Translate this page into:
Design, synthesis, in silico and antibacterial evaluation of curcumin derivatives loaded nanofiber as potential wound healing agents
⁎Corresponding authors. sasikala@ucsiuniversity.edu.my (Sasikala Chinnappan), shaik@ucsiuniversity.edu.my (Shaik Ibrahim Khalivulla)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objective
Drug resistance can result in mortality and morbidity due to treatment failures. Therefore, researchers are actively involved in the urgent development of new antibacterial agents which are more effective against drug-resistant bacteria. This study is aimed to prepare curcumin derivatives (CD) nanofiber and test its antibacterial activity by disk diffusion test and evaluate their role in wound healing when potentially binds with transforming growth factor-beta type 1 kinase domain (TGF-β) and Glycogen synthase kinase-3 beta (GSK3-β).
Method
Molecular docking and molecular dynamic have been used to select the most potent dual-action molecules on both TGF-β and GSK3-β, most active 2 compounds have been synthesized and characterized by 1H and 13C (NMR), IR spectroscopy, Electrospinning of CD in ethanol were studied by adding Poly vinyl alcohol (PVA) as a polymer. The properties of the CD\PVA solutions measured, morphology of electrospun CD\PVA was tested by Scanning Electrode Microscope (SEM), antibacterial activity of electrospun nanofiber against Staphylococcus aureus (S. aureus) was tested by disk diffusion method.
Results and Conclusions: CD nanofiber were synthesized successfully. The results showed that CD were successfully synthesized with good yield (72% and 78%) for compound1 and 2 respectively, Disc diffusion antibacterial test demonstrated an increased inhibition zone from 0.5 to 5.0 mg loaded CD with the largest inhibition zone of 8.5 ± 0.71 and 9.67 ± 0.29 mm for compound1 and 2 respectively. Hence, it can be concluded that these CD\PVA could be a new source in designing new antibacterial wound dressing for diabetic patients.
Keywords
Antibacterial
Curcumin derivatives
Nanofiber
Electrospinning
Molecular dynamic
1 Introduction
Several reports point out the serious impact of foot ulcer on life, they stated that the mortality rate from chronic wounds in the last 5 years are more than the breast and prostate cancer rates (Armstrong et al., 2020). Among the most important factors that impaired the wound healing in a diabetic person is; suppression of growth factor production, angiogenesis (Martin and Nunan, 2015). In the wound healing process, the interactions of different cells, proteins from extracellular matrix (ECM), as well as their receptors are entailed throughout these organic significant processes, which are moderated by cytokines as well as growth factors. Amongst the various cytokines that involved in injury healing, transforming growth factor beta type 1 kinase domain (TbetaR-I) and Glycogen synthase kinase-3 beta (GSK3-β) possesses the widest sphere of impacts (Cheon et al., 2006; Guo et al., 2017; Kim et al., 2021). TbetaR-I protein play numerous roles in the body, its function depends on the cell type and the level of differentiation the target cell. However, its play a critical role in every step during wound healing process. Glycogen synthase kinase-3 (GSK-3) has also been implicated in wound healing process, it’s a multifunctional serine-threonine protein kinase which found in all mammal cells that responsible for cell metabolism regulation, fate of cell, cellular division, as well as cell death (Doble and Woodgett, 2003).
Curcumin is one of the potent natural products that has important roles in wound healing which exerts its anti-inflammatory, antioxidant activities throughout the different stages of the wound healing procedure (Alabdali, 2022) and may promote the formation of collagen, fibroblasts differentiation and migration as well as epithelialization and for apoptotic processes that throw the inflamed tissues coming from the injured site (Alabdali et al., 2021; Ibrahim et al., 2018; Panchatcharam, Miriyala, Gayathri, and Suguna, 2006). Curcumin derivatives showed antibacterial activities towards drug-sensitive microbes (Staphylococcus aureus, Salmonella enterica, Enterococcus faecalis, and Escherichia coli). Researchers also mentioned that these derivatives showed the ability in preventing bacteria to develop into drug-induced resistance (Gunes et al., 2016).
Nanofibers were invented for delivering antibacterial agents. Studies have shown that it is beneficial in fields such as wound dressing (Rasouli and Barhoum, 2018). Nanofibers loaded with antibacterial agents are useful in the field of packaging, textiles, water purification, and filtration (Alabdali, 2022; Song, Wu, Qi, and Kärki, 2017). Natural polymer or synthetic polymer can be used to produce nanofibers which will give different properties, thus, nanofibers that are made by natural polymers usually have better biocompatibility and are less likely to cause immunogenic reactions, however, synthetic polymers allow researchers to make modifications in synthesizing nanofibers as they give better flexibility (Stojanov and Berlec, 2020).
Since drug design is very difficult process because many parameters should be reliable at once such as drug safety, potency, bioavailability, selectivity, clearance. Today molecular docking software is being used widely among the drug discovery community because it is considered an effective method in finding the potentially valuable and best hits drugs in a time and cost-effective approach, by enhance screening efficiency of drug development (Eweas, Maghrabi, and Namarneh, 2014). Accordingly, molecular docking has numerous applications in drug development and discovery, such as structure–activity of the lead compounds, virtual screening for potential lead identification, hypothesized ligand-target interaction, assist in the prediction of the ligand-binding site, improvement of lead compounds, and combinatorial chemistry study, from this view, molecular docking and molecular dynamics make the synthesize of new chemicals reliable, because it gives an idea about the protein–ligand interactions and shows the selectivity and binding affinity between the compound and its target (Aminpour, Montemagno, and Tuszynski, 2019), however these finding must follow by the in vivo and in vitro tests to be confirmed. The present paper studies the biological activities of more than 275 curcumin derivatives that have potential activities against wound healing process through the GSK-3β and TbetaR-I signaling pathways and their mechanism in wound recovery as well as decrease scarring. These activities have been tested by using structure-based drug design followed by synthesis and in vitro study of the most active compounds.
2 Materials and methods
2.1 Molecular modeling
2.1.1 Ligand’s preparation
In order to study the molecular docking for the ligands; the new CD have been prepared before, first the 3D structures of curcumin and CD have been drawn by Chemdraw16.0 software, then geometrical optimization of molecules was done by carrying MM + field force method by using HyperChem8.0 software for all ligands and then saved as mol. format file, Recife Model 1(RM) calculations have been performed to obtain extra structural optimization for the selected derivatives, these result have been saved as a separate file (sdf format) in order to carry out the final optimization step by Spartan 14.0 software. This molecular docking study has been performed by using Schrodinger (Maestro 11.1, licensce (INCREMENT BIOLUMINATE_PROTEIN_DOCKING 999 HOSTID = 14jan7a6ck4d)) software Glide docking tool, Maestro version 11.5. have been used in the grid generation for the CD, the grid box set at 1.00 Å with an atomic charge of 0.25 (Noha et al., 2015).
2.1.2 Protein structure preparation
TbetaR-I (PDB code: 6B8Y) and GSK-3β (PDB code: 5K5N) structures have been obtained from protein data bank (PDB) (Studio, 2009), out of 88 crystal structure of GSK-3β and 627 crystal structure of TbetaR-I available, one structure was chosen for each protein to perform molecular docking study with the selected ligand and resolution of 2.20 Å and 1.65 Å, respectively (Dallakyan and Olson, 2015). These protein structure files have been prepared before being used by protein preparation wizard in Schrodinger software, to eliminate any water molecule, metals ions or even co-crystal ligand from protein structure (Morris and Lim-Wilby, 2008).
2.2 Chemistry
2.2.1 Chemicals and reagents
Chemicals and reagents were purchased from Sigma Aldrich, Shimadzu FT-IR 8400S infrared spectrophotometer was used to determine the infrared spectra by KBr pellet technique, 1H and 13C NMR spectra for the two products ((E)-3-(2,3-dimethoxyphenyl)-1-(5-methylfuran-2-yl)prop-2-en-1-one and (E)-3-(2,5-dimethoxyphenyl)-1-(5-methylfuran-2-yl)prop-2-en-1-one) were analyzed by a Bruker DPX-400 and DPX-600 FT-NMR spectrometer (Bruker BioSpin AG, Switzerland) internal standard was tetramethylsilane and methanol as a solvent. Regarding the splitting pattern; s for singlet; d for doublet; m for multiplet.
2.2.2 Synthesis of CD
Synthesis of CD was illustrated in Scheme1. A solution of 2.33 mL of 5-methylfuran-2-carbaldehyde 1 was dissolved in ethanol 20 mL then added to a different aldehyde 2, in 20 mL HCl with 12 h stirring at room temperature. Then, this mixture reaction was transferred into a 100 mL of distilled water then left for the next day. The precipitate was collected after one day then further purified by recrystallization from ethanol (Baell and Holloway, 2010).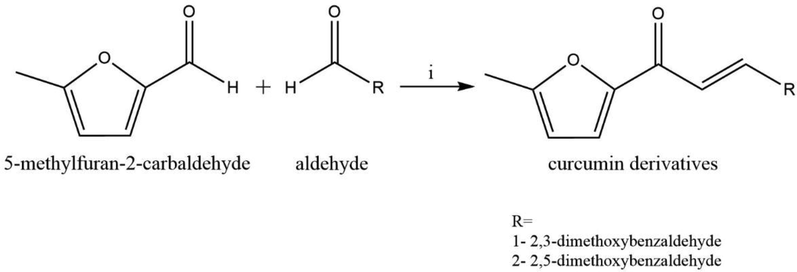
Synthesis of compound 1& 2, i = 40%NaOH, ethanol 6.
2.3 Synthesis of nanofiber
10 mg of CD (5% drug) obtained from the synthesis was dissolved in ethanol and combined with 5 mL of 4% PVA solution. The CD\PVA solutions were loaded into a syringe fitted with a 21-G needle and fitted at the syringe pump. The baking paper-coated collector was rotated by drum controller and maintained around 150 revolutions per minute (rpm). The electrode on the needle was fixed. Electric field (approximately 19.4 kV) was applied by using a high voltage power source. Tyndall effect of the solution was seen constantly. The distance between the needle and the baking paper coated- collector was 10 cm. The extrusion rate was 2 mL/hr, and the target was 5 mL.
2.3.1 Characterization of nanofiber by scanning electron microscope (SEM)
The surface morphology and average diameter distribution of 5 wt% CD-loaded electrospun PVA nanofiber was characterized by using field emission scanning electron microscopy (of nanofiber under SEM (Manufacturer: Hitachi, Model: SU 8030).
2.4 Antibacterial testing of CD\PVA
Antibacterial activity of CD\PVA against S. aureus were evaluated using agar diffusion method by determining the inhibition zone. Three small pieces of the nanofiber with areas of 1 cm2 have been removed and placed on the Muller Hinton Agar medium carefully and incubated at 37 °C for 24 h. The inhibition zone were measured in centimetres (cm) by using a ruler. The Polyvinyl alcohol only nanofiber is used as the control in this test. Calculate the average for the three zones of inhibition.
3 Results and discussion
3.1 Molecular docking and virtual screening
Since several papers reported the activity of CD against wound healing activity(Abbas et al., 2019; Kant et al., 2015; Tejada et al., 2016), thus, aiming to discover and synthesis new derivatives that possess powerful and significant wound healing activity for patients with diabetic wound ulcer, molecular scaffold of CD was drawn by using Chemdraw16.0 software. A list of 146 available structures that related to this scaffold were obtained from ChEMBL database (https://www.ebi.ac.uk/chembl); which is a large scale open bioactive database that contains more than 2,105,464 distinct molecules(Davies et al., 2015). Those chosen molecules have been exposed for virtual screening with molecular docking by Schrodinger (Maestro 11.1) software, a series of promising CD with highest binding affinity are listed in Table 1.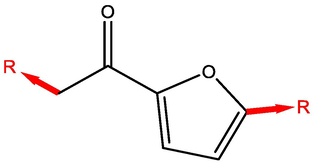
Cpd
R1
R2
Binding Energy with 5K5N (Kcal/mol)
Binding Energy with 6B8Y (Kcal/mol)
1

−8.27
−7.18
2
CH3
−8.22
−6.5
3
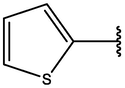
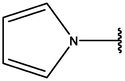
−8.17
−7.32
4
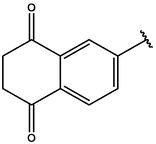
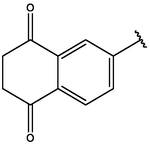
−7.83
−7.42
5
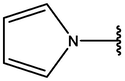
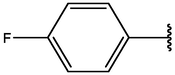
−7.43
−6.54
6


−7.23
−6.35
7
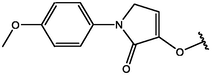
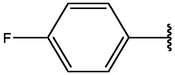
−7.20
−5.31
8
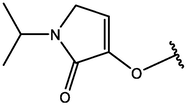
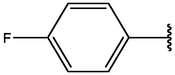
−6.30
−6.72
9
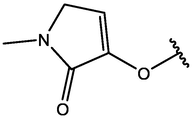

−6.21
−5.44
10
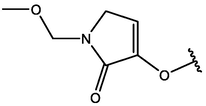
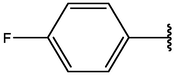
−5.04
−6.80
Molecular modeling with QSAR study applied from Table 1 result to specify the impact of efficient substitution related with increasing of binding affinity. (Fig. 1) shows the highest binding compounds in 3D structure surrounded by colored contours regrading desirable, undesirable, positive and negative charges.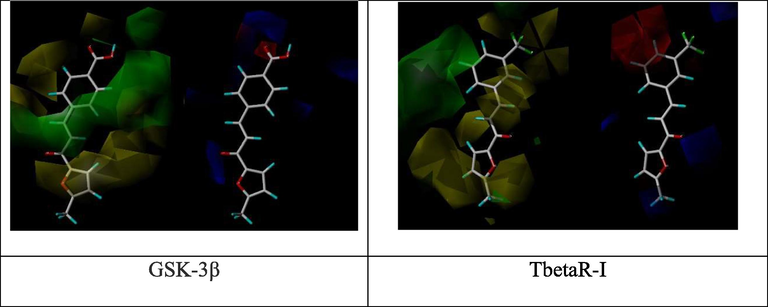
Molecular modeling and QSAR study of curcumin derivatives 8 based on their wound healing docking scores.
To improve the activity of the listed compounds that showed promising biological activity toward GSK-3β and TbetaR-I proteins, several chemical group replacements have been done on the basic scaffold at R1. Interestingly, by changing and adding different chemical groups on curcumin scaffold, the affinity ranking scores have been increased (Table 2), as well as the affinity between ligand and proteins, (Fig. 2) showed the most active compounds.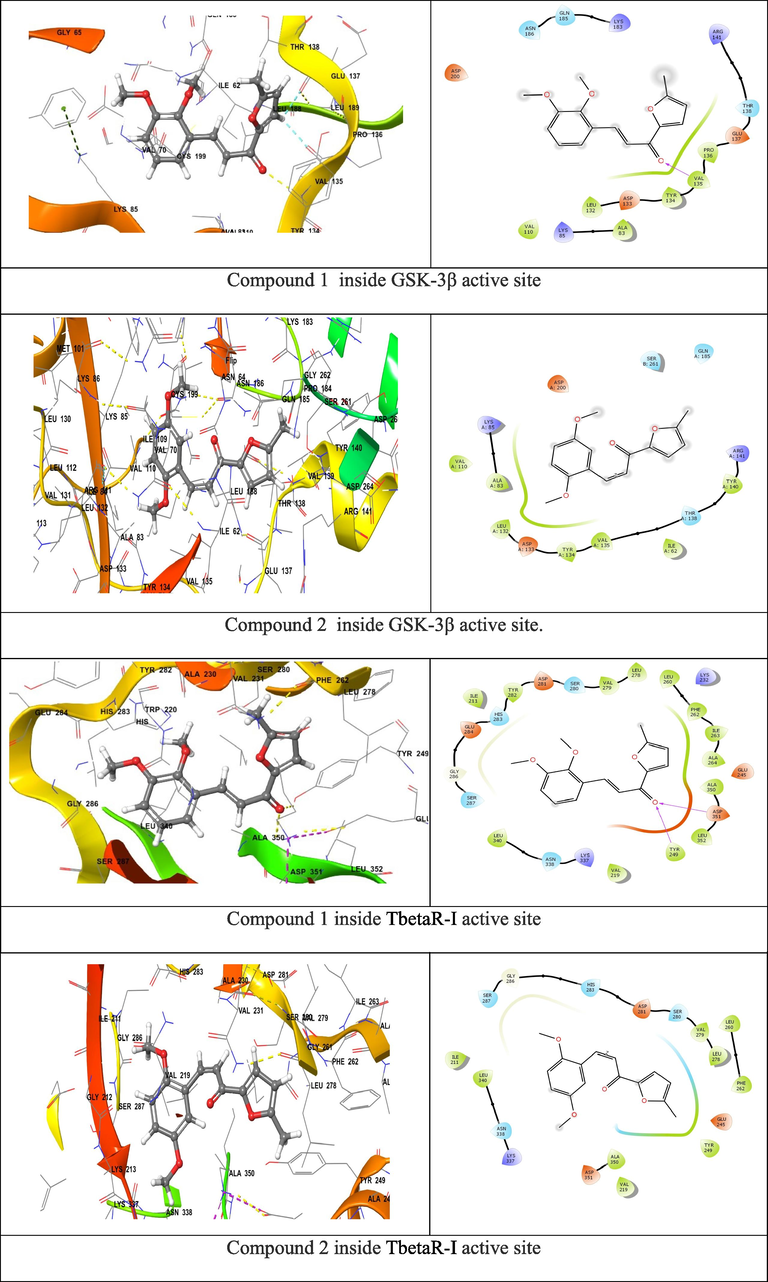
Compounds 1&2 binding surface and ligand interaction diagram 9.
Table 2 shows compounds 1&2, it represents a very high binding affiant inside both proteins. Because of this ability, docking analysis and molecular dynamic studies have been applied to evaluate the stability of these two compounds inside active site of each protein. Molecular docking study have been used to predict the biological activities of the new CD and the development of topical wound healing medicine, locate the best binding position of these derivatives in GSK-3β and TbetaR-I enzymes, find stable binding conformation of these ligand-receptor, and determine the residues in the protein binding region. Comparing with curcumin binding affinity inside selected active sites (−7.99 with GSK-3β and −7.62 with TbetaR-I). More details of surrounding amino acids and binding interactions are listed in Table 3.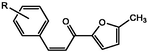
Cpd
CD structure
Binding Energy with 5K5N (Kcal/mol)
Binding Energy with 6B8Y (Kcal/mol)
1

−10.641
−11.06
2
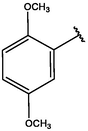
−10.68
−11.27
2
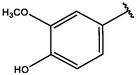
−10.138
−10.21
4
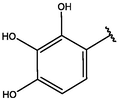
−10.136
−10.019
5
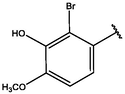
−9.95
−9.75
6
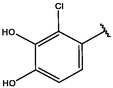
−9.935
−9.95
7
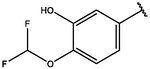
−9.75
−7.52
8
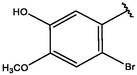
−9.12
−8.32
9
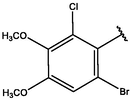
−8.75
−7.63
10
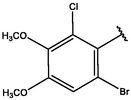
−8.28
−8.76
Curcumin
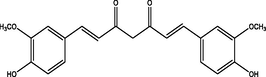
−7.99
−7.62
Cpd No.
Interacting residues inside GSK-3β
H-bonding with amino acids inside GSK-3β
Interacting residues inside TbetaR-I
H-bonding with amino acids inside TbetaR-I
01
Arg141, Thr138, Glu137, Pro136, Val135, Tyr134, Asp133, Leu132, Ala83, Lys85, Val110, Asp200, Asn186, Gln185, Lys183
Val135
Ser287, Gly286, Glu284, His283, Tyr282, Asp281, Ser280, Val279, Leu278, Leu260, Phe262, Ile263, Ala264, Ala350, Asp351, Leu352, Tyr249, Val219, Lys337, Asn338, Leu340.
Tyr249, Asp351
02
Thr138, Glu137, Val135, Tyr134, Asp133, Leu132, Val110, Ala83, Lys85, Cys199, Asp200, Asn186, Gln185, Lys183
Val135
Ser287, Glt286, His283, Tyr282, Asp281, Ser280, Val279, Leu278, Ala230, Val231, Lys232, Glu2245, Tyr249, Leu352, Asp351, Ala350, Val219, Ile211
Asp351, Tyr249
Binding affinity between ligand and receptor calculated with a scoring function which is a good indicator for the binding strength and stability, high negative value indicates strong binding energy between investigated derivatives and the receptors. Docking results with TbetaR-I protein, compounds 1,2 showed the highest docking score at binding site of protein and record high wound healing activity. Those compounds display good docking scores (−11.06, −11.27) combined with the active site of TbetaR-I protein by forming hydrogen bonds with Tyr249, Asp351 residue with oxygen (Figs. 1 and 2). Each compound has different nonbonding interacting that stabilize the coordination bond. The highest affinity molecule binds by hydrogen bonds inside TGF-β receptor type 1 kinase domain enzyme active site, this H-bond interact with Tyr249, Asp351 with hydroxyl chemical groups, it also surrounded by non-bonded amino acid that illustrated in Table 3.
Regarding molecular dynamic study, both compounds 1&2 were applied for 5 nano second simulation time to evaluate the stability inside active site. (Fig. 3) shows the RMSD and interaction percent of compounds 1&2 inside active site of GSK-3β & TbetaR-I.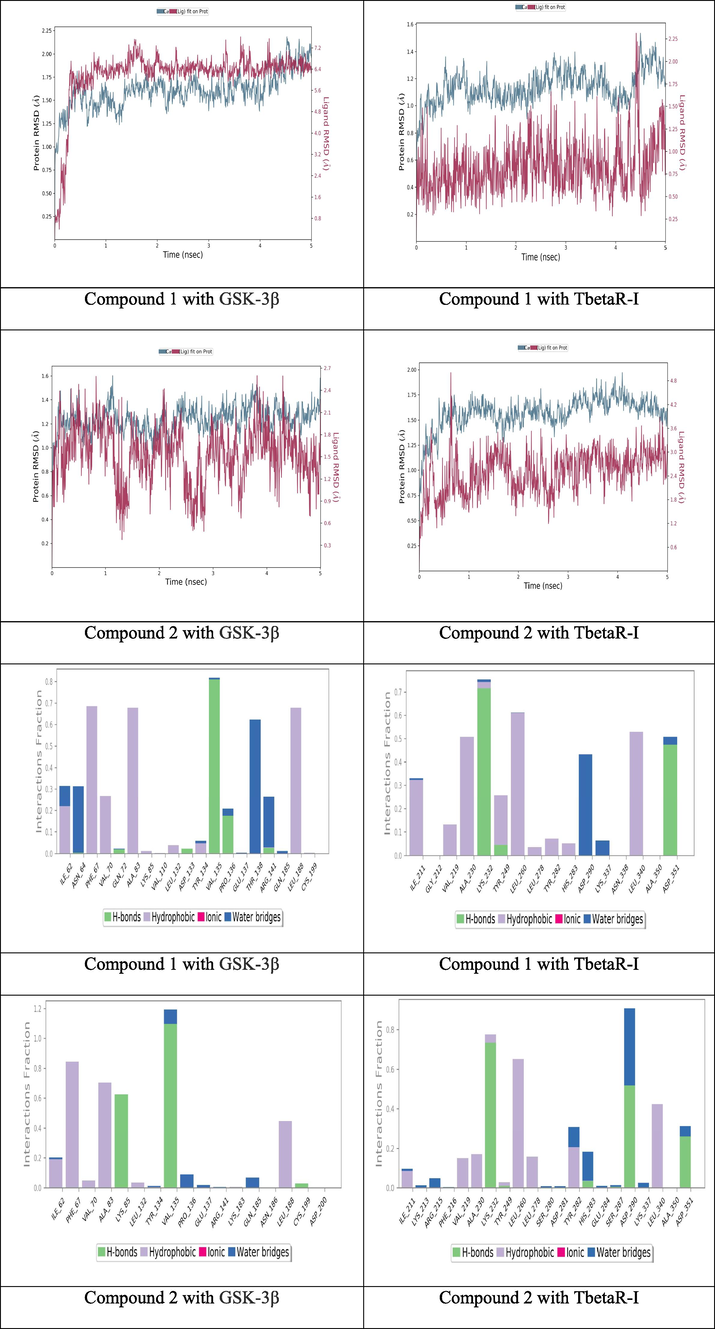
RMSD of compounds 1&2 and the interaction percent of compounds 9 1&2 with surrounding amino acids.
The observation of RMSD reveal the equilibrium of fluctuations around the selected molecular structure till the end of the simulation duration. During simulation time, the receptor variations were excellent and well below the permitted stated ranges. It has been observed that modifications of 1&2 are deemed totally acceptable. However, if the changes are greater than this amount, the protein is undergoing a significant conformation change during the simulation time. For compounds 1&2, they show a very good fluctuations ranges with protein till the end of simulation time. At the same time, (Fig. 3) shows the interaction percent of compounds 1&2 with surrounding amino acids. Because of the unique structure of the suggested new molecules, surrounding amino acids inside the active site pocket were kept bound to both compound throughout simulation with various ratios of percentage contribution of each amino acid, supporting the stability of ligand binding inside the active site with effective interactions. Inside GSK-3β active site, the H-bonds interaction of VAL135 with both compounds is significantly stabile during more than 80% of simulation time. The same interaction is appeared inside TbetaR-I active site for both compounds with ASP351. After computational study and with all promising result, these two compounds have been selected for synthesis.
3.2 Chemistry
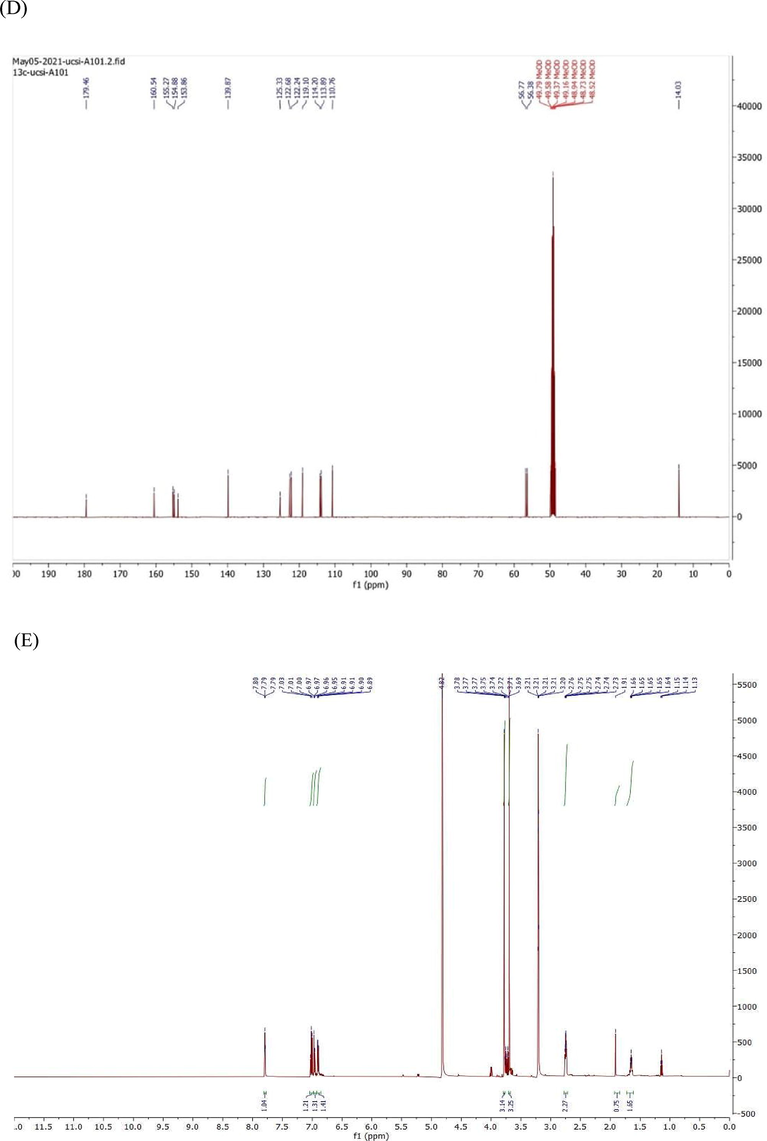
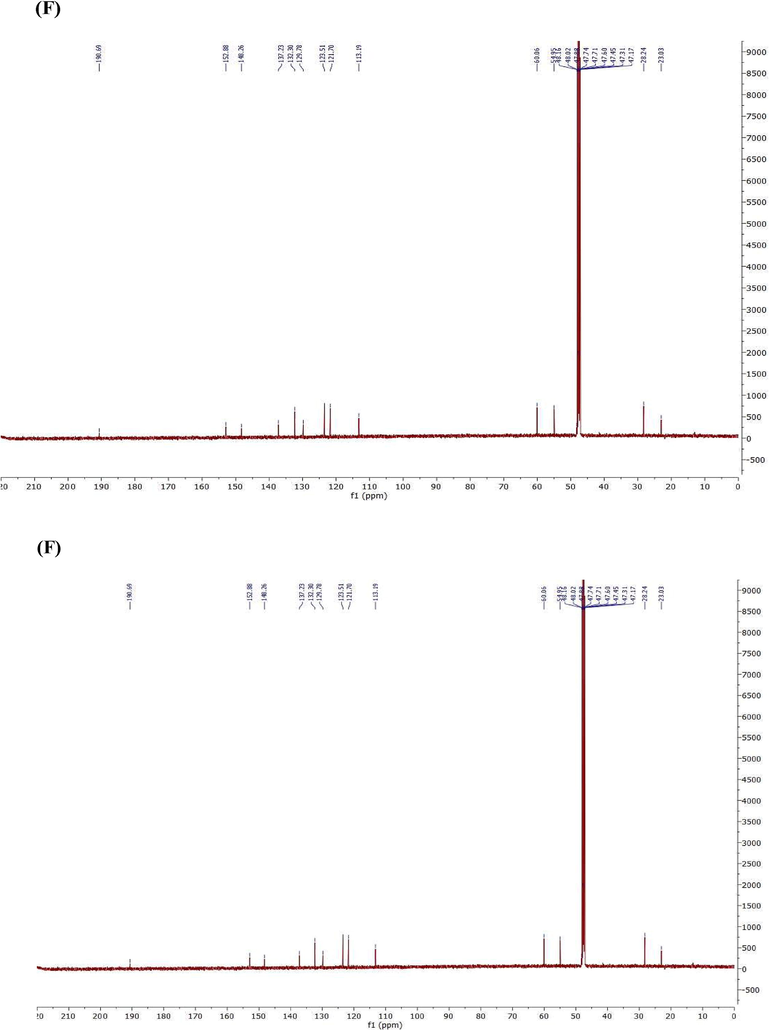
Two CD were synthesized (Scheme 1) and confirmed by using Fourier-transform infrared spectroscopy (FTIR) and 1H and 13C-nuclear magnetic resonance (NMR) spectroscopy. FTIR spectroscopy was used to characterize the specific functional groups present in the pure products. The FTIR spectrum of the synthesized CD, was demonstrated in (Fig. 4A,B).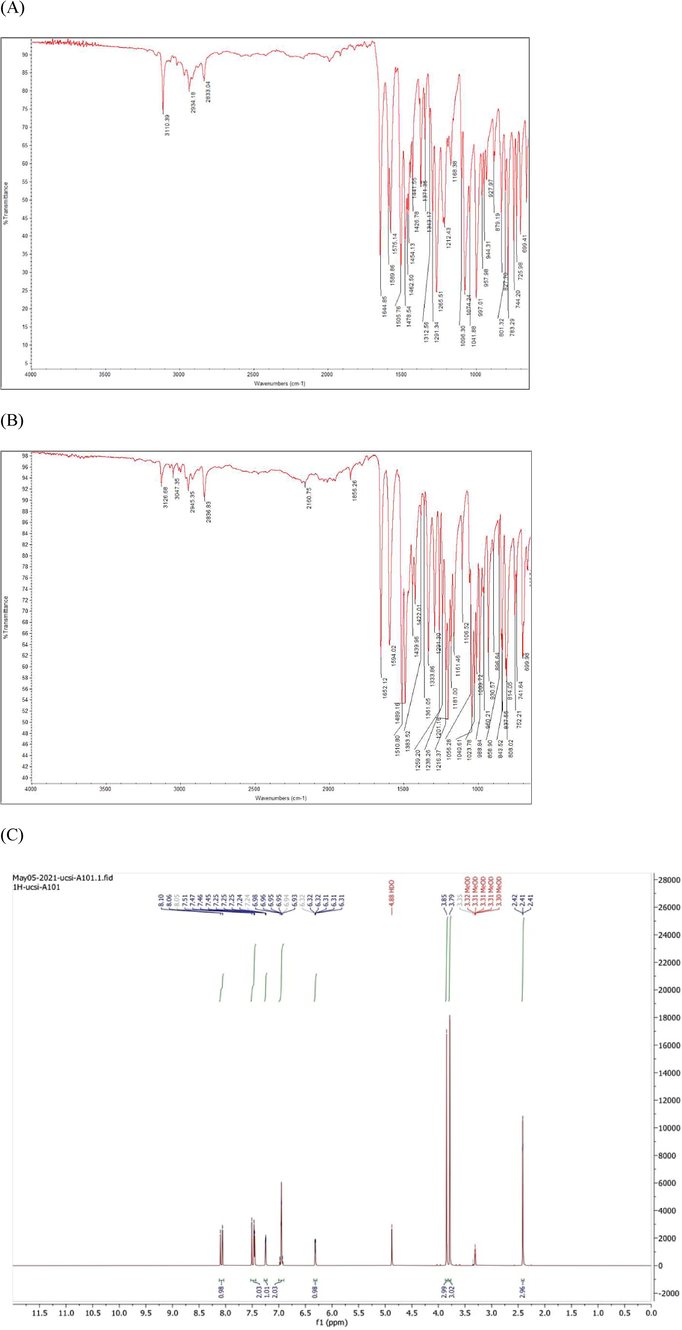
FTIR & NMR spectrum (A) FTIR of compound 1, 11 (B) FTIR of compound 2 (C) 1H NMR of compound 1 (D) 13C NMR of compound 1 (E) 1H NMR of compound 2 (F) 13C NMR of compound 1 in Chloroform-d.
FTIR & NMR spectrum (A) FTIR of compound 1, 11 (B) FTIR of compound 2 (C) 1H NMR of compound 1 (D) 13C NMR of compound 1 (E) 1H NMR of compound 2 (F) 13C NMR of compound 1 in Chloroform-d.
FTIR & NMR spectrum (A) FTIR of compound 1, 11 (B) FTIR of compound 2 (C) 1H NMR of compound 1 (D) 13C NMR of compound 1 (E) 1H NMR of compound 2 (F) 13C NMR of compound 1 in Chloroform-d.
Fig. 4C–F show 1H NMR and 13C-nuclear magnetic resonance (NMR) spectroscopy of both compound 1&2. Compound1: 3-(2,3-dimethoxyphenyl)-1-(5-methylfuran-2-yl) prop-2-en-1-one (2j); Yield: 76%; yellow crystals; IR(KBr) mmax:2934 (C–H stretch), 1651 (C=O), 1596 (C=C), 1512 (C=C), 1267 (C–O aromatic), 1075, 999, 1H NMR (500 MHz, Chloroform-d) δ 7.93–7.86 (m, 1H), 7.28 (d, J = 17.6 Hz, 1H), 7.24–7.16 (m, 2H), 7.16–7.08 (m, 2H), 6.38 (dq, J = 5.3, 0.9 Hz, 1H), 3.86 (s, 6H), 2.38 (s, 1H). 13C NMR (125 MHz, Chloroform-d) δ 181.64, 158.26, 153.38, 151.35, 145.87, 139.11, 130.39, 129.73, 125.97, 123.30, 119.09, 114.07, 109.78, 61.01, 55.95, 14.18.
Compound 2: 3-(2,5-dimethoxyphenyl)-1-(5-methylfuran-2-yl)prop-2-en-1-one, Yield: 84%; yellow powder, IR(KBr) mmax:2934 (C–H stretch), 1644 (C=O), 1596 (C=C), 1513 (C=C), 1268 (C–O aromatic), 1073, 999, 1H NMR (500 MHz, Chloroform-d) δ 7.49 (d, J = 12.1 Hz, 1H), 7.18 (d, J = 5.1 Hz, 1H), 7.14–7.06 (m, 2H), 6.89 (d, J = 2.3 Hz, 1H), 6.78–6.73 (m, 1H), 6.38 (dq, J = 5.3, 0.9 Hz, 1H), 3.84 (s, 2H), 3.79 (s, 2H). 13C NMR (125 MHz, Chloroform-d) δ 181.80, 158.26, 153.54, 152.66, 151.35, 139.06, 129.45, 124.29, 119.09, 115.99, 114.77, 112.54, 109.78, 55.82, 55.71, 14.18.
3.3 Antibacterial activity of curcumin derivative-loaded nanofiber
Evaluation of the antibacterial activity proved that 5 wt% CD loaded electrospun nanofiber mat for both derivatives exhibited antibacterial efficiency against S. aureus in our study. A clear inhibition zone around the sample nanofibers was observed and the average diameter from the triplicate test was measured to be 10.00 ± 1.10 and 10.67 ± 1.53 mm for compound1 and 2 respectively (Fig. 5). Pure PVA nanofiber was acted as control with no inhibition zone observed after incubation for 24 h. However, the results were thought to be an underestimation due to the handling of nanofiber pieces during the test. The nanofiber was easily sticking to each other when removed from the non-sticking baking paper and applied onto the agar surface. Therefore, one of the nanofiber sample pieces was shrank dimensionally when placed onto the agar surface and affecting the total area of the fiber mat in contact with the S. aureus inoculated agar surface. Consequently, the inhibition zone diameter was also be affected and considered to be decreased in the measurement.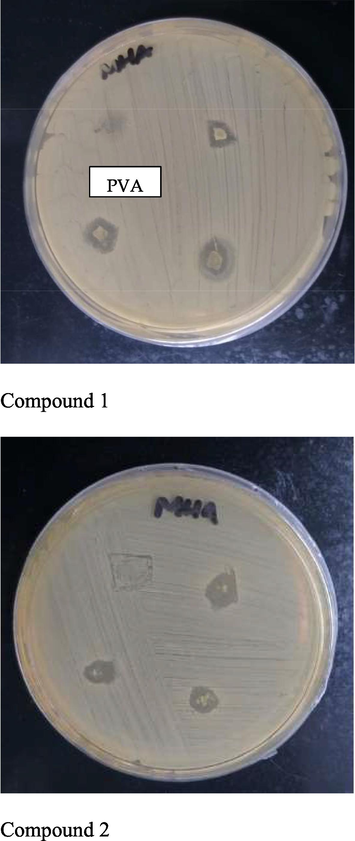
Antibacterial testing with pure PVA nanofiber as control 12 and 5 wt% CD loaded nanofiber in triplicate.
As antibacterial activity was proved with the observation of clear circular growth of inhibition zone, thereby we proposed that: (1) CD was successfully loaded into the PVA polymeric matrix; (2) electrospinning process did not affect the chemical composition of the loaded drug and retained its chemical structures and individual properties as an antibacterial agent; (3) the drug was able to diffuse out from the PVA polymeric matrix onto the agar plate and showed its antibacterial activity against S. aureus.
3.3.1 Characterization of nanofiber under SEM
Characterization of nanofiber has been done by SEM (Fig. 6). The results indicated that the nanofiber possessed a fine, smooth and porous cross-sectional structure. The fiber was free from beads with a uniform diameter distribution observed in the structure. In addition, no CD crystals were observed on the nanofiber surface which indicate it was completely incorporated into the PVA polymeric matrix which mean CD has been completely incorporated into the fibers. A random sample of 10 nanofibers was selected from the SEM image of the fiber mat at two different locations and measured using SEM image analysis software.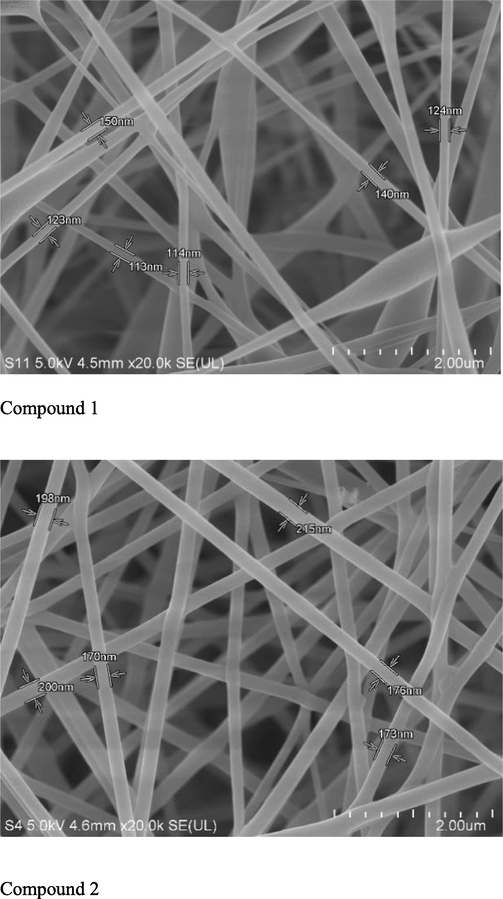
Morphology of electrospun curcumin derivative-loaded PVA 12 nanofiber compound 1 &2 (Magnification: 20 000 x).
4 Conclusion
CD (E)-3-(2,3-dimethoxyphenyl)-1-(5-methylfuran-2-yl)prop-2-en-1-one and (E)-3-(2,5-dimethoxyphenyl)-1-(5-methylfuran-2-yl)prop-2-en-1-one) have been successfully synthesized and showed that these ligands are well settled inside the protein cavity. Also, showed a very good fluctuations ranges with protein till the end of simulation time. Curcumin derivatives 5 wt% nanofiber have been proved to show antibacterial activity against Staphylococcus aureus by disc diffusion method. Hence, it can be concluded that these compounds nanofiber could be a new source of antibacterial agent against S. aureus, and treatment for wound healing treatment.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Wound healing potential of curcumin cross-linked chitosan/polyvinyl alcohol. Int. J. Biol. Macromol.. 2019;140:871-876.
- [Google Scholar]
- APPLICATION OF NANOANTIBIOTICS APPROACH AGAINST ANTI-BACTERIAL RESISTANCE. International Journal of Applied Pharmaceutics 2022
- [CrossRef] [Google Scholar]
- Research Journal of Pharmacy and Technology 2022
- [CrossRef]
- Impact of covid-19 on multiple body organ failure: a review. Indian J. Forensic Med. Toxicol. 2021:835-844.
- [Google Scholar]
- An overview of molecular modeling for drug discovery with specific illustrative examples of applications. Molecules. 2019;24(9):1693.
- [Google Scholar]
- Five year mortality and direct costs of care for people with diabetic foot complications are comparable to cancer. J. Foot Ankle Res.. 2020;13(1):1-4.
- [Google Scholar]
- New substructure filters for removal of pan assay interference compounds (PAINS) from screening libraries and for their exclusion in bioassays. J. Med. Chem.. 2010;53(7):2719-2740.
- [Google Scholar]
- Beta-catenin regulates wound size and mediates the effect of TGF-beta in cutaneous healing. FASEB J.. 2006;20(6):692-701.
- [Google Scholar]
- Dallakyan, S., Olson, A. J. (2015). Small-molecule library screening by docking with PyRx. In Chemical Biology (pp. 243-250): Springer.
- ChEMBL web services: streamlining access to drug discovery data and utilities. Nucleic Acids Res.. 2015;43(W1):W612-W620.
- [Google Scholar]
- GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci.. 2003;116(7):1175-1186.
- [Google Scholar]
- Advances in molecular modeling and docking as a tool for modern drug discovery. Der Pharma Chem.. 2014;6(6):211-228.
- [Google Scholar]
- Antibacterial effects of curcumin: an in vitro minimum inhibitory concentration study. Toxicol. Ind. Health. 2016;32(2):246-250.
- [Google Scholar]
- Entanglement of GSK-3β, β-catenin and TGF-β1 signaling network to regulate myocardial fibrosis. J. Mol. Cell. Cardiol.. 2017;110:109-120.
- [Google Scholar]
- Wound healing properties of selected natural products. Int. J. Environ. Res. Public Health. 2018;15(11):2360.
- [Google Scholar]
- Curcumin-induced angiogenesis hastens wound healing in diabetic rats. J. Surg. Res.. 2015;193(2):978-988.
- [Google Scholar]
- Adenosine and cordycepin accelerate tissue remodeling process through adenosine receptor mediated Wnt/β-catenin pathway stimulation by regulating GSK3b activity. Int. J. Mol. Sci.. 2021;22(11):5571.
- [Google Scholar]
- Cellular and molecular mechanisms of repair in acute and chronic wound healing. Br. J. Dermatol.. 2015;173(2):370-378.
- [Google Scholar]
- Molecular docking. In: Molecular Modeling of Proteins. Springer; 2008. p. :365-382.
- [Google Scholar]
- Discovery of novel, non-acidic mPGES-1 inhibitors by virtual screening with a multistep protocol. Bioorg. Med. Chem.. 2015;23(15):4839-4845.
- [Google Scholar]
- Curcumin improves wound healing by modulating collagen and decreasing reactive oxygen species. Mol. Cell. Biochem.. 2006;290(1):87-96.
- [Google Scholar]
- Advances in nanofibers for antimicrobial drug delivery. In: Barhoum A., Bechelany M., Makhlouf A., eds. Handbook of Nanofibers. Cham: Springer International Publishing; 2018. p. :1-42.
- [Google Scholar]
- Electrospun nanofibers with antimicrobial properties. Electrospun Nanofibers 2017:551-569.
- [Google Scholar]
- Electrospun nanofibers as carriers of microorganisms, stem cells, proteins, and nucleic acids in therapeutic and other applications. Front. Bioeng. Biotechnol.. 2020;8:130.
- [Google Scholar]
- Studio, D. (2009). Version 2.5.5; Accelrys, Inc. San Diego, CA, USA, 26.
- Wound healing effects of curcumin: A short review. Curr. Pharm. Biotechnol.. 2016;17(11):1002-1007.
- [Google Scholar]







