Translate this page into:
Quartz tuning fork-based biosensor for the direct detection of human cytomegalovirus
⁎Corresponding author at: Department of Biomedical Technology, College of Applied Medical Sciences, King Saud University, Riyadh 11451, Saudi Arabia. aassaifan@ksu.edu.sa (Abdulaziz K. Assaifan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University. Production and hosting by Elsevier.
Abstract
Human cytomegalovirus (HCMV) possess great threat to immunocompromised patients and pregnant women since It can cause disability if left untreated. Especially for unborn babies, if the virus was not detected at early stages, it can cause disabilities as the baby develops. Furthermore, the virus can be asymptomatic, hence, low-cost and rapid detection techniques are desirable. Currently available detection techniques of the virus are labor intensive and demand experienced technicians. For these reasons, new detection techniques are needed to overcome the current challenges associated with conventional techniques. In this work, quartz tuning fork (QTF)-based biosensor was developed for the detection of UL83-antigen of HCMV for the first time. Firstly, QTF coated with gold was functionalized with cysteamine and glutaraldehyde for UL83-antibody immobilization at the QTF surface. Then, the biosensor was tested against a variety of UL83-antigen concentrations. As the UL83-antigen concentration increased, the measured resonance frequency decreased due to increased mass loading at the QTF surface. The sensitivity of the biosensor is 15.91 Hz/ln(ng/mL). Whereas the limit of detection is 0.36 ng/mL. The biosensor showed comparable biosensing performances to those available in the literature. Furthermore, the biosensor demonstrated its selectivity towards UL83-antigen when tested against samples containing a mixture of biomarkers. The reported work demonstrates a platform for the direct and low-cost mass screening of diseases.
Keywords
Quartz tuning fork
Biosensor
Cytomegalovirus
Label-free
Disability
1 Introduction
Human cytomegalovirus (HCMV) is a widely spread virus that can infect almost anyone. It was estimated that more than 80% of world’s population may be infected with this virus (Britt 2008, Rybachuk 2009, Turner et al., 2014, Zuhair et al., 2019). HCMV is considered as the largest herpesvirus in comparison to other types (MacDonald et al., 1997, Brown and Abernathy 1998, Britt 2010). It exhibits the usual characteristics of other herpesviruses in terms of the gene expression, the viral structure and life-long latency and persistence. It consists of double helix DNA genome entrapped in a capsid which is surrounded by a proteinaceous matrix called the tegument (Hasan et al., 2021). The tegument and capsid are covered by a glycoprotein-containing lipid bilayer, known as envelope (Loomis et al., 2003, Mettenleiter et al., 2006, Liu et al., 2021). The glycoproteins embedded in the viruses envelop play a crucial rule in facilitating their interactions with the host cells (Crough and Khanna 2009). The virus usually doesn't cause any symptoms, but it can be dangerous and life-threatening in some cases such as in pregnant women and people with weakened immune systems (Demmler-Harrison 2009). If a pregnant woman contracts HCMV, there is an increased risk of childhood disabilities. The elimination of HCMV by the immune system is relatively difficult, and the virus may stay dormant in the host until reactivation (Griffiths and Reeves 2021). The diagnosis of the virus based on its symptoms is not trustworthy. Therefore, it is urgently necessary to expand on existing diagnostic techniques and step up the fight against this virus.
Conventionally, the diagnosis of HMCV rely on serological or nucleic acid–based techniques (Gerna et al., 2000, Liu et al., 2017, Razonable et al., 2020). One of the most prominent HCMV serological tests is enzyme-linked immunosorbent assay (ELISA). Furthermore, polymerase chain reaction (PCR) is a commonly used nucleic acid-based assay. However, the application of these techniques is hindered by a number of issues, including the generation of false-positives, the demand for expertise and the challenging procedure of DNA extraction (Huang et al., 2022). Biosensors have opened new avenues of viral diagnosis that may replace the traditional techniques.
Biosensors are promising analytical techniques that can be used in several fields such as clinical diagnostics, environmental monitoring and other disciplines that require quick and reliable results. Some biosensing-based techniques have been successfully commercialized. However, the majority require further improvement to address flaws. Quartz crystal microbalance (QCM) was used in the past as HCMV immunosensor (Susmel et al., 2000). The human cytomegalovirus glycoprotein B (gB) epitope was detected with 1 μg/ml limit of detection (LOD). A magnetic particle-based enzyme method was proposed as a sensor for determining gB of HCMV (gB-HCMV) in real samples (Pires et al., 2018). Magnetic particles were modified with protein G followed by monoclonal antibody (mAb1) to facilitate the attachment of gB-HCMV. The biosensor showed a linear response between 90 and 700 pg/ml and a limit of detection of 90 pg/ml. Glycoprotein B was further employed for HCMV diagnosis using an electrochemical sensor (Pires et al., 2015). The electrode surface was functionalized with anti-human cytomegalovirus glycoprotein B and then immersed into a solution containing gB-HCMV and gold nanoparticles functionalized with anti-HCMV glycoprotein B. The identification of gB-HCMV using electrochemical stripping analysis resulted in an LoD of 3.3 ng/ml. Azek et al. proposed an electrochemical biosensor for detecting DNA sequences of HCMV and demonstrated an LoD of 0.6 amol/ml (Azek et al., 2000). Huang et al. utilized an electrochemical biosensor for the diagnosis of pp65-antigen of HCMV utilizing pp65-antibodies (Huang et al., 2016). The biosensor’s dynamic range was between 0.1 and 80 ng/ml with an LoD of 30 pg/ml. Furthermore, a microelectromechanical-based affinity biosensor was used to detect UL83-antigen. The biosensor exhibited an LoD of 84 pg/ml and a dynamic range between 0.3 and 300 ng/ml (Alzahrani et al., 2022). Despite the sensitivity and good sensing performances demonstrated by the abovementioned biosensing techniques, there are disadvantages associated with these techniques. Electrochemical biosensors mainly require the use of redox agents and multiple electrodes during detection which will add an extra step towards detection, hence, limiting the direct detection of HCMV. Furthermore, QCM-based biosensors are bulky and sensitive to surrounding vibrations and extraneous factors. Therefore, on-site detection using the QCM technique is challenging. In this regard, easy to use, direct and rapid detection techniques of HCMV are highly desirable. Quart tuning fork (QTF) simple structure and detection means allow on-site and direct detection of HCMV. The technique does not require extra steps during detection and can be easily made portable. Hence, facilitating mass screening of the virus and subsequently manage the spread of the virus or other diseases in case of epidemics.
Quartz tuning fork (QTF) is an acoustic resonator which consists of quartz crystal-made two-pronged forks. Applying an oscillating voltage causes the QTF to vibrate. The QTF resonance frequency is mainly governed by its prong dimensions and quartz crystal properties (Lin et al., 2022). Usually, the commercially available QTF resonance frequency is approximately 32.7 KHz with a high quality factor as high as 10,000 in the atmosphere (Friedt and Carry 2007). QTF-based methods can potentially become powerful sensors in a variety of applications due to their low cost, high precision, excellent stability, low power consumption and compact size. QTFs can be employed as mechanical sensors in different areas including force spectroscopy (Barbic et al., 2007, Yamada et al., 2019), gas sensing (Lang et al., 2021, Zhou et al., 2021), biological sensing (Su et al., 2002, Wei et al., 2021) and measurements of density and viscosity (Gonzalez et al., 2017). QTF was employed as affinity biosensor to detect antibody-antigen binding events. It was utilized to sense the binding events between anti-human immunoglobulin G (IgG) and human IgG (Su et al., 2002).
In this work, a low-cost, fast, sensitive, specific and simple QTF-based biosensor was prepared and used for the detection of HCMV. The QTF prongs were modified with UL83- antibody of HCMV through standard bio-functionalization steps. Then, the biosensor was tested against different concentrations of UL83-antigen. Furthermore, the biosensor sensitivity, limit of detection (LoD), stability and selectivity were investigated. In addition, the performance of the QTF-based biosensor was compared to others reported in literature.
2 Materials and methods
2.1 Materials
QTFs coated with 100 nm thick gold (Au) layer (see Fig. 1) were supplied by Forien Inc., Edmonton, AB, Canada. The QTF load capacitance, spring constant and resonance frequency are 12.5 pF, 20 kN/m and 32.768 kHz, respectively. Cysteamine and glutaraldehyde were purchased from Sigma Aldrich. UL83-antibody was purchased from Virusys-Corporation, USA. UL83-protein (antigen) was purchased from Miltenyl Biotec Ltd., UK. Phosphate buffer saline (PBS) (pH 7.4) was obtained from Fisher scientific (USA). LDL-antigen (Sino Biological, Chin), vitamin c and vitamin D were used for selectivity experiments.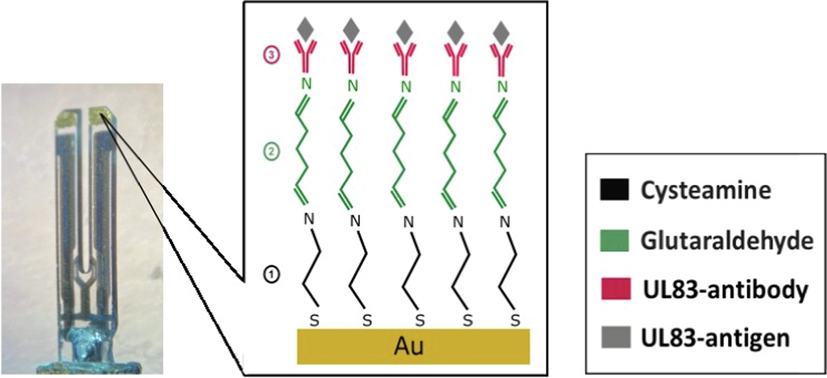
UL83-antibody biofunctionalization steps applied at the Au-coating on the QTF.
2.2 QTF biofunctionalization
For biofunctionalization, The Au coating at the QTF (see Fig. 1) was functionalized with cysteamine by immersing the QTF in an ethanolic solution of 100 µL of 10 nM cysteamine for 60 min. Then, the surface was subsequently washed using ethanol and followed by drying with air. The QTF was then immersed in 2.5% of 100 µL glutaraldehyde in PBS for 60 min. Rinsing with DI water was then applied at the QTF surface. Finally, UL83-antibody attachment was conducted by immersing the QTF in 100 µL of 10 µg/mL UL83-antibody diluted in DI water for a time period of 60 min. Rinsing with DI water was then performed at the Au-coated QTF followed by drying. Fig. 1 shows the functionalization steps performed at the QTF’s Au coating.
2.3 Sensor testing
After biofunctionalization, the QTF-based biosensor was tested against four concentrations of UL83-antigen (0.3 ng/mL, 3 ng/mL, 30 ng/mL and 300 ng/mL). Firstly, the biosensor was immersed in 100 µL of PBS solution for 15 min. Then, it was connected to Quester Q10 instrument (Fouiren Inc., Edmonton, AB, Canada) and submerged in 100 µL of DI water where the baseline resonance frequencies of the QTF were measured. Note that during measurements, the QTFs were submerged at a 25 µm depth into 100 µL DI water. Hence, only the Au-coated part of the QTF (see Fig. 1) is immersed in the PBS droplet. After measuring the baseline, the QTFs were disconnected from the Quester Q10 and washed with DI water, followed by incubation in UL83-antigen for 15 min. Then, the same process performed to measure the baseline resonance frequencies was performed in order to determine the resonance frequencies of the different concentrations of UL83-antigens. Similarly, each biofunctionalization step resonance frequency was measured using the same process applied to measuring the resonance frequencies for both, the baseline and the UL83-antigen concentrations. Frequency sweeps were carried out via integrated impedance analyzer in the Quester Q10. Furthermore, the real part and the imaginary part of the impedance response are measured by the impedance analyzer. To resonate the QTF at specific frequencies, the proportional integrated differential technique was utilized. MATLAB or Origin Lab program were utilized to analyze the output data. All experiments were repeated three times.
3 Results and discussion
3.1 Biofunctionalization
In this study, the effect of each functionalization step on the resonance frequency was investigated. The QTF frequency was swept between 31,000 to 39000 Hz and each cycle lasted for 30 s.
Fig. 2a shows the resonance frequency responses as a result of functionalization steps. Initially, the Au-coated QTF resonance frequency was 32.832 Hz. Then, the resonance frequency decreased to 32.790 Hz after cysteamine functionalization. Similarly, the resonance frequency decreased after functionalization with glutaraldehyde and UL83-antibody to 32.724 Hz and 32.655 Hz, respectively. The resonance frequency shift (
for each functionalization step is presented in Fig. 2b. The resonance frequency shift (
for each functionalization step is presented in Fig. 2b and was calculated by subtracting the measured resonance frequency of baseline from the functionalization steps measured resonance frequency.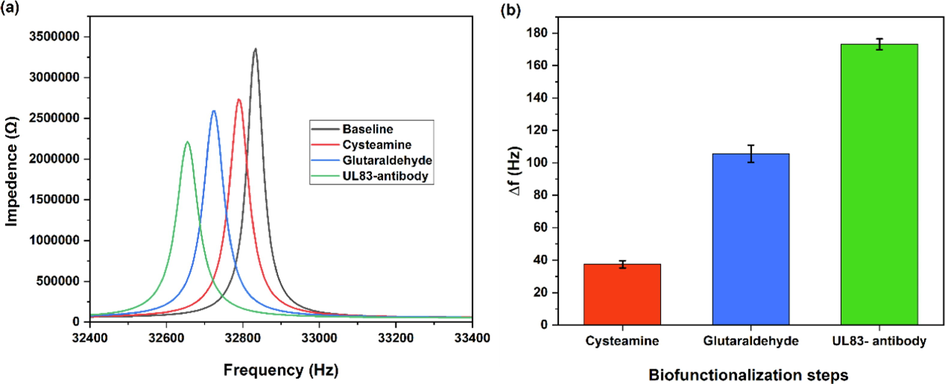
(a) resonance frequency responses with functionalization steps. (b)
f as a result of biofunctionalization steps.
The change in resonance frequency of QTF is mainly attributed to mass loading change and effective stiffness of the substance which are denoted as (
) and (
), respectively, as illustrated in Eq. (1) (Alshammari et al., 2020).
3.2 QTF-based biosensor performance
After biofunctionalization, the QTF-based biosensor was tested against different concentrations of UL83-antigen. Firstly, the baseline frequency was measured. The QTF-based biosensor was then incubated with 0.3 ng/mL, 3 ng/mL 30 ng/mL and 300 ng/mL of UL83-antigen. The frequency decreased due to UL83-antigen concentrations incubations at the QTF-biosensor (see Fig. 3). The measured frequency due to testing the biosensor against UL83-antigen was normalized to the measured frequency of baseline. Fig. 3b shows the shift in resonance frequency (
) of the UL83-antigen concentrations. Incubating the biosensor against 0.3 ng/mL of UL83-antigen resulted in
of 19.1 ± 1.5 Hz.
increased to 41 ± 2.3 as the biosensor was incubated in 3 ng/mL of the antigen. The values of
were 72 ± 2.8 Hz and 131 ± 3.5 Hz when the biosensor was tested against 30 ng/mL and 300 ng/mL of UL83 antigen, respectively. This increase in
is related to the increased mass loading at the QTF surface with increased UL83-antigen concentration. Similarly,
against series incubations of PBS was calculated.
is 7.2 ± 2.8 Hz after introducing the biosensor with PBS four times. The change in
as a result of testing against the PBS (blank) sample could be due to the deposition of PBS contents at the surface of the QTF. The biosensor demonstrated larger change in
when tested against the UL83-antigen as compared to testing against only blank samples (PBS).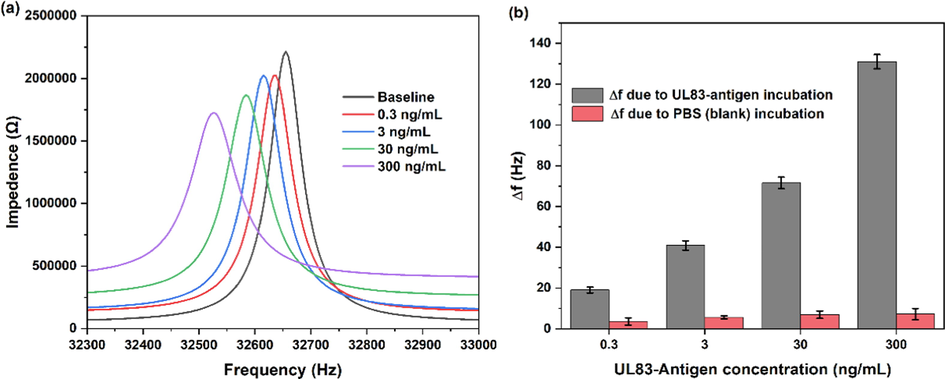
(a) Resonance frequency measurements with different UL83-antigen concentrations. (b)
f when biosensing was performed against UL83-antigen and blanks samples.
Fig. 4 illustrates the reported biosensor calibration curve. The linear fitting of the of the UL83-antigen concentrations from 0.3 ng/mL to 300 ng/mL is represented by the red solid line. The sensitivity of the QTF-based biosensor which can be found from the calibration curve is 15.91 Hz/ln(ng/mL), whereas the LoD is 0.36 ng/mL. The LoD was obtained using Eq. (2) (Chiavaioli et al., 2017, Cardona-Maya et al., 2018).
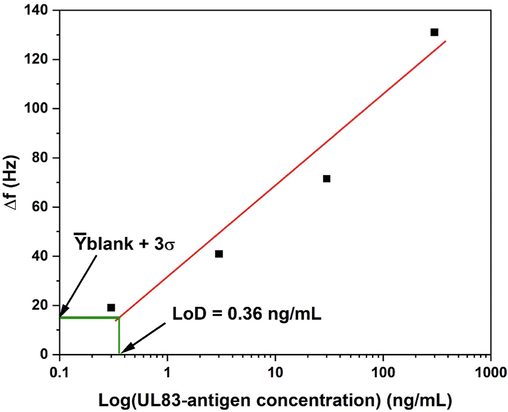
Calibration curve of the QTF-based biosensors.
HCMV Biosensor
LoD
Refs.
Imaging Ellipsometry
0.024 ng/mL
(Sun et al., 2015)
Differential pulse voltammetry
0.03 ng/mL
(Huang et al., 2016)
Colorimetric biosensor
0.03 ng/mL
(Alba-Patiño et al., 2020)
Microelectromechanical system‐based biosensor
0.084 ng/ml
(Alzahrani et al., 2023)
Enzymatic biosensor
0.09 ng/mL
(Pires et al., 2018)
QTF-based biosensor
0.36 ng/mL
This work
Anodic stripping voltammetry
2.225 ng/mL
(Authier et al., 2001)
Selectivity is an important factor in biosensors. The biosensor reported here was tested against two samples, positive and negative samples. The positive sample consists of UL83-antigen, LDL-antigen, vitamin D and vitamin C. The negative sample, however, consists of LDL-antigen, vitamin D and vitamin C without UL83-antigen. All biomarkers were diluted in PBS and the concentration of each was 30 ng/mL. The change in measured resonance frequency against the positive sample (83 Hz) is larger than the change in measured frequency against the negative sample (20.08 Hz) which demonstrates the selectivity of the biosensor against UL83-antigen (see Fig. 5).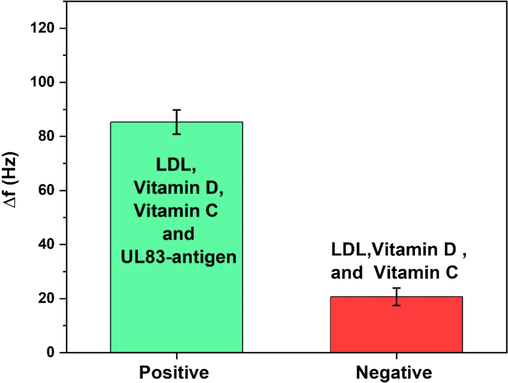
Biosensor selectivity when tested against positive and negative samples.
4 Conclusion
In this work, a simple QTF-based biosensor for the simple detection of HCMV UL83-antigen is demonstrated. The study demonstrates that QTF can be used in future for the mass screening of diseases. The biosensor demonstrated here is suitable for on-site detection of biomarkers without the need to send samples to trained personnel in central laboratories. Furthermore, the simplicity of the device working principle allow early detection of HCMV and hence allow clinicians to make early medical interventions to avoid any disability to new-born babies in future. The detection limit obtained for the QTF-based biosensor is 0.36 ng/mL with a sensitivity of 15.91 Hz/ln (ng/mL). In addition, the biosensor demonstrated its selectivity towards UL83-antigen which is an important factor to consider in diagnostic devices.
Acknowledgement
The authors extend their appreciation to the King Salman Center for Disability Research for funding this work through Research Group no KSRG-2022-019
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Nanoparticle reservoirs for paper-only immunosensors. ACS Sensors. 2020;5(1):147-153.
- [CrossRef] [Google Scholar]
- Detection of chemical host-guest interactions using a quartz tuning fork sensing system. IEEE Sens. J.. 2020;20(21):12543-12551.
- [CrossRef] [Google Scholar]
- Microelectromechanical system-based biosensor for label-free detection of human cytomegalovirus. IET Nanobiotechnol. 2022
- [Google Scholar]
- Microelectromechanical system-based biosensor for label-free detection of human cytomegalovirus. IET Nanobiotechnol.. 2023;17(1):32-39.
- [CrossRef] [Google Scholar]
- Gold nanoparticle-based quantitative electrochemical detection of amplified human cytomegalovirus DNA using disposable microband electrodes. Anal. Chem.. 2001;73(18):4450-4456.
- [CrossRef] [Google Scholar]
- Hybridization assay at a disposable electrochemical biosensor for the attomole detection of amplified human cytomegalovirus DNA. Anal. Biochem.. 2000;284(1):107-113.
- [Google Scholar]
- Femto-Newton force sensitivity quartz tuning fork sensor. Sens. Actuators, A. 2007;136(2):564-566.
- [Google Scholar]
- Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Hum. Cytomegalovirus 2008:417-470.
- [Google Scholar]
- Britt, W.J., 2010. Human cytomegalovirus: propagation, quantification, and storage. Curr. Protocols Microbiol. 18 (1) 14E.13.11-14E.13.17.
- Brown, H.L., Abernathy, M.P., 1998. Cytomegalovirus infection. Seminars in perinatology, Elsevier.
- Label-free wavelength and phase detection based SMS fiber immunosensors optimized with cladding etching. Sens. Actuators B. 2018;265:10-19.
- [CrossRef] [Google Scholar]
- Towards a uniform metrological assessment of grating-based optical fiber sensors: from refractometers to biosensors. Biosensors. 2017;7(2)
- [CrossRef] [Google Scholar]
- Immunobiology of human cytomegalovirus: from bench to bedside. Clin. Microbiol. Rev.. 2009;22(1):76-98.
- [Google Scholar]
- Congenital cytomegalovirus: public health action towards awareness, prevention, and treatment. J. Clin. Virol.. 2009;46:S1-S5.
- [Google Scholar]
- Human cytomegalovirus immediate-early mRNA detection by nucleic acid sequence-based amplification as a new parameter for preemptive therapy in bone marrow transplant recipients. J. Clin. Microbiol.. 2000;38(5):1845-1853.
- [Google Scholar]
- Gonzalez, M., Seren, H., Buzi, E., et al., 2017. Fast downhole fluid viscosity and density measurements using a self-oscillating tuning fork device. In: 2017 IEEE Sensors Applications Symposium (SAS), IEEE.
- Pathogenesis of human cytomegalovirus in the immunocompromised host. Nat. Rev. Microbiol.. 2021;19(12):759-773.
- [Google Scholar]
- Analytical methods for detection of human cytomegalovirus clinched biosensor a cutting-edge diagnostic tool. Biomed. Eng. Adv.. 2021;1:100006
- [Google Scholar]
- Recent progresses on biosensors for Escherichia coli detection. Food Anal. Methods 2022:1-29.
- [Google Scholar]
- Electrochemical immunoassay for cytomegalovirus antigen detection with multiple signal amplification using HRP and Pt-Pd nanoparticles functionalized single-walled carbon nanohorns. Electroanalysis. 2016;28(5):1126-1133.
- [CrossRef] [Google Scholar]
- Quartz tuning fork-based demodulation of an acoustic signal induced by photo-thermo-elastic energy conversion. Photoacoustics. 2021;22:100272
- [Google Scholar]
- Application of standard and custom quartz tuning forks for quartz-enhanced photoacoustic spectroscopy gas sensing. Appl. Spectrosc. Rev. 2022:1-23.
- [Google Scholar]
- Detection of congenital cytomegalovirus in newborns using nucleic acid amplification techniques and its public health implications. Virol. Sin.. 2017;32:376-386.
- [Google Scholar]
- Structure of human cytomegalovirus virion reveals host tRNA binding to capsid-associated tegument protein pp150. Nat. Commun.. 2021;12(1):5513.
- [Google Scholar]
- Binding partners for the UL11 tegument protein of herpes simplex virus type 1. J. Virol.. 2003;77(21):11417-11424.
- [Google Scholar]
- Late expression of a beta chemokine homolog by murine cytomegalovirus. J. Virol.. 1997;71(2):1671-1678.
- [Google Scholar]
- Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol.. 2006;9(4):423-429.
- [Google Scholar]
- Disposable immunosensor for human cytomegalovirus glycoprotein B detection. Talanta. 2015;136:42-46.
- [Google Scholar]
- A rapid magnetic particle-based enzyme immunoassay for human cytomegalovirus glycoprotein B quantification. J. Pharm. Biomed. Anal.. 2018;156:372-378.
- [Google Scholar]
- Clinical diagnostic testing for human cytomegalovirus infections. J Infect Dis. 2020;221(Supplement_1):S74-S85.
- [Google Scholar]
- Antiviral chemotherapeutic agents against equine herpesvirus type 1: the mechanism of antiviral effects of porphyrin derivatives. Louisiana State University and Agricultural & Mechanical College; 2009.
- Detection of cytomegalovirus antibodies using a biosensor based on imaging ellipsometry. PLoS One. 2015;10(8):e0136253.
- [Google Scholar]
- Human cytomegalovirus detection by a quartz crystal microbalance immunosensor. Enzyme Microb. Technol.. 2000;27(9):639-645.
- [Google Scholar]
- Incidence and impact of CMV infection in very low birth weight infants. Pediatrics. 2014;133(3):e609-e615.
- [Google Scholar]
- Simultaneous detection of vertical and lateral forces by bimodal AFM utilizing a quartz tuning fork sensor with a long tip. Jpn. J. Appl. Phys.. 2019;58(9):095003
- [Google Scholar]
- Absorption spectroscopy gas sensor using a low-cost quartz crystal tuning fork with an ultrathin iron doped cobaltous oxide coating. Sens. Actuators B. 2021;326:128951
- [Google Scholar]
- Estimation of the worldwide seroprevalence of cytomegalovirus: a systematic review and meta-analysis. Rev. Med. Virol.. 2019;29(3):e2034.
- [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.102703.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







