Translate this page into:
Tomato leaf disease detection and management using VARMAx-CNN-GAN integration
⁎Corresponding author. srinivasareddy.k@vitap.ac.in (Srinivasa Reddy K.)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Abstract
In contemporary agriculture, farmers confront substantial challenges in maintaining crop yields and mitigating agricultural losses attributable to diseases. The existing methods for diagnosing and managing tomato leaf diseases often exhibit deficiencies in terms of accuracy, robustness, and interpretability. Typically, these methods are reactive, addressing symptoms after the disease has already impacted the plants, resulting in delayed and often ineffective interventions. The precision of disease localization and severity estimation plays a crucial role in efficient disease treatment; regrettably, existing post-processing techniques frequently fall short in this regard. While these methods have their flaws, our proposed method uses the best parts of deep learning and vector autoregressive moving average processes with eXogenous regressors (VARMAx processes) to quickly and accurately find tomato leaf diseases. Our approach represents an innovative solution to the challenges currently confronting the agriculture sector, thanks to its proactive attributes, improved categorization capabilities, and advanced post-processing stages. Convolutional neural networks (CNNs) and generative adversarial networks (GANs), built upon the “VARMAx-CNN-GAN Integration” framework, form the core of our method. In this integrated model, convolutional neural networks serve the purpose of extracting features and performing early disease classification, whereas generative adversarial networks come into play for generating synthetic images, expanding the dataset, and enhancing the model’s ability to generalize. The “VARMAx-CNN-GAN Integration” model improves disease classification and decision-making for farmers and agronomists by providing insights into critical leaf images. Compared to traditional methods, it improves precision, accuracy, recall, AUC, and delay in identifying tomato diseases. The approach also shows potential for disease prevention, revolutionizing tomato leaf disease identification and management.
Keywords
Disease detection
Deep learning
Convolutional neural networks
Generative adversarial networks
1 Introduction
Tomatoes are a vital global crop, crucial for food security, but various leaf diseases threaten their productivity. Traditional detection methods lack accuracy and timeliness, leading to significant crop and economic losses. Current approaches are reactive and often misdiagnose diseases due to reliance on visible symptoms. Deep learning models like the Modified-Xception-based Multi-Level Feature Fusion (MXF) can address these issues. Deploying such models on resource-constrained agricultural devices presents challenges like limited processing power and memory. However, optimization techniques, hardware acceleration, and efficient inference strategies can make deployment feasible. Memory management through model caching and compression, and energy-efficient strategies can further improve performance. A federated learning framework can facilitate collaborative training, maintaining data localization and privacy. Traditional post-processing in disease detection lacks precision and interpretability, hindering effective disease management. Our research integrates IoT, deep learning, and “VARMAx-CNN-GAN Integration” mechanisms for early detection and management of tomato leaf diseases. This approach uses a fusion of CNNs, GANs, and autoencoders for accurate disease classification and saliency mapping for better disease localization and severity estimation. Extensive experiments show the model's effectiveness over existing methods.
The paper includes a literature review, methodology, experimental setup, results, comparative analysis, and concludes with implications for enhancing agricultural practices and reducing tomato leaf disease losses.
2 Literature review
The detection and management of tomato leaf diseases have been extensively researched, utilizing methods ranging from traditional visual inspection to advanced machine learning models. Historically, visual inspection by trained agronomists and farmers has been the primary method, relying on physical symptoms of plants (Nandhini and Ashokkumar, 2021; Thanammal et al., 2022; Huang et al., 2023; Moussafir et al., 2022). While simple and equipment-free, this approach is often subjective, time-consuming, and inaccurate, especially in early disease stages (Gurubelli et al., 2022; Nawaz et al., 2022; Vengaiah and Konda, 2023; Hanamasagar et al., 2021). Spectral analysis techniques have improved detection accuracy and timeliness by leveraging light properties reflected or transmitted by leaves (Lee et al., 2022; Thangaraj et al., 2021,2023; Djimeli-Tsajio et al., 2022). However, these require expensive, specialized equipment, limiting their use in resource-limited settings (Wang et al., 2021; Novak et al., 2021; Guo et al., 2023; Gadekallu et al., 2021).
Traditional machine learning models like Support Vector Machines, Decision Trees, Random Forests, and k-Nearest Neighbors have also been applied to detect diseases (Martins et al., 2021; Balafas et al., 2023; Lobin et al., 2022; Ahmed et al., 2021,2023). Despite their advancements, these models struggle with the variability and complexity of disease symptoms (Sahu et al., 2023; Sunil et al., 2023; Kaur et al., 2023; Al Hashimi et al., 2022; Hosny et al., 2023).Deep learning models, particularly CNNs like Multivariate Normal Deep Learning Neural Network (MNDLNN), have been employed to address these limitations, enhancing classification accuracy by learning intricate patterns from images (Bora et al., 2023; Gajjar et al., 2022; Vengaiah and Konda, 2023; Mondal et al., 2022; Joseph, et al., 2024). However, these models require large datasets and may struggle with generalizing to new disease variants, as well as with interpretability issues (Kurmi et al., 2021; Moupojou et al., 2023; Garg and Singh, 2023; Liu and Wang, 2020. Table 1 describes the review of existing models used to estimate the tomato leaf diseases.
Method
Findings
Limitations
Research Gaps
SVM (Aishwarya et al., 2023)
High accuracy in classifying tomato diseases.
Limited by feature extraction and training data.
Kernel selection and adaptation to large datasets.
CNN (Russel and Selvaraj, 2022)
Exceptional feature learning.
Need significant computational resources and data.
Transfer learning potential and architecture optimization.
RF (Zhang and Chen, 2023)
Robust against noisy datasets.
Struggles with imbalanced datasets and interpretability.
Parameter tuning and addressing class imbalance.
DT (Bhagat and Kumar, 2023)
Simple and interpretable.
Prone to overfitting and instability.
Ensemble methods for stability and generalization.
KNN (Uppada and Kumar, 2023)
Effective proximity-based grouping.
Performance drops with high-dimensional data.
Tailored distance metrics and handling high-dimensional data.
NB (Zhou et al., 2022)
Efficient probabilistic categorization.
Assumes feature independence.
Advanced probabilistic methods for complex relationships.
MLP (Zhou et al., 2021)
Adapts to intricate patterns.
Prone to overfitting without regularization.
Regularization techniques and hybrid models.
GAN (Roy, 2023)
Produces diseased leaf images.
Vulnerable to mode collapse.
GAN training stability and conditional GANs.
PCA (Ahmed et al., 2022)
Enables dimensionality reduction.
Loss of interpretability and sensitivity to scaling.
Advanced PCA variants for nonlinear extraction.
LDA (Özbılge et al., 2022)
Facilitates multiclass discrimination.
Assumes class homoscedasticity.
Robust LDA variants and integration with feature selection.
HMM (Shafik et al., 2023)
Captures sequential progression.
Depends on data sequence quality.
Hybrid models with deep learning for sequence learning.
SVM-RFE (Wu et al., 2022)
Highlights key features.
Computationally intensive.
Efficient feature selection and noise resilience.
CNN-LSTM (Sarma et al., 2022)
Proficient in sequential feature extraction.
Prone to vanishing gradients and overfitting.
Improved mechanisms within hybrid architecture.
DT-RF (Ashwathappa et al., 2021)
Enhanced classification accuracy.
Potential redundancy and complexity.
Pruning strategies and complexity mitigation.
SAE (Rahman et al., 2023)
Unsupervised feature learning.
Sensitive to hyperparameters.
Hyperparameter optimization and regularization techniques.
ELM (Tian et al., 2023)
Rapid training and generalization.
Limited interpretability.
Enhancing interpretability and optimizing hidden neurons.
CRF (Thangaraj et al., 2022)
Sequential pattern recognition.
Dependent on feature engineering.
Deep learning integrations and parameter calibration.
ESVM (Ashwathappa et al., 2022)
Improved categorization.
Computationally intense.
Hybrid ensembles with lightweight classifiers.
GMM (Alzahrani and Alsaade, 2023)
Probabilistic clustering.
Sensitive to initialization.
Robust initialization strategies and alternative models.
RNN (Daniya and Vigneshwari, 2023)
Excels in sequential learning.
Vanishing gradient issues.
New activation functions and transfer learning methods.
Preemptive disease detection systems use weather, soil, and historical data to predict potential disease outbreaks. However, data availability and quality can limit these models, and they may struggle with real-world variability. Validation on a dataset involving diverse conditions, tomato varieties, and disease types shows precision, accuracy, recall, and specificity values. Current methods for diagnosing and managing tomato leaf diseases have shortcomings, including accuracy, robustness, and interpretability. Post-processing methods often fail to meet the need for accurate disease detection. Some of the more advanced systems integrate IoT devices with machine learning models for continuous monitoring and disease detection. These systems, along with (Chavan and Balani, 2022; Balani et al., 2022; Vengaiah and Konda, 2024) can provide real-time alerts and recommendations for disease management, enhancing the timeliness of interventions. However, existing models can be complex to set up and maintain, particularly in large-scale farming operations.
3 Proposed model
After conducting an extensive review of existing models for tomato leaf disease identification, we discovered that their efficiency diminishes when applied to multiple datasets, and their complexity further constrains their scalability levels. To overcome these issues, this section discusses the design of a “VARMAx-CNN-GAN Integration” for early detection and management of tomato leaf diseases. As shown in Fig. 1, the proposed model integrates CNNs for feature extraction and initial disease classification with GANs for generating synthetic images, augmenting the dataset, and enhancing generalization. The GAN model augments leaf images by upsampling via bicubic sampling and iterating through the GAN model. Initially, the model estimates cross-entropy between image samples of different classes, as detailed in equation (1).,
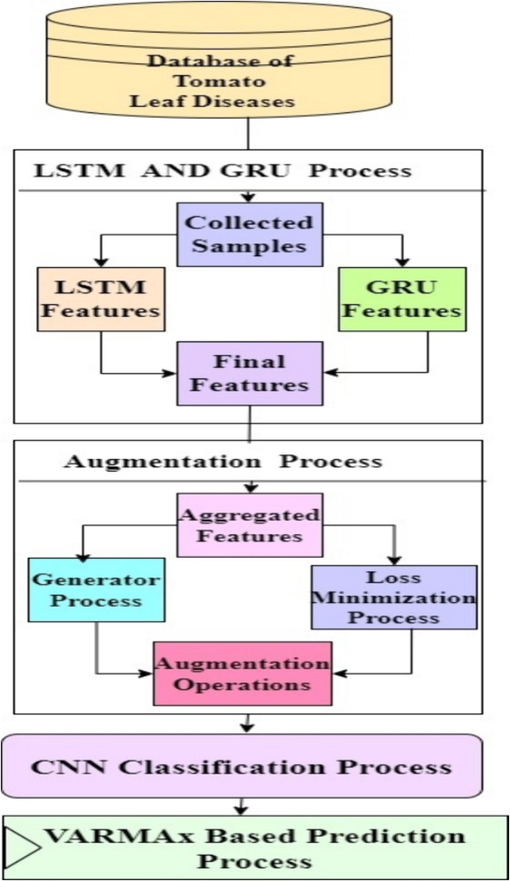
Design of the proposed model for prediction of Tomato Diseases from Leaf Image Sets.
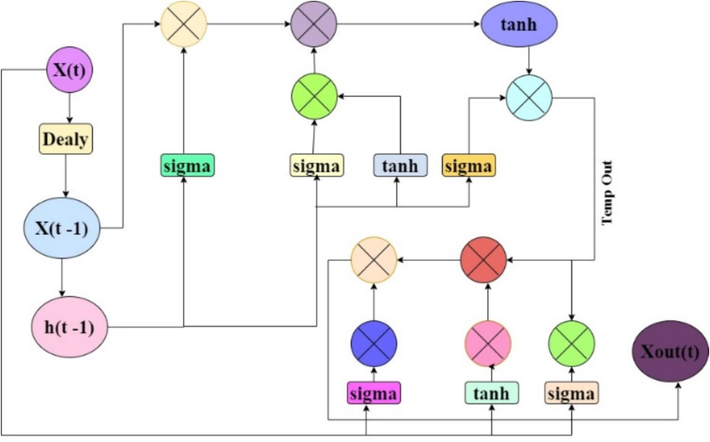
Cascade of LSTM & GRU for extraction of GAN Features.
The cascaded model estimates LSTM output features via equation (2),
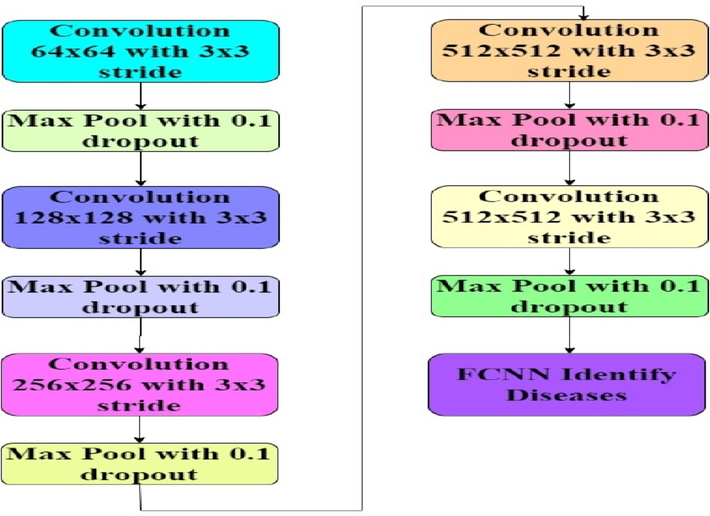
Design of the 2D CNN Process for identification of tomato leaf diseases.
The 2D CNN process initially extracts Convolutional features via equation (17),
An efficient “VARMAx-CNN-GAN Integration” model has been designed to predict tomato plant diseases from feature patterns. We leverage the strengths of LSTM and GRU models to extract high-density temporal features from surveillance data. The fused outputs of these models, capturing intricate spatio-temporal patterns, serve as inputs for the “VARMAx-CNN-GAN Integration” model, a robust time series forecasting process. This model uses the history of fused LSTM and GRU outputs to predict future values, considering interactions between variables and exogenous influences. The Vector AutoRegressive (VAR) component plays a crucial role by regressing each variable in the fused outputs against its past values to find temporal dependencies, as described in Equation (20) for variable i at time t.
Exogenous inputs, which represent external factors (including temperature, rainfall, pH levels, and soil parameters) affecting the disease behavior, are integrated into the “VARMAx-CNN-GAN Integration” framework process via equation (21),
4 Result analysis and experimentation
The proposed model combines CNN and GAN operations to improve tomato leaf disease classification and prediction. Using a dataset of 350,000 images, it was divided into training (65 %), validation (15 %), and testing (20 %) sets. Preprocessing involved resizing, data augmentation, and normalization. Models such as MXF[5], MIR[23], MNDLNN[24], and custom designs were evaluated using cross-entropy loss and the Adam optimizer, focusing on specificity, precision, and accuracy. Experiments ran on high-end GPUs with TensorFlow and PyTorch. Efficiency was enhanced by optimizing architecture, reducing parameters, and using pruning, quantization, data pipeline optimization, parallelization, and transfer learning to ensure robust performance.
4.1 Performance of the classification process
Based on this strategy, the Precision, Accuracy, Recall, and Specificity levels were estimated via equations (22), 23, 24 & 25 as follows,
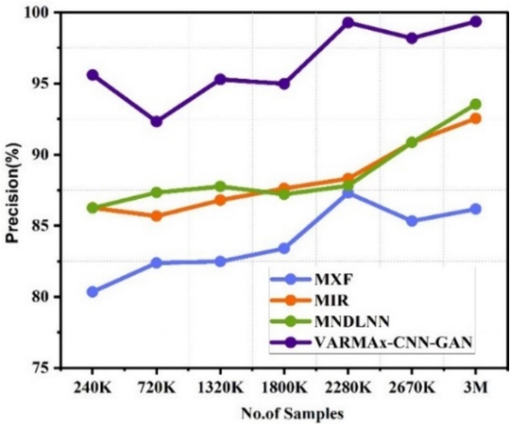
Precision observed for identification of different diseases.
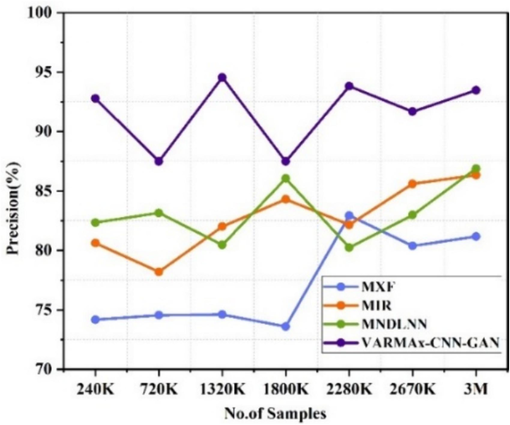
Precision for pre-emption of diseases.
The Fig. 5(a) shows the accuracy of various disease identification methods at different data sizes. MXF[5], MIR[23], and MNDLNN[24] show varying accuracy rates at different data sizes (ranging from 240 K to 3 M). VARMAx-CNN-GAN consistently outperforms these methods, with an accuracy of 97.61 % at 3 M data, and demonstrating superior accuracy across all data sizes. The Fig. 5(b) shows the accuracy observed for the pre-emption of diseases using various methods at different data sizes (ranging from 240 K to 3 M). MXF [5] shows accuracy ranging from 81.41 % to 89.79 %, with the highest accuracy of 89.79 % at 1800 K data. MIR [23] demonstrates an increasing trend in accuracy from 73.51 % at 720 K to 89.70 % at 3 M. MNDLNN [24] shows fluctuating accuracy, starting at 68.22 % at 240 K and achieving its highest accuracy of 83.23 % at 2280 K. VARMAx-CNN-GAN consistently outperforms the other methods, with accuracy values between 85.61 % and 97.61 %, reaching the highest accuracy of 97.61 % at 3 M data. Overall, VARMAx-CNN-GAN demonstrates superior accuracy for the pre-emption of diseases across all data sizes compared to the other methods.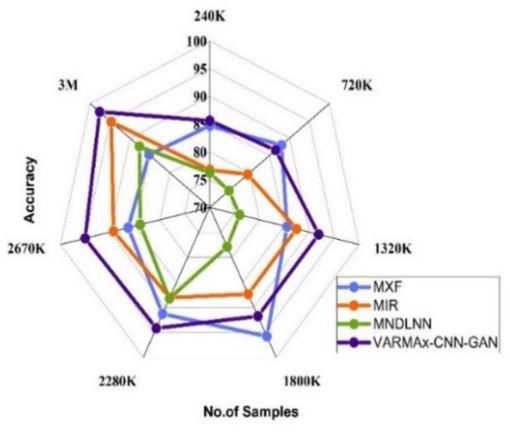
Accuracy observed for identification of different diseases.
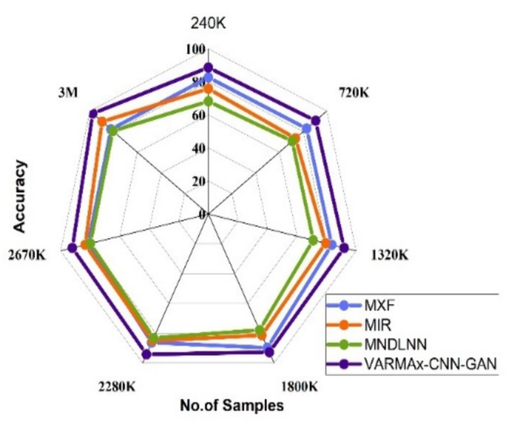
Accuracy for pre-emption of diseases.
The Fig. 6(a) shows the recall observed for the identification of different diseases using various methods at different data sizes (ranging from 240 K to 3 M). MXF [5] shows recall values ranging from 78.65 % to 86.66 %, with the highest recall of 86.66 % at 3 M data. MIR [23] demonstrates recall values ranging from 80.39 % to 89.58 %, peaking at 89.58 % at 2670 K data. MNDLNN [24] shows recall values fluctuating between 82.35 % and 92.05 %, achieving the highest recall of 92.05 % at 3 M data. VARMAx-CNN-GAN consistently outperforms the other methods, with recall values between 89.04 % and 99.55 %, reaching the highest recall of 99.55 % at 3 M data and demonstrates the highest recall for the identification of different diseases across all data sizes compared to the other methods. Similarly, the Fig. 6(b) shows the recall observed for the observed for the pre-emption of diseases using various methods at different data sizes (ranging from 240 K to 3 M). VARMAx-CNN-GAN consistently outperforms the other methods, with recall values between 89.04 % and 99.55 %, reaching the highest recall of 99.55 % at 3 M data.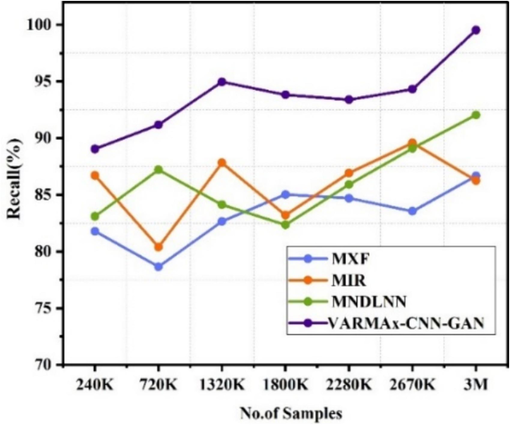
Recall observed for identification of different diseases.
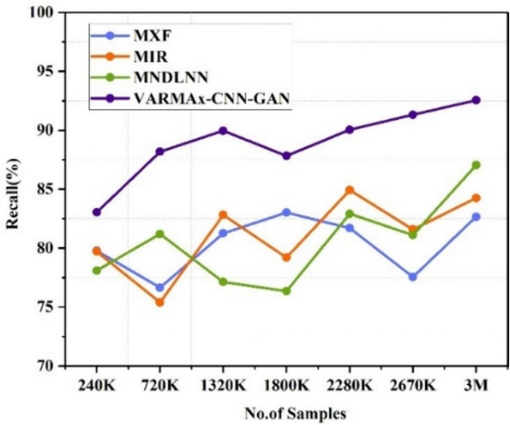
Recall for preemption of diseases.
Fig. 7(a) shows the delay in milliseconds for identifying different diseases using various methods across data sizes from 240 K to 3 M. MXF [5] delays range from 173.09 ms to 192.73 ms, peaking at 2670 K data. MIR [23] delays range from 154.27 ms to 185.66 ms, peaking at 2280 K data. MNDLNN [24] delays are more consistent, from 141.06 ms to 149.83 ms, peaking at 3 M data. VARMAx-CNN-GAN shows the lowest delays, from 127.94 ms to 142.77 ms, peaking at 3 M data, and is the fastest across all data sizes. Fig. 7(b) shows the delay for disease pre-emption, where VARMAx-CNN-GAN again shows the lowest delays, from 125.94 ms to 138.77 ms, peaking at 3 M data, consistently outperforming other methods. VARMAx-CNN-GAN also achieves the highest AUC values for disease identification (85.14 to 98.70) and pre-emption (81.09 to 92.70), peaking at 98.70 and 92.70, respectively. Comparatively, MXF [5] ranges from 75.90 to 85.73 in identification and 75.90 to 80.73 in pre-emption; MIR [23] ranges from 76.17 to 90.60 in identification and 76.17 to 84.87 in pre-emption; MNDLNN [24] ranges from 75.13 to 87.83 in identification and 75.13 to 80.15 in pre-emption.
Delay observed for identification of different diseases.
Delay for preemption of diseases.
The VARMAx-CNN-GAN consistently demonstrates high specificity in both disease identification and prevention across varying data sizes (240 K to 3 M). For disease identification, MXF [5] achieves a maximum specificity of 86.32 % at 2670 K data, MIR [23] peaks at 89.00 % at 1800 K data, and MNDLNN [24] reaches 88.84 % at 2280 K data. In disease prevention, MXF [5] peaks at 83.32 % at 2670 K data, MIR [23] at 86.13 % at 3 M data, and MNDLNN [24] at 82.61 % at 2670 K data. VARMAx-CNN-GAN outperforms with a maximum specificity of 96.44 % for disease identification and 87.44 % for disease prevention at 3 M data. Fig. 8(a) displays the specificity observed in the detections of various diseases, whereas Fig. 8(b) illustrates the specificity in the preemption of diseases.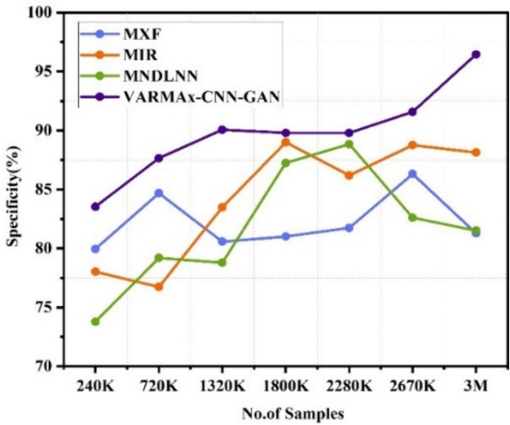
Specificity observed for identification of different diseases.
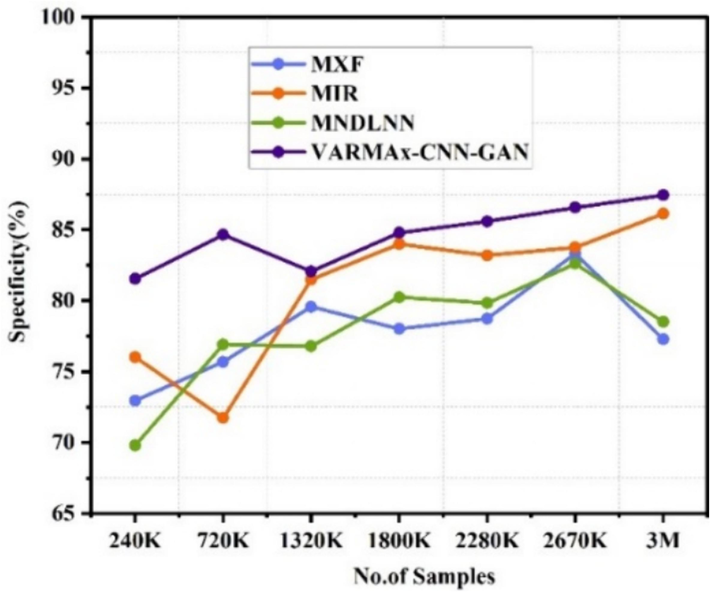
Specificity for preemption of diseases
Table 2 shows improvements across precision, recall, accuracy, and F1-score as additional techniques (GAN and VARMAx) are incorporated into the base CNN model.
Model Variant
Precision
Recall
Accuracy
F1-score
CNN
0.85
0.82
0.88
0.83
CNN + GAN
0.87
0.84
0.89
0.85
CNN + VARMAx
0.86
0.83
0.89
0.84
CNN + GAN + VARMAx
0.90
0.88
0.92
0.88
The proposed model for disease classification faces errors like false positives and false negatives due to inadequate feature representation, class imbalance, and environmental factors. To address these issues, the model may need to augment training data, improve feature representation, or incorporate advanced techniques. Strategies include data augmentation, transfer learning, and ensemble learning. Further validation and refinement are needed for real-life agricultural situations.
5 Conclusion
The approach suggested herein creates a CNN-based model for disease detection in tomato crops. Each of the three convolution and max pooling layers in the proposed CNN-based architecture contains a different number of filters. The present research undertook a thorough evaluation and comparison of four distinct classification models, assessing them across various criteria such as delay, AUC levels, and specificity. This in-depth analysis has yielded valuable insights, enriching our understanding of the effectiveness and efficiency of each model in performing classification tasks. The proposed work consistently showcased superior performance across a spectrum of metrics. Whether examining delay values, where it excelled in minimizing processing time, AUC levels, where it demonstrated impressive discrimination ability, or specificity levels, where it displayed remarkable proficiency in accurately identifying negative instances, the proposed method consistently outperformed the existing models, including MXF, MIR, and MNDLNN. This performance underscores the strength and effectiveness of the proposed approach in comparison to its counterparts. The methodology adopted in the proposed work, has proven effective in attaining optimized results. The comparative analysis reflects a holistic approach where various aspects of the classification process were taken into consideration, allowing for a multifaceted understanding of the models' behaviors. The improved classification performance demonstrated by the proposed method may pave the way for new opportunities in applications where accurate, efficient, and dependable classification is crucial. This encompasses domains such as medical diagnosis, and disease detection. The enhanced model performance offers the potential to significantly benefit real-world scenarios where precise and timely classification plays a pivotal role.
CRediT authorship contribution statement
Vengaiah Cheemaladinne: Methodology. Srinivasa Reddy K.: Supervision.
Acknowledgement
We acknowledge VIT-AP University for its support in providing the necessary resources to carry out the proposed work.
References
- First report of Tomato yellow leaf curl virus and associated Tomato leaf curl betasatellite infecting eggplant (Solanum melongena) and in Pakistan. Australasian. Plant. Dis. Notes. 2023;18:15.
- [CrossRef] [Google Scholar]
- Less is More: Lighter and Faster Deep Neural Architecture for Tomato Leaf Disease Classification. IEEE. Access. 2022;10:68868-68884.
- [CrossRef] [Google Scholar]
- Tomato leaf curl Oman virus and associated Betasatellite causing leaf curl disease in tomato in Pakistan. Eur. J. Plant. Pathol.. 2021;160:249-257.
- [CrossRef] [Google Scholar]
- Smart farming for detection and identification of tomato plant diseases using light weight deep neural network. Multimed. Tools. Appl.. 2023;82:18799-18810.
- [CrossRef] [Google Scholar]
- Al Hashimi, A.H., Al Aamir, A.F., Janke, R. et al. Effect of tomato yellow leaf curl disease on yield, height and chlorophyll of open field grown tomato genotypes in Oman. Vegetos 35, 269–275(2022).doi: 10.1007/s42535-021-00285-z.
- Transform and deep learning algorithms for the early detection and recognition of tomato leaf disease. Agronomy. 2023;13(5):1184.
- [CrossRef] [Google Scholar]
- Association of Tomato leaf curl Karnataka virus and satellites with enation leaf curl disease of Pseuderanthemum reticulatum (Radlk.) a new ornamental host for begomovirus infecting tomato in India. Indian. Phytopathol.. 2021;74:1065-1073.
- [CrossRef] [Google Scholar]
- Fenugreek plants showing the severe leaf curl disease are associated with tomato leaf curl Kerala virus, DNA-B molecule of tomato leaf curl New Delhi virus and a novel betasatellite. Australasian. Plant. Dis. Notes. 2022;17:24.
- [CrossRef] [Google Scholar]
- Machine learning and deep learning for plant disease classification and detection. IEEE. Access 2023
- [Google Scholar]
- Nisha Balani, Pallavi Chavan, and Mangesh Ghonghe. 2022. Design of high-speed blockchain-based sidechaining peer to peer communication protocol over 5G networks. Multimedia Tools Appl. 81, 25 (Oct 2022), 36699–36713. doi: 10.1007/s11042-021-11604-6.
- Efficient feature selection using BoWs and SURF method for leaf disease identification. Multimed. Tools. Appl.. 2023;82:28187-28211.
- [CrossRef] [Google Scholar]
- A detection of tomato plant diseases using deep learning MNDLNN classifier. SIViP 2023
- [CrossRef] [Google Scholar]
- Chavan, P.V., Balani, N. (2022). Design of heuristic model to improve block-chain-based sidechain configuration. In International Journal of Computational Science and Engineering (Vol. 1, Issue 1, p. 1). Inderscience Publishers. doi: 10.1504/ijcse.2022.10050704.
- Daniya, T., Vigneshwari, S. (2023). Rider Water Wave-enabled deep learning for disease detection in rice plant. In Advances in Engineering Software (Vol. 182, p. 103472). Elsevier BV. doi: 10.1016/j.advengsoft.2023.103472.
- Improved detection and identification approach in tomato leaf disease using transformation and combination of transfer learning features. J. Plant. Dis. Prot.. 2022;129:665-674.
- [CrossRef] [Google Scholar]
- A novel PCA–whale optimization-based deep neural network model for classification of tomato plant diseases using GPU. J. Real-Time. Image. Proc. 2021;18:1383-1396.
- [CrossRef] [Google Scholar]
- Real-time detection and identification of plant leaf diseases using convolutional neural networks on an embedded platform. Vis. Comput. 2022;38:2923-2938.
- [CrossRef] [Google Scholar]
- An aggregated loss function based lightweight few shot model for plant leaf disease classification. Multimed. Tools. Appl. 2023;82:23797-23815.
- [CrossRef] [Google Scholar]
- Antiviral effect of the cotton plant-derived gossypol against tomato yellow leaf curl virus. J. Pest. Sci. 2023;96:635-646.
- [CrossRef] [Google Scholar]
- Colour texture descriptor for CBIR of diseased tomato leaf images using modified local zigzag pattern. Multimed. Tools. Appl 2022
- [CrossRef] [Google Scholar]
- Characterization of Tomato leaf curl Palampur virus associated with leaf curl and yellowing disease of watermelon from India. Ind. Phytopathol.. 2021;74:1075-1088.
- [CrossRef] [Google Scholar]
- Multi-class classification of plant leaf diseases using feature fusion of deep convolutional neural network and local binary pattern. IEEE. Access. 2023;11:62307-62317.
- [CrossRef] [Google Scholar]
- Tomato Leaf Disease Detection System Based on FC-SNDPN. Multimed. Tools. Appl. 2023;82:2121-2144.
- [CrossRef] [Google Scholar]
- Real-time plant disease dataset development and detection of plant disease using deep learning. IEEE. Access 2024
- [Google Scholar]
- Performance analysis of segmentation models to detect leaf diseases in tomato plant. Multimed. Tools. Appl 2023
- [CrossRef] [Google Scholar]
- A novel transfer deep learning method for detection and classification of plant leaf disease. J. Ambient. Intell. Human. Comput. 2023;14:12407-12424.
- [CrossRef] [Google Scholar]
- Leaf image analysis-based crop diseases classification. SIViP. 2021;15:589-597.
- [CrossRef] [Google Scholar]
- First report of tomato spotted wilt virus infecting parlor palm (Chamaedorea elegans) with leaf mosaic and ring spot disease in Korea. J. Plant. Pathol. 2022;104:415.
- [CrossRef] [Google Scholar]
- Early recognition of tomato gray leaf spot disease based on MobileNetv2-YOLOv3 model. Plant. Methods. 2020;16:1-16.
- [Google Scholar]
- A meta-analysis of climatic conditions and whitefly Bemisia tabaci population: implications for tomato yellow leaf curl disease. JoBAZ. 2022;83:57.
- [CrossRef] [Google Scholar]
- Nanopore sequencing of tomato mottle leaf distortion virus, a new bipartite begomovirus infecting tomato in Brazil. Arch. Virol. 2021;166:3217-3220.
- [CrossRef] [Google Scholar]
- Deep learning-based approach to detect and classify signs of crop leaf diseases and pest damage. SN. Comput. Sci.. 2022;3:433.
- [CrossRef] [Google Scholar]
- FieldPlant: A dataset of field plant images for plant disease detection and classification with deep learning. IEEE. Access. 2023;11:35398-35410.
- [Google Scholar]
- Design of efficient techniques for tomato leaf disease detection using genetic algorithm-based and deep neural networks. Plant. Soil. 2022;479:251-266.
- [CrossRef] [Google Scholar]
- Improved crossover based monarch butterfly optimization for tomato leaf disease classification using convolutional neural network. Multimed. Tools. Appl. 2021;80:18583-18610.
- [CrossRef] [Google Scholar]
- A robust deep learning approach for tomato plant leaf disease localization and classification. Sci. Rep. 2022;12:18568.
- [CrossRef] [Google Scholar]
- Characterization of tomato leaf mould pathogen, Passalora fulva. Croatia. J. Plant. Dis. Prot. 2021;128:1041-1049.
- [CrossRef] [Google Scholar]
- Tomato disease recognition using a compact convolutional neural network. IEEE. Access. 2022;10:77213-77224.
- [CrossRef] [Google Scholar]
- Image processing based system for the detection, identification and treatment of tomato leaf diseases. Multimed. Tools. Appl. 2023;82:9431-9445.
- [CrossRef] [Google Scholar]
- Detection of tomato leaf diseases for agro-based industries using novel PCA DeepNet. IEEE. Access. 2023;11:14983-15001.
- [CrossRef] [Google Scholar]
- Leaf species and disease classification using multiscale parallel deep CNN architecture. Neural. Comput. Applic. 2022;34:19217-19237.
- [CrossRef] [Google Scholar]
- Classification of crop leaf diseases using image to image translation with deep-dream. Multimed. Tools. Appl 2023
- [CrossRef] [Google Scholar]
- Learning aided system for agriculture monitoring designed using image processing and IoT-CNN. IEEE. Access. 2022;10:41525-41536.
- [CrossRef] [Google Scholar]
- A Systematic Literature Review on Plant Disease Detection: Motivations, Classification Techniques, Datasets, Challenges, and Future Trends. IEEE. Access. 2023;11:59174-59203.
- [CrossRef] [Google Scholar]
- Systematic study on deep learning-based plant disease detection or classification. Artif. Intell. Rev 2023
- [CrossRef] [Google Scholar]
- Crossover-based wind-driven optimized convolutiosnal neural network model for tomato leaf disease classification. J. Plant. Dis. Prot. 2022;129:559-578.
- [CrossRef] [Google Scholar]
- Automated tomato leaf disease classification using transfer learning-based deep convolution neural network. J. Plant. Dis. Prot. 2021;128:73-86.
- [CrossRef] [Google Scholar]
- Artificial intelligence in tomato leaf disease detection: a comprehensive review and discussion. J. Plant. Dis. Prot. 2022;129:469-488.
- [CrossRef] [Google Scholar]
- A deep convolution neural network model based on feature concatenation approach for classification of tomato leaf disease. Multimed. Tools. Appl 2023
- [CrossRef] [Google Scholar]
- Identification of Tomato Leaf Diseases based on a Deep Neuro-fuzzy Network. J. Inst. Eng. India. Ser. A. 2022;103:695-706.
- [CrossRef] [Google Scholar]
- Computer-aided fusion-based neural network in application to categorize tomato plants. SIViP 2023
- [CrossRef] [Google Scholar]
- A review on tomato leaf disease detection using deep learning approaches. Int. J. Recent Innovat Trends Comput. Commun.. 2023;11(9s):647-664.
- [CrossRef] [Google Scholar]
- Improving tomato leaf disease detection with DenseNet-121 architecture. Int. J. Intellig. Syst. Appl. Eng.. 2023;11(3):442-448.
- [Google Scholar]
- A comparative study of convolutional neural network architectures for enhanced tomato leaf disease classification using refined statistical features. Traitement. Du. Signal. 2024;41(1)
- [Google Scholar]
- Early real-time detection algorithm of tomato diseases and pests in the natural environment. Plant. Methods. 2021;17:43.
- [CrossRef] [Google Scholar]
- Plant Leaf Diseases Fine-Grained Categorization Using Convolutional Neural Networks. IEEE. Access. 2022;10:41087-41096.
- [CrossRef] [Google Scholar]
- An IoT-enabled energy-efficient approach for the detection of leaf curl disease in tomato crops. Wireless. Netw. 2023;29:321-329.
- [CrossRef] [Google Scholar]
- Early warning and diagnostic visualization of Sclerotinia infected tomato based on hyperspectral imaging. Sci. Rep. 2022;12:21140.
- [CrossRef] [Google Scholar]
- Tomato leaf disease identification by restructured deep residual dense network. IEEE. Access. 2021;9:28822-28831.
- [CrossRef] [Google Scholar]







