Translate this page into:
Sero-RAPD makers based evaluation of chilli pepper germplasm for resistance to chilli veinal mottle virus
⁎Corresponding author at: Plant Pathology, IPP, MNS-University of Agriculture, Multan, Pakistan. mashfaq@mnsuam.edu.pk (Muhammad Ashfaq)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Background
Chilli veinal mottle virus is a remarkable threat to chilli crops worldwide including Pakistan.
Methods
In the present study, 18 chilli pepper genotypes were catalogued against ChiVMV ATIPK isolate by using DAS-ELISA with virus-specific polyclonal antisera.
Results
Out of 18 genotypes, four genotypes viz., Loungi pepper, Sanam, Red Pepper, and CV-1 were categorized as highly susceptible followed by three susceptible (Soofi, Royal Wonder, Pepper PLSQ) and four (Yolo wonder, Bhakkar Local, CIM-506, CR-25) were moderately susceptible. The plants of these genotypes exhibited characteristic symptoms of ChiVMV and accumulated a high level of virus titer. The rest of three (California wonder, Gola Peshawar, Red top) and four (CV-7, Hot Pepper, Doni Pepper, Ghotki) genotypes were grouped as resistant and moderately resistant, respectively. Three resistant and three highly susceptible genotypes were further appraised for genetic diversity by RAPD markers and a total of 75 bands were scored, 76% of them unveiled polymorphism with an amplification range of 5–9 bands. Genetic analysis revealed 44–78.67% similarities with Mean Similarity Index (MSI) ranging from 59.46 to 65.86%. Most of the primers were highly informative with PIC value fluctuated from 0.388 to 0.666, with an average value of 0.512. Cluster analysis further confirmed and clustered both groups of genotypes in a separate clade.
Conclusion
It is concluded that genotypes; California Wonder and Red Top showed diverse response from other genotypes, not only based on symptoms and ELISA but also on markers' basis. Therefore, these genotypes could be useful as ChiVMV resistant sources in the chilli breeding program.
Keywords
Chilli
RAPD
Genetic diversity
ELISA
ChiVMV
1 Introduction
Capsicum (Capsicum annuum and Capsicum frutescens) is a most consumed and famous solanaceous vegetable after potato and tomato across the globe including Pakistan and its main usage is as sauces and spices (Berke, 2002; Riaz et al., 2022). It has a level-headed amount of phosphorus, calcium, iron, capsaicinoids, and high in vitamins also i.e. tocopherols, carotenoids and folate (Altaey, 2018; Jimenez-garcia et al., 2014; Kantar et al., 2016; Wahyuni et al., 2011). In Pakistan, chilli’s cultivation practised on a large scale as a cash crop for small farmers (Ashfaq et al., 2014). Capsicum cultivation area is increasing day by day in Pakistan especially in Punjab due to transition from traditional to non-traditional farming. Furthermore, when compared with world chilli’s production, the gap is too high where production in Pakistan is ten times less (1–2 tons/ha) compared to China (20 tons/ha) and other chilli growing countries (Amjad et al., 2007).
Viral diseases are known to badly affect successful solanaceous crops production in Pakistan (Ahmad and Ashfaq, 2017). Several viruses have been reported to heavily attack and results in colossal losses in South Asian countries including Pakistan (Ahsan et al., 2020; Green and Kim, 1991; Hameed et al., 1995; Riaz et al., 2022; Shah et al., 2009). Among the viral diseases, Chilli veinal mottle virus affects many solanaceous crops worldwide (Zhao et al., 2014) including Pakistan (Ahmad and Ashfaq, 2017). This virus was first time reported to infect Capsicum (C. frutescens) from Malaysia on visual symptoms basis followed by detection using serological methods in 1979 (Ong et al., 1979). In Pakistan, ChiVMV was detected based on symptomatology and serology in 1993 (Hameed et al., 1995) and subsequently confirmed by sequencing of capsid protein cistron (Ahmad and Ashfaq, 2018, 2017; Riaz et al., 2018). Some workers have also reported ChiVMV disease from pepper fields of Pakistan with an incidence of 40–44.7 % (Shah et al., 2009; Shah and Khalid, 1999). ChiVMV is flexuous filamentous particle (Genus: Potyvirus, Family: Potyviridae) present mainly in hot pepper cultivars of Asian countries (Ahmad and Ashfaq, 2018; Green and Kim, 1991; King et al., 2012). ChiVMV contains a 9.7-kb + ssRNA genome (flanked by UTR at the 5′ and 3′ends) that translates a 350-kDa polyprotein while virus-encoded proteinases cleaved it into ten functional proteins (Anindya et al., 2004).
The ChiVMV has a narrow host range, transmitted by sap and aphids in non-persistent manner (Raccah et al., 1985). ChiVMV could be successfully transmitted by mechanical inoculation and grafting techniques but evidence of transmission through seed is not reported (Abu Kassim, 1986). The virus produces characteristic symptoms of leaf mottling, dark green and vein-banding on capsicum leaves (Ong et al., 1980). Random Amplified Polymorphic DNA (RAPD) markers have proved to be useful in assessing genetic diversity in many crops (Abbas et al., 2008). Molecular markers were deployed for the detection of variation among DNA sequences of cultivars and ultimately save the varietal developmental resources and showed promise in expediting plant-breeding procedures (De Mattia et al., 2012). Furthermore, this method is cheaper and simpler compared to other methods and there is no need for prior knowledge of sequence (Ward, 1995; Yasmin et al., 2006). Genetic variability of breeding materials and genotypes is prerequisite knowledge for the breeders to improve the plant's genetic makeup. The study of genetic variability to detect groups within morphologically identical genotypes is imperative for well-organized preservation, proper evaluation and better exploitation of genetic materials. Selection and cultivation of virus-resistant crop genotypes are considered as an efficient, manageable, durable and economical method for managing plant diseases especially plant viruses (Ashfaq et al., 2014). Keeping into account the devastating nature of this virus, the present study is aimed to determine the resistant/ susceptible response of the chilli pepper genotypes against ChiVMV Pakistani isolate.
2 Materials and methods
2.1 Virus source and maintenance
Pakistani chilli veinal mottle virus (ChiVMV) ATIPK isolate (Accession No. KJ472764) was obtained from M. Ashfaq's Plant Virology Laboratory. The ChiVMV was revived, propagated and maintained on Capsicum annuum cv., Loungi pepper plants and assayed by double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) using ChiVMV specific monoclonal antisera (Ashfaq et al., 2014; Iqbal et al., 2011). The inoculated Loungi pepper plants were retained in screen house (insect free environment) and renewed after two to three weeks. The infection of ChiVMV in Loungi pepper plants was confirmed by using specific DAS-ELISA.
2.2 Plant material
The chilli pepper genotypes consisting of 18 sets of cultivars (Bhakkar Local, Pepper PLSQ, Loungi pepper, CIM-506, Gola Peshawar, CV-7, Red Pepper, Sanam, CV-1, Hot Pepper, Doni Pepper, Ghotki, Yolo wonder, CR-25, Soofi, Royal Wonder, California wonder, Red Top) were obtained from the Federal Seed Certification & Registration Department, Islamabad and Horticultural Research Institute, NARC, Islamabad. Fifteen seeds of each cultivar were sown in germination trays containing peat moss under greenhouse conditions (relative humidity 70 % and temperature 25 °C). The capsicum seedlings at two to three-leaf stage were transplanted @ two plants per plastic pot (3″x3″) filled with sterilized soil mixture (sand, clay and compost @ 1:1:1) under greenhouse conditions and one to two plants per pot were maintained. Each experiment was replicated thrice consisting of 10 plants per replication.
2.3 Mechanical inoculation
Symptomatic and ELISA positive leaf samples of Loungi pepper inoculated with ChiVMV ATIPK isolate at 14 days of post-inoculation (dpi) were used for screening chilli pepper genotypes. One gram infected Loungi pepper leaves were harvested and homogenized in 0.05 M Phosphate buffer (pH 7.2) containing 1 % Na2SO3 @ 1/3 w/v and resultant sap extract was subsequently used to mechanically inoculate pepper plants at 2–3 leaf stage (Ashfaq et al., 2014, Ashfaq et al., 2010). The inoculated plants were washed with distilled water to get rid of excess material and placed under greenhouse conditions (relative humidity 70 % and temperature 25 °C) while healthy plants of test genotypes served as control. The genotypes were categorized as highly resistant (symptoms less), resistant (Infected plants showing mild symptoms i.e. 1–10 %), moderately resistant (infected plants showing 11–20 % symptoms), moderately susceptible (infected plants representing 21–30 % symptoms), susceptible (infected plants showing 31–40 % symptoms) and highly susceptible (Infected plants showing severe symptoms i.e. >40 %) based on host reaction (Ashfaq et al., 2007).
2.4 Serological assay
Leaf sample from ChiVMV-inoculated (diseased) plants was taken at 21 dpi and subsequently subjected to ChiVMV-specific DAS-ELISA (Loewe Biochemicals, Germany Cat. No. 07185) as per manufacturer’s instructions. An Automatic ELISA reader (HER-480 HT Company (Ilford) ltd., UK) was used to measure absorbance value at 405 nm and the samples with two times or higher reading compared to the average of healthy tissue and negative control were categorized as ChiVMV positive.
2.5 Molecular analysis
Total DNA from six chilli pepper genotypes (three susceptible and three resistant) was extracted by (i) Proteinase K method (Sambrook and Ruseel, 2001) and (ii) Plant Genomic DNA Purification Mini Kit (Cat No. K0791, Thermo Scientific Gene JET, USA). Twenty RAPD primers were deployed in the current study and primers having polymorphic amplification were used for molecular characterization studies. Polymerase chain reaction (PCR) of extracted DNA was carried out by following the protocol as described by Devos and Gale (1992). Furthermore, the amplified products were separated by electrophoresis and polymorphism was detected by using agarose gel (3 %) pre-stained with ethidium bromide.
2.6 Statistical analysis
UV Transilluminator was used to observe the amplified bands in electrophoresed gels of RAPD markers. SynGene Gel Documentation System was used to take the image. The absence or presence of amplified bands was recorded as 0 or 1, respectively for all tested genotypes with eleven primers. For analysis, data was obtained by the counting of polymorphic bands and subjected to Popgene32 software (Yeh et al., 2000). The value of polymorphism information content (PIC) was measured with the below given equation (Botstein et al., 1980).
3 Results
3.1 Germplasm screening
Eighteen genotypes were screened against ChiVMV ATIPK isolate based on visual symptoms (Fig. 1) by following the disease rating scale and serological assay. All the 18 genotypes of chilli pepper showed characteristic symptoms' viz., dark green vein banding and leaf mottling upon mechanical inoculation with infective sap of ChiVMV. Individual plants of Loungi pepper, Sanam, Red Pepper and CV-1 showed dark green vein banding and leaf mottling at 14 dpi while the rest of the genotypes showed symptoms between 18 and 26 dpi. All the plants of these genotypes exhibited >50 % ChiVMV infection based on 0–5 disease rating scale (DRS) and DAS-ELISA revealed relatively high titer (>1.0), so all of these genotypes were considered as highly susceptible to ChiVMV ATIPK isolate. Similarly, other three genotypes viz., Soofi, Royal Wonder, Pepper PLSQ and four genotypes viz., Yolo wonder, Bhakkar Local, CIM-506 and CR-25 exhibited <50 % infection and DAS-ELISA showed relatively low titer (>0.5 %), were catalogued as susceptible and moderately susceptible, respectively. On the other hand, three genotypes (California wonder, Gola Peshawar, Red top) showed 5–10 % ChiVMV infection based on DRS with relatively low titer (<0.25), therefore, grouped as resistant showing light green banding along veins followed by the rest of the four genotypes viz., CV-7, Hot Pepper, Dane Pepper, and Ghotki with a moderately resistant response showing symptoms like moderate green banding along veins with light mottling (Table 1).
Chilli pepper (cv., Loungi pepper) plants showing characteristic symptoms (A, B) upon sap inoculation with ChiVMV isolate ATIPK.
Sr. No
Reaction Grade
Disease rating scale (DRS)
Disease Response
Genotypes
ELISA Reading at 405 nm
1
Highly Resistant
0
0 %
Nil
0.122–0.201
2
Resistant
1
1–10 %
Gola Peshawar, California wonder, Red Top
0.229–0.311
3
Moderately Resistant
2
11–20 %
CV-7, Hot Pepper, Doni Pepper, Ghotki
0.517–0.638
4
Moderately Susceptible
3
21–30 %
Yolo wonder, Bhakkar Local, CIM-506, CR-25
0.651–0.723
5
Susceptible
4
31–40 %
Soofi, Royal Wonder, Pepper PLSQ
1.073–1.251
6
Highly Susceptible
5
>40 %
Loungi pepper, Red Pepper, Sanam, CV-1
1.753–1.997
3.2 Genetic diversity
Six chilli pepper genotypes (Loungi pepper, Sanam, CV1, California Wonder, Gola Peshawar, Red Top) were selected because of their diverse response against ChiVMV to genetically evaluate them using twenty RAPD primers. Eleven primers were used for final analysis out of twenty primers which produced clear, distinct, and readily detectable fragments having a high rate of polymorphism. Considering all the primer amplification in total tested genotypes, 75 bands were amplified in PCR reactions. Eleven primers generated 57 polymorphic fragments which showed 76 % of polymorphism with a maximum polymorphism percentage by primers GLD-19 and GLH-13 (85.71 %), whereas primers GLA-06, GLB-02, and GLC-10 showed the minimum percentage of polymorphism 66.66 % (Table 2). The average statistics of total bands per primer were 6.8, whereas, average amplification of polymorphic bands of each primer was 5.18 (bands) (Fig. 2). The score of amplified fragments per variety varied from 28 to 39, with an average number of 36.33 fragments per variety. The highest score of fragments was generated by Loungi pepper (39), Sanam (39), and CV-1 (39), followed by Red Top (38) and Gola Peshawar (35) genotypes. Whereas, California Wonder genotype produced minimum numbers of bands i.e., 28 (Fig. 3). The fragment amplification varied from 5 to 9; the highest score of bands was recorded for primer GLF-06 i.e., (9), while minimum numbers of fragments (5) were produced by primer GLD-13 (Fig. 4). The PIC value for 11 primers ranged from 0.388 to 0.666 with an average value of 0.512. Out of eleven primers, the maximum PIC value (0.66) was shown by primer GLH-13 while the minimum PIC value (0.38) was shown by primer GLA-06. Whereas, primer GLB-02 showed the PIC value (0.41), followed by GLA-02 (0.42), GLC-10 (0.44), GLB-07 (0.45), GLC-09 (0.54), GLD-13 (0.56), GLD-19 (0.57), GLF-06 (0.57) and GLE-02 (0.58) (Table 2). Ranges of markers PIC values showed that out of eleven markers, 6 were moderately informative having a share of 54.54 % while 5 were highly informative having a share of 45.45 % (Table 3). The size of securable bands produced by all primers ranged between 250 and 1500 bps. Primer GLA-06 produced bands within a range of 1000 to 1500 bps, whereas three primers GLA-02, GLB-02, and GLC-10 produced securable bands ranged from 750 to 1500 bps. Two primers GLC-09 and GLD-19 produced bands within a range of 500 to 1500 bps, primer GLD-13 produced bands between 400 and 1500 bps (Fig. 5), while single primer GLB-07 produced bands between 750 and 1000 bps. Primers GLE-02 and GLF-06 produced bands between a range of 500 to 1000 bps, while primer GLH-13 produced bands ranging from 250 to 1000 bps.
Sr. No.
Primer name
Sequence
No. of Alleles
PIC Value
Polymorphism (%)
Amplified product range (bp)
1
GLA-02
TGCCGAGCTG
07
0.428
71.42
750–1,500
2
GLA-06
GGTCCCTGAC
06
0.388
66.66
1,000–1,500
3
GLB-07
GGTGACGCAG
08
0.458
75.00
750–1,000
4
GLB-02
TGATCCCTGG
06
0.416
66.66
750–1,500
5
GLC-09
CTCACCGTCC
08
0.541
75.00
500–1,500
6
GLC-10
TGTCTGGGTG
06
0.444
66.66
750–1,500
7
GLD-19
CTGGGGACTT
07
0.571
85.71
500–1,500
8
GLD-13
GGGGTGACGA
05
0.566
80.00
1,000–1,500
9
GLE-02
GGTGCGGGAA
06
0.583
83.33
500–1,000
10
GLF-06
GGGAATTCGG
09
0.574
77.77
500–1,000
11
GLH-13
GACGCCACAC
07
0.666
85.71
250–1,000
Average
6.81
0.512
76
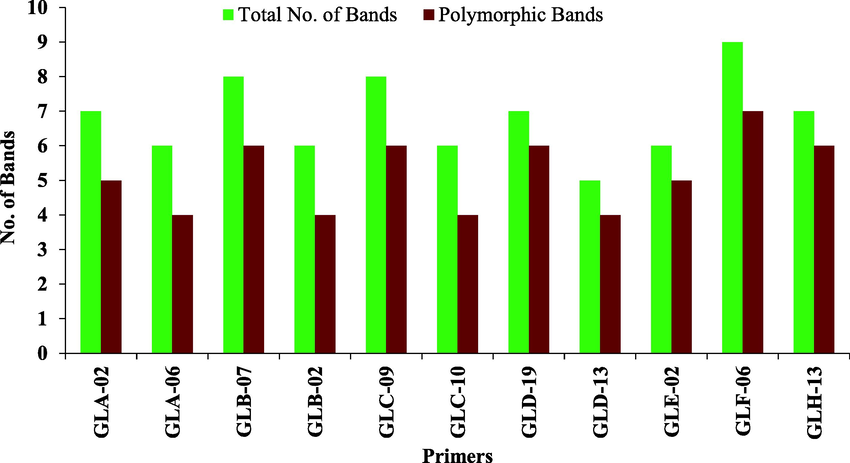
Total No of bands and polymorphic bands per RAPD primer.
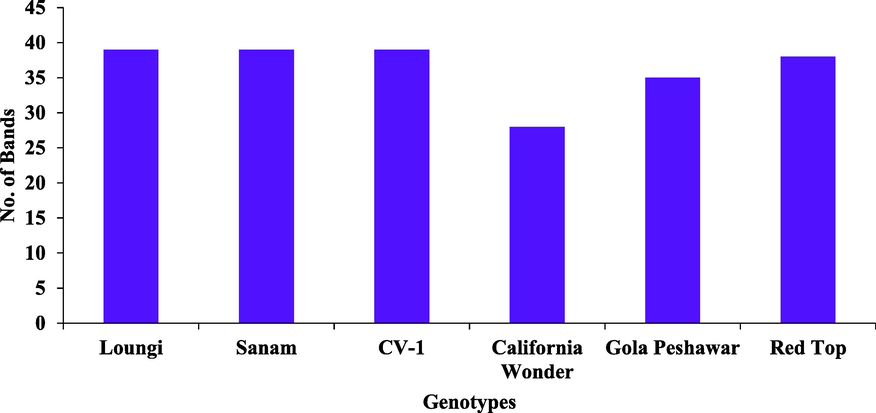
Number of amplified bands per genotypes with 10-base nucleotide RAPD primers.
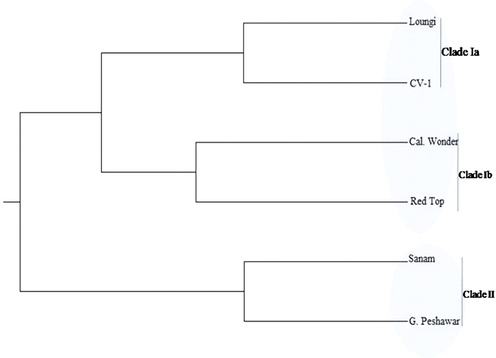
Dendrogram of six chilli pepper genotypes constructed based on genetic distance using 11 RAPD primers.
Ranges of PIC Values
Information level
≤0.30 Uninformative
0.30–0.55 Moderately Informative
≥0.56 Highly Informative
No. of Primers
0
06
05
%age of Primers
0.00 %
54.54 %
45.45 %
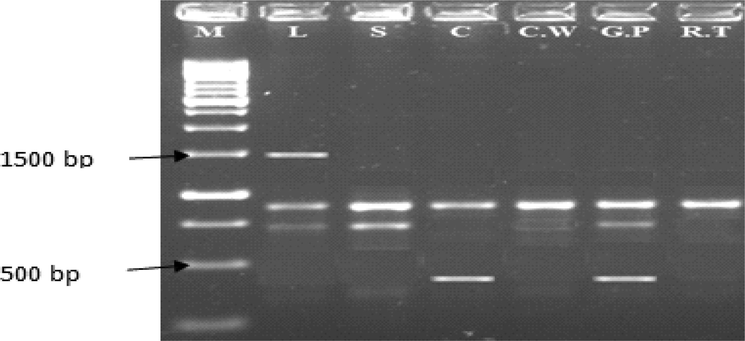
PCR amplification profile of chilli pepper genotypes (L = Loungi pepper, S = Sanam, C = CV-I C.W = California Wonder, G.P = Gola Peshawar, R.T = Red Top) using the primer GLD-13.
3.3 Genetic similarity matrix
Estimation of genetic similarity and distance of capsicum genotypes was done using multivariate analysis based on Nei’s UPGMA dendrogram. Genetic distance analysis showed the similarity coefficient varied from 44 % to 78 % within tested chilli pepper genotypes. The highest similarity 78.67 % was recorded between two sets of genotypes i.e., CV-1 and Loungi pepper, Sanam and Gola Peshawar, whereas the lowest similarity was noticed within genotypes CV-1 and Sanam. Red Top and California Wonder genotypes showed the similarity of 73.33 %, while California Wonder and Sanam, Red Top and Loungi pepper showed 69.33 % similarity. Three sets of genotypes i.e., Gola Peshawar and California Wonder, CV-1 and Red Top, California Wonder and Loungi pepper revealed 64 % similarity. Gola Peshawar showed 62.67 % similarity with Loungi pepper and CV-1 while 58.67 % similarity was observed between CV-1 and California Wonder. Whereas, similarities of 56 %, 50.67 %, and 49.33 % were evident in Red Top and Sanam, Red Top and Gola Peshawar, and Sanam and Loungi pepper. Maximum mean similarity index (65.86 %) was shown by California Wonder with other five genotypes followed by Loungi pepper with 64.80 %, Gola Peshawar (63.73 %), Red Top (62.66 %), CV-1 (61.60 %), and minimum mean similarity index (59.46 %) was shown by Sanam (Table 4). Nei's genetic identity (above diagonal).
Pop ID
Loungi pepper
Sanam
CV1
California Wonder
Gola Peshawar
Red Top
Loungi pepper
1
0.4933
0.7867
0.6400
0.6267
0.6933
Sanam
–
1
0.4400
0.6933
0.7867
0.5600
CV1
–
–
1
0.5867
0.6267
0.6400
California Wonder
–
–
–
1
0.6400
0.7333
Gola Peshawar
–
–
–
–
1
0.5067
Red Top
–
–
–
–
–
1
Cluster analysis catalogued understudied six genotypes into two main clades. Clade I further comprised of two sub-clades each consisting of two genotypes, sub-clade (Ia) contained Loungi pepper and CV-1 which are closely related to each other, while sub-clade (Ib) contained Red Top and California Wonder which are closely related to each other. Clade II comprises of two genotypes, Sanam, and Gola Peshawar which also showed their close relationship with each other (Fig. 4).
4 Discussion
Chilli pepper cultivars mostly grown in farmer’s field are susceptible to ChiVMV. Therefore, the importance of chilli pepper germplasm screening against ChiVMV on a regular basis and resistance gene based identification has been increased. Conventional breeding programs based on the phenotypic and symptomatic response of genotypes against specific disease are too much time-consuming. Whereas, molecular markers assisted breeding for varietal development of resistance is more reliable, authentic and time-saving. Variations between the cultivars concerning biochemical characters, molecular characters, and resistance response represents direct or indirect variation at the DNA level. Genetic diversity assessment of a crop is extremely useful for its improvement and efficient germplasm conservation. Genetic diversity between and within the populations and species have been characterized by the molecular markers while different classes of variations may be observed with different markers (Powell et al., 1996; Russell et al., 1997). With the advent of PCR, different molecular techniques such as sequence-tagged sites (STS), simple sequence repeats (SSR), inter simple sequence repeat (ISSR) polymorphic DNA, random amplified microsatellite polymorphism (RAMP), and random amplified polymorphic DNA (RAPD) have been developed (Wu et al., 1994). RAPD is easy to use as it requires small amount of plant material and is relatively cheaper (Devos and Gale, 1992; Joshi and Nguyen, 1993).
Chilli pepper genotypes having a different response against ChiVMV were evaluated and characterized on molecular basis by deploying RAPD markers. Briefly, eighteen chilli pepper genotypes screened against ChiVMV and showed a diverse response; genotypes belonging to a most diverse group based on disease response were selected for molecular characterization i.e., California wonder, Gola Peshawar, Red Top (resistant genotypes) and Loungi pepper, Sanam, CV-1 (highly susceptible genotypes). Genotypes selected from diverse reaction group in response to ChiVMV were genetically analyzed and they showed different levels of similarity and diversity and generate different clusters. RAPD markers have been employed successfully in various studies for polymorphism assessment in chilli which is an important parameter for evaluation and measurement of genetic distance in chilli germplasm (Bahurupe et al., 2013; Rai et al., 2013; Thul et al., 2012). Out of 20 RAPD primers used, 11 primers generated highly polymorphic and reproducible bands. Out of 75 bands, 57 (76 %) were polymorphic. In the present study, the polymorphic bands percentage is much higher compared to the other reports where scientists reported 44.4 %, 73.57 %, 45.2 %, 48.76 %, 50.8 %, 34.2 %, polymorphic bands in 23, 39, 47, 24, 10, 15, and 34 chilli pepper genotypes, respectively (Bahurupe et al., 2013; Lefebvre et al., 2001; Litoriya et al., 2009; Oyama et al., 2006; Paran et al., 1998; Rad et al., 2010; Subramanyam et al., 2012).
Bands produced by primers used in this study ranged from 5 to 9 bands and these are comparable with the results of other scientists where they reported 1–15, 3–12, and 9–20 bands amplification by RAPD primers (Bahurupe et al., 2013; Bhadragoudar and Patil, 2011; Moses and Umaharan, 2012; Subramanyam et al., 2012). Average numbers of amplified bands per primer in the current study are 6.8 and it is slightly lower than other reports i.e., 8–9 bands and 7.5 bands per primer (Lefebvre et al., 2001; Paran et al., 1998). While, other study revealed more average bands per primer i.e. 12.6 compared to the present study (Litoriya et al., 2009). Our results exhibited that percentage of polymorphism among primers ranged from 66.66 to 85.71 % which is highly significant and comparable to the findings of other studies where the same was observed in varying percentages i.e., 0–88 %, 14.29–66.67 %, and 16.16–81.81 % (Bahurupe et al., 2013; Bhadragoudar and Patil, 2011; Subramanyam et al., 2012).
Number of expressed alleles and their frequency were taken into account to assess the discriminatory power of a locus or loci using polymorphism information content (PIC) value. The PIC value of the current study ranged from 0.388 to 0.666, with a mean polymorphic information content of 0.512. Out of eleven primers, six were moderately informative whereas five primers were highly informative according to the PIC value. The PIC values of the current study showed a more informative nature of 11 primers compared to previous studies where a range of PIC values (0.31 to 0.43) with a mean PIC value (0.368) of 10 RAPD markers from 26 capsicum genotypes was reported (Singh et al. 2013). Results of current study PIC value are also comparable with earlier studies where PIC value range from 0.0 to 0.93 & 0.27–0.82 with average PIC value was 0.54 & 0.52 among 24 and 56 Capsicum genotypes, respectively (Rana et al., 2014; Yumnam et al., 2012).
Genetic similarity index generated by RAPD markers showed that genetic similarities among six genotypes vary between 44 and 78.67 % indicating a high level of genetic distance between the genotypes. These results are consistent with previous studies where 23–88 %, 20–89 %, and 38–85 % similarity among chilli pepper genotypes was observed (Baral and Bosland, 2004; Sitthiwong et al., 2005; Thul et al., 2012). The findings of previous studies exhibited 32–88 %, 8–85 %, and 40–96 % similarity among 24, 25, and 24 Capsicum genotypes, respectively whereas, the latter showed a mean similarity index of 0.68, predicting a significant level of diversity in genotypes. These are similar to the outcome of the present study.
5 Conclusions
The ChiVMV is an important viral pathogen that results in colossal losses in chilli production around the world including Pakistan. Among 18 genotypes challenged to ChiVMV, three resistant genotypes viz., California wonder, Gola Peshawar, Red top were found to have diverse genetic makeup compared to susceptible genotypes (Loungi pepper, Sanam, CV1) and these resistant genotypes might have resistance gene against ChiVMV. Gola Peshawar and Sanam genotypes exhibited resistant and highly susceptible responses, respectively and were grouped in a separate clade II. The maximum resemblance on genetic basis was observed within these two genotypes that urged us to believe that they might belong to common ancestors because of their repeated use in chilli pepper germplasm improvement program. RAPD primers deployed in the present study are expected to be a very useful tool for DNA fingerprinting of closely related chilli pepper genotypes. These results are very useful, effective, and prolific for the better management of ChiVMV menace in Pakistan because resistant genotypes are considered as one of the best options to manage the viral diseases.
Acknowledgement
This work was funded by International Foundation for Science (IFS), Sweden under the Research Grant No. C/5259-1 to M. Ashfaq. This project was also supported by Researchers Supporting Project Number (RSP-2023R7) King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Virus diseases of horticultural crops in Malaysia. In: FFTC Book Series. Taipei, Taiwan: Food and Fertilizer Technology Centre for the Asian and Pacific Region; 1986. p. :3-6.
- [Google Scholar]
- First report of chilli veinal mottle virus in tomato in Pakistan. J. Plant Pathol.. 2017;99:287-304.
- [Google Scholar]
- Genetic diversity and recombination analysis based on capsid protein gene of Chilli veinal mottle virus isolates from Pakistan. Eur. J. Plant Pathol.. 2018;151:891-900.
- [CrossRef] [Google Scholar]
- Current status and genetic variability of cucumber mosaic cucumovirus (CMV) isolates infecting major cucurbits andsolanaceous vegetables in Pothwar region of Pakistan. Pakistan J. Agric. Sci.. 2020;57:1353-1361.
- [Google Scholar]
- Complete genomic sequence of Pepper vein banding virus (PVBV): a distinct member of the genus Potyvirus. Arch. Virol.. 2004;149:625-632.
- [Google Scholar]
- Evaluation of urdbean germplasm for resistance against urdbean leaf crinkle virus. Pakistan J. Bot.. 2007;39:2103-2111.
- [Google Scholar]
- Effect of urdbean leaf crinkle virus infection on total soluble protein and antioxidant enzymes in blackgram plants. Pakistan J. Bot.. 2010;42:447-454.
- [Google Scholar]
- Screening for resistance to cucumber mosaic cucumovirus in Chilli pepper. J. Anim. Plant Sci.. 2014;24:791-795.
- [Google Scholar]
- Genetic diversity analysis in chilli (Capsicum annuum L.) using RAPD markers. Bioscan. 2013;8:915-918.
- [CrossRef] [Google Scholar]
- Berke, T., 2002. The Asian vegetable research development center pepper project, In: Proceeding of the 16th International Pepper Conference Tampico. Tamaulipas, Mexico.
- Construction of a Genetic Linkage Map in Man Using RestrictionFragment Length Polymorphisms. Am. J. Hum. Genet.. 1980;32:314-331.
- [Google Scholar]
- A multi-marker DNA barcoding approach to save time and resources in vegetation surveys. Bot. J. Linn. Soc.. 2012;169:518-529.
- [CrossRef] [Google Scholar]
- Green, S.K., Kim, J.S., 1991. Characteristics and Control of Viruses Infecting Peppers : A Literature Review Characteristics and Control of Viruses Infecting Peppers : A Literature Review, Asian Vegetable Research and Development Center.
- Biological characterization of Pakistani isolates of cucumber mosaic cucumovirus (CMV) Pakistan J. Bot.. 2011;43:3041-3047.
- [Google Scholar]
- Journal of chemical, biological and physical sciences changes in the quantity of phenolic compounds in peppers (Capsicum annuum L.) sprinkled with elicitors under cold. Stress. 2014;4:10-17.
- [Google Scholar]
- RAPD (random amplified polymorphic DNA) analysis based intervarietal genetic relationships among hexaploid wheats. Plant Sci. 1993:95-103.
- [Google Scholar]
- Virus taxonomy classification and nomenclature of viruses. In: International Committee on Taxonomy of Viruses (9th ed.). UK: Academic Press; 2012.
- [Google Scholar]
- Evaluation of genetic distances between pepper inbred lines for cultivar protection purposes: comparison of AFLP, RAPD and phenotypic data. Theor. Appl. Genet.. 2001;102:741-750.
- [CrossRef] [Google Scholar]
- Varietal identification of chilli (Capsicum annuum L.) using randomly amplified polymorphic DNA markers. Indian J. Agric. Biochem.. 2009;22:83-86.
- [Google Scholar]
- Genetic structure and phylogenetic relationships of Capsicum chinense. J. Am. Soc. Hortic. Sci.. 2012;137:250-262.
- [CrossRef] [Google Scholar]
- Aetiological investigation on a Chilli veinal mottle virus of chili (Capsicum annuum L.) newly recorded from peninsular Malaysia. MARDI Res. Bull.. 1979;7:78-88.
- [Google Scholar]
- The effect of chilli veinal mottle virus on yield of chilli (Capsicum annuum L.) MARDI Res. Bull.. 1980;8:74-78.
- [Google Scholar]
- Genetic structure of wild and domesticated populations of Capsicum annuum (Solanaceae) from Northwestern Mexico analyzed by RAPDs. Genet. Resour. Crop Evol.. 2006;53:553-562.
- [CrossRef] [Google Scholar]
- Variation in Capsicum annuum revealed by RAPD and AFLP markers. Euphytica. 1998;99:167-173.
- [CrossRef] [Google Scholar]
- The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis Wayne. Mol. Breed.. 1996;2:225-238.
- [Google Scholar]
- The role of flying aphid vectors in the transmission of cucumber mosaic virus and potato virus Y to peppers in Israel. Ann. Appl. Biol.. 1985;106:451-460.
- [CrossRef] [Google Scholar]
- Evaluation of genetic diversity in Capsicum spp. as revealed by RAPD markers. Acta Hortic.. 2010;829:275-278.
- [CrossRef] [Google Scholar]
- Genetic diversity in Capsicum germplasm based on microsatellite and random amplified microsatellite polymorphism markers. Physiol. Mol. Biol. Plants. 2013;19:575-586.
- [CrossRef] [Google Scholar]
- Estimation of genetic diversity in Capsicum annuum L. germplasm using PCR-based molecular markers. Natl. Acad. Sci. Lett.. 2014;37:295-301.
- [CrossRef] [Google Scholar]
- An insight into genetic variability and host response of Pakistani isolate of Chilli veinal mottle virus (ChiVMV) infecting chilli pepper. Int. J. Biosci.. 2018;12:302-312.
- [CrossRef] [Google Scholar]
- Evaluation of the chilli veinal mottle virus cp gene expressing transgenic Nicotiana benthamiana plants for disease resistance against the virus. Brazilian J. Biol.. 2022;82
- [CrossRef] [Google Scholar]
- Molecular Cloning: A Laboratory Manual. NY, USA, Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory; 2001.
- Prevalence, occurrence and distribution of chili veinal mottle virus in Pakistan. Pakistan J. Bot.. 2009;41:955-965.
- [Google Scholar]
- Classification of pepper (Capsicum annuum L.) accessions by RAPD analysis. Biotechnology. 2005;4:305-309.
- [Google Scholar]
- Subramanyam, Koona, Subramanyam, Kondeti, Rajasekhar, P., Chirra, @bullet, Reddy, S., 2012. Assessment of Genetic Relationships among South Indian Chilli (Capsicum annum L.) Cultivars Using RAPD and ISSR Markers.
- Molecular profiling for genetic variability in Capsicum species based on ISSR and RAPD markers. Mol. Biotechnol.. 2012;51:137-147.
- [CrossRef] [Google Scholar]
- Metabolite biodiversity in pepper (Capsicum) fruits of thirty-two diverse accessions: Variation in health-related compounds and implications for breeding. Phytochemistry. 2011;72:1358-1370.
- [CrossRef] [Google Scholar]
- PopGene32, Microsoft windows-based freeware for population genetic analysis. Version 1.32. In: Molecular Biology and Biotechnology Centre. University of Alberta: Edmonton, Canada; 2000.
- [Google Scholar]
- First Report of Chilli veinal mottle virus Infecting Tomato (Solanum lycopersicum) in China. Plant Dis.. 2014;98:1589.
- [Google Scholar]







