentheses, indicating the corresponding chain identifiers. The diagram showcases the catalytic triad comprising Lys319, Phe101, His458, and Thr204, along with the ligand's TRP residue snuggled within the enzyme's highly hydrophobic specificity pocket in the active site. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The aromatic amines are produced through the catalytic action of ubiquitous aromatic amino acid decarboxylases (AADCs), a group of PLP-dependent enzymes. Despite having similarities in terms of extensive amino acid residue, subunit structures, and kinetic properties, AADCs demonstrate significant variations in their substrate specificities. (Facchini et al., 2000). In the case of AADC in plant, they have developed with changes in function and substrate preference. (Günther et al., 2019). The naturally occurring amines obtained from plants are known for wide range of metabolic pathways, mostly contributing to the synthesis of alkaloids (Torrens-Spence et al., 2020). The identified and functionally characterized one or more AADC-encoding genes from a single plant, suggesting the increase in gene families of whole-genome duplication in the process of evolution (Kang et al., 2021).
The protein–ligand docking is to predict the orientation and positioning of a ligand in the binding pocket of a particular receptor. It involves the formation of hydrogen bond interactions between the protein and the ligand (Fig. 6b). The docking simulations with Trp revealed a complex network of interactions involving multiple amino acids besides the binding site residues. Fig. 6c showed the amino acid residues of the RtTDC binding pocket involved in the interaction with the substrate Trp according to the results of molecular docking simulations. The analysis of C. roseus TDC binding to L-tryptophan was also performed (Facchiano et al., 2019), showing the same amino acid residues (Lys319) as those of RtTDC interacting with Trp in the docking simulations.
3.8 TDC enzymatic assay
The enzymatic decarboxylation activity of R. tetraphylla were analysed to investigate the role of TDC in the terpenoid indole alkaloid biosynthetic pathway. The assays involved the incubation of L-tryptophan, PLP and enzyme extract. HPLC analyses revealed the presence of an enzymatic reaction product in the RtTDC catalyzed reaction mixture (Fig. 7). The HPLC retention time of the enzymatic reaction product was found to be identical to that of authentic tryptamine. It demonstrates that the enzymatic product generated by RtTDC is indeed tryptamine, confirming the enzyme’s role in the decarboxylation of L-tryptophan, a crucial step in the biosynthesis of alkaloids in R. tetraphylla.
In vitro decarboxylation of tryptophan to tryptamine. HPLC traces of decarboxylation reaction mixture catalysed by RtTDC (panel I) and panel II represent standard tryptamine.
The investigations into TDC activity in various plant systems reveal its pivotal role in the biosynthesis of indole alkaloids. In Catharanthus roseus transformed root culture, TDC demonstrates a coordinated increase in activity with the accumulation of alkaloids ajmalicine and catharanthine, emphasizing its regulatory function in the indole alkaloid pathway (Islas et al., 1994). Additionally, TDC activity in Citrus x limon seedlings, evidenced by the conversion of deuterium-labelled tryptophan to tryptamine and subsequent methylation, expands our understanding of TDC across plant genera (De Masi et al., 2017). The CaTDC3 in C. acuminata further emphasizes the enzyme’s versatility, showing strict stereoselectivity for L-tryptophan and catalytic promiscuity towards various tryptophan derivatives (Qiao et al., 2022). These inferences shed light on the multifaceted nature of TDC, providing insights into its catalytic capabilities and broadening its applications in plant alkaloid metabolism.
4 Conclusion
The wide range of tryptophan-indole metabolism in plants leads to the production of indole phytochemicals in different plant genera. Specifically, within the Apocynaceae family, Rauvolfia species such as R. tetraphylla, R. serpentina, and R. verticillata are known for producing indole alkaloids, with mechanisms involved in their synthesis. TDC is catalytic enzyme plays a crucial role in conversion of tryptophan to tryptamine tailored to the biosynthetic pathway of different plant species for production of various indole alkaloids. Understanding the catalytic properties of TDC from R. tetraphylla is essential in elucidating these processes. This study represents a total genomic DNA of TDC gene from R. tetraphylla with 1500 base pair. The potential activity of TDC were analysed by molecular docking and enzymatic assay to conform the alkaloid biosynthesis of R. tetraphylla. Based on this new study, it's conceivable that RtTDC could play a significant role in the biosynthesis of indole alkaloids, paving the way for novel approaches in the drug development of fast tissue regeneration. It facilitates an alternative means of producing valuable indole alkaloids for wound healing drugs.
CRediT authorship contribution statement
Lavanya Nallasamy: Investigation, Methodology, Writing – original draft. Najat A. Bukhari: Data curation, Project administration, Visualization, Writing – review & editing. Girija Sangari Murugavelu: Data curation, Writing – review & editing. Deepika Krishnamoorthy: Data curation, Writing – review & editing. S. Mahalakshmi: Methodology, Visualization. Amutha Swaminathan: Conceptualization, Investigation, Supervision, Validation, Writing – review & editing. Appunu Chinnaswamy: Conceptualization, Methodology, Validation, Writing – review & editing.
Acknowledgments
The authors would like to thank ICAR—Sugarcane Breeding Institute, Coimbatore for providing the necessary infrastructure. Our sincere thanks to Avinashilingam Institute for Home Science and Higher Education for Women for providing moral support to complete our project. Many thanks to Tamil Nadu, Forest genetics Division, Coimbatore for collecting this plant. My special thanks to my supervisor for their extraordinary support. The authors extend their appreciation to the Researchers supporting project number (RSP2024R229) King Saud University, Riyadh, Saudi Arabia.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Stimulation of reserpine biosynthesis in the callus of Rauvolfia tetraphylla L. by precursor feeding. Afr. J. Biotechnol.. 2006;5(8):659-661.
- [Google Scholar]
- Dopa decarboxylase from Drosophila melanogaster: Purification, characterization and an analysis of mutants. Mol. Gen. Genet.. 1978;162:287-297.
- [Google Scholar]
- The potential of alkaloids in drug discovery. Phytother. Res.. 2001;15(3):183-205.
- [CrossRef] [Google Scholar]
- Guidelines for Sanger sequencing and molecular assay monitoring. J. Vet. Diagn. Invest.. 2020;32(6):767-775.
- [Google Scholar]
- Molecular cloning and analysis of cDNA encoding a plant tryptophan decarboxylase: comparison with animal dopa decarboxylases. Proc. Natl. Acad. Sci.. 1989;86(8):2582-2586.
- [Google Scholar]
- Experimental evidence and in silico identification of tryptophan decarboxylase in Citrus genus. Molecules. 2017;22(2):272.
- [Google Scholar]
- Isolation and characterization of nuclear localized abiotic stress responsive cold regulated gene 413 (SsCor413) from Saccharum spontaneum. Plant Mol. Biol. Report.. 2020;38:628-640.
- [Google Scholar]
- Bioinformatic resources for the investigation of proteins and proteomes. Peptidomics. 2017;3(1):1-10.
- [Google Scholar]
- Structure and ligands interactions of Citrus tryptophan decarboxylase by molecular modeling and docking simulations. Biomolecules. 2019;9(3):117.
- [Google Scholar]
- Plant aromatic L-amino acid decarboxylases: evolution, biochemistry, regulation, and metabolic engineering applications. Phytochemistry. 2000;54(2):121-138.
- [Google Scholar]
- Gene discovery in gelsemium highlights conserved gene clusters in monoterpene indole alkaloid biosynthesis. Chembiochem. 2019;20(1):83-87.
- [Google Scholar]
- Transformation of plasmid DNA into E. coli using the heat shock method. JoVE (J. Vis. Exp.) (6):e253.
- [Google Scholar]
- Goddijn, O. J., Lohman, F. P., de Kam, R. J., hilperoort, R. A., & Hoge, J. H. C. (1994). Nucleotide sequence of the tryptophan decarboxylase gene of Catharanthus roseus and expression of tdc-gus A gene fusions in Nicotiana tabacum. Molecular and General Genetics MGG, 242, 217-225.
- Overexpression of a tryptophan decarboxylase cDNA in Catharanthus roseus crown gall calluses results in increased tryptamine levels but not in increased terpenoid indole alkaloid production. Transgenic Res.. 1995;4:315-323.
- [Google Scholar]
- Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis. 2009;30(S1):S162-S173.
- [Google Scholar]
- Separate pathways contribute to the herbivore-induced formation of 2-phenylethanol in poplar. Plant Physiol.. 2019;180(2):767-782.
- [Google Scholar]
- Tryptophan decarboxylase activity in transformed roots from Catharanthus roseus and its relationship to tryptamine, ajmalicine, and catharanthine accumulation during the culture cycle. In Vitro Cell. Develop. Biol.-Plant. 1994;30:81-83.
- [Google Scholar]
- Withania coagulans tryptophan decarboxylase gene cloning, heterologous expression, and catalytic characteristics of the recombinant enzyme. Protoplasma. 2017;254:181-192.
- [Google Scholar]
- Molecular cloning and quantitative real-time PCR analysis to study the expression of tryptophan decarboxylase gene from chillies (Capsicum annuum L.) against whitefly. Pharmacogn. Mag.. 2022;18(78):476-493.
- [Google Scholar]
- A chromosome-level Camptotheca acuminata genome assembly provides insights into the evolutionary origin of camptothecin biosynthesis. Nat. Commun.. 2021;12(1):3531.
- [Google Scholar]
- Characterization of rice tryptophan decarboxylases and their direct involvement in serotonin biosynthesis in transgenic rice. Planta. 2007;227:263-272.
- [Google Scholar]
- A systematic review on popularity, application and characteristics of protein secondary structure prediction tools. Curr. Drug Discov. Technol.. 2019;16(2):159-172.
- [Google Scholar]
- Glutamic acid decarboxylase cDNA: nucleotide sequence encoding an enzymatically active fusion protein. J. Neurosci.. 1987;7(9):2768-2772.
- [Google Scholar]
- MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol.. 2018;35(6):1547.
- [Google Scholar]
- A simple method for displaying the hydropathic character of a protein. J. Mol. Biol.. 1982;157(1):105-132.
- [Google Scholar]
- Laskowski, R. A., & Swindells, M. B. (2011). LigPlot+: multiple ligand–protein interaction diagrams for drug discovery. In: ACS Publications.
- Laskowski, R. A., Furnham, N., & Thornton, J. M. (2013). The Ramachandran plot and protein structure validation. In Biomolecular forms and functions: a celebration of 50 years of the ramachandran map (pp. 62-75). World Scientific.
- Tryptophan decarboxylase plays an important role in ajmalicine biosynthesis in Rauvolfia verticillata. Planta. 2012;236:239-250.
- [Google Scholar]
- Terpenoid indole alkaloid biosynthesis in Catharanthus roseus: effects and prospects of environmental factors in metabolic engineering. Biotechnol. Lett.. 2021;43(11):2085-2103.
- [CrossRef] [Google Scholar]
- Tryptophan decarboxylase is encoded by two autonomously regulated genes in Camptotheca acuminata which are differentially expressed during development and stress. Plant J.. 1997;11(6):1167-1175.
- [Google Scholar]
- Spatial localization of monoterpenoid indole alkaloids in Rauvolfia tetraphylla by high resolution mass spectrometry imaging. Phytochemistry. 2023;209:113620
- [Google Scholar]
- Rauvolfia tetraphylla L. (Apocynaceae)-a comprehensive review on its ethnobotanical uses, phytochemistry and pharmacological activities. Int. J. Pharm. Biol. Sci.. 2019;9(2):664-682.
- [Google Scholar]
- Cloning and sequence analysis of recombinant plasmodium vivax merozoite surface protein 1 (PvMSP-142 kDa) in pTZ57R/T vector. Iran. J. Parasitol.. 2015;10(2):197.
- [Google Scholar]
- Morris, G. M., Huey, R., & Olson, A. J. (2008). Using autodock for ligand‐receptor docking. Current protocols in bioinformatics, 24(1), 8.14. 11-18.14. 40.
- A systematic review of analytical methods for quantification of natural indole alkaloids from Catharanthus and Rauvolfia species. Res. J. Pharmacogn.. 2023;10(1):57-66.
- [Google Scholar]
- Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat. Prod. Rep.. 2006;23(4):532-547.
- [Google Scholar]
- Induction of serotonin biosynthesis is uncoupled from the coordinated induction of tryptophan biosynthesis in pepper fruits (Capsicum annuum) upon pathogen infection. Planta. 2009;230:1197-1206.
- [Google Scholar]
- Functional characterization of a catalytically promiscuous tryptophan decarboxylase from camptothecin-producing Camptotheca acuminata. Front. Plant Sci.. 2022;13:987348
- [Google Scholar]
- Ligand docking and binding site analysis with pymol and autodock/vina. Int. J. Basic Appl. Sci.. 2015;4(2):168.
- [Google Scholar]
- Sambrook, J., Fritsch, E. F., & Maniatis, T. (1989). Molecular cloning: a laboratory manual. Cold spring harbor laboratory press.
- Clustal omega for making accurate alignments of many protein sequences. Protein Sci.. 2018;27(1):135-145.
- [Google Scholar]
- The Rauvolfia tetraphylla genome suggests multiple distinct biosynthetic routes for yohimbane monoterpene indole alkaloids. Commun. Biol.. 2023;6(1):1197.
- [CrossRef] [Google Scholar]
- Structural basis for divergent and convergent evolution of catalytic machineries in plant aromatic amino acid decarboxylase proteins. Proc. Natl. Acad. Sci.. 2020;117(20):10806-10817.
- [Google Scholar]
- Medicinal plant rauvolfia tetraphylla l its medicinal uses and pharmacological activities. J. Med. Plants. 2022;10(5):119-121.
- [Google Scholar]
- SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res.. 2018;46(W1):W296-W303.
- [Google Scholar]
- Camptothecin biosynthetic genes in hairy roots of Ophiorrhiza pumila: Cloning, characterization and differential expression in tissues and by stress compounds. Plant Cell Physiol.. 2003;44(4):395-403.
- [Google Scholar]







