Exploring potential anti-chikungunya virus activity of phytocompounds: Computational docking and in vitro studies
⁎Corresponding author at: Central Research Laboratory, Meenakshi Academy of Higher Education and Research (Deemed to be University), Chennai, India. n_arunagiri@yahoo.co.in (Narasingam Arunagirinathan) acadeicofficer@maher.ac.in (Narasingam Arunagirinathan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
Objective
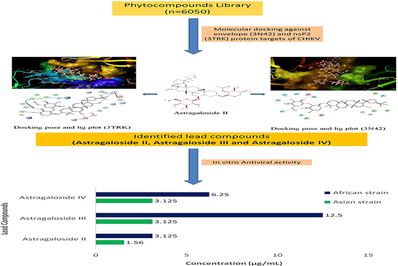
This study aimed to identify phytocompounds that possess anti-chikungunya virus activity using computational docking and in vitro studies.
Methods
A total of 6050 phytocompounds were retrieved from PubChem and other online databases. Compounds were virtually screened against E (3N42) and nsP2 (3TRK) proteins of CHIKV using iGEMDOCK. Molecular docking was performed for the screened compounds using the Maestro Glide module. Finally, screened lead compounds were studied for their in vitro antiviral activity against the Asian and African strains of CHIKV.
Results
Among the three lead phytocompounds screened, astragaloside II showed the highest binding energy value of −10.603 kcal/mol, followed by astragaloside IV (-9.007 kcal/mol) and astragaloside III (-6.197 kcal/mol) against the 3TRK target protein whereas astragaloside II showed the highest binding energy value of −10.603 kcal/mol, followed by astragaloside IV (-10.548 kcal/mol) and astragaloside III (-9.539 kcal/mol) against the 3N42 target protein. ADMET analysis revealed that all three lead compounds have non-mutagenic and non-carcinogenic properties. Antiviral studies showed that astragaloside II exhibited antiviral activity at 1.56 µg/mL and 3.12 µg/mL, astragaloside IV at 3.12 µg/mL and 6.25 µg/mL, and astragaloside III at 3.12 µg/mL and 12.5 µg/mL against Asian and African strains of CHIKV, respectively.
Conclusion
From the findings of this study, it is concluded that the phytocompounds such as astragaloside II, astragaloside III, and astragaloside IV possess promising in vitro antiviral activity against Asian and African strains of CHIKV.
Keywords
Chikungunya virus
Antiviral activity
Phytocompounds
Astragaloside
Molecular docking
1 Introduction
Chikungunya virus (CHIKV) is an enveloped virus belonging to the genus Alphavirus and the family Togaviridae. It contains positive-sense single-stranded RNA with a genome size of ∼11.8 kb (Mohan et al., 2010; Rougeron et al., 2015). CHIKV causes chikungunya in humans and it is transmitted by the Aedes aegypti and Aedes albopictus mosquitoes. This virus is prevalent in the tropical and sub-tropical regions (Fischer and Staples, 2014; Wahid et al., 2017) and its infection affects neonates and elderly persons, and the death rate was five times more in persons aged above 45 years (Economopoulou et al., 2009). Symptoms of CHIKV infection generally arise after an incubation time of 4–7 days, and high fever, arthralgia, myalgia, headache, and rash are the common clinical symptoms (Couderc and Lecuit, 2015). Eventhough chikungunya is a self-limiting disease, long-lasting chronic symptoms are observed in nearly 50% of patients (Brito et al., 2016; Ganesan et al., 2017). Despite many complications associated with chikungunya, still there are no approved vaccines or antiviral drugs available. Hence, chikungunya is being treated symptomatically with analgesic, antipyretic and anti-inflammatory drugs (Pialoux et al., 2007).
The CHIKV genome comprises two terminal open reading frames (ORFs). The first ORF at the 3′-terminal encodes for five structural proteins viz: capsid protein, envelop proteins (E1, E2, E3), and RNA protein (6K). The second ORF at the 5′- terminal encodes for four non-structural proteins (nsP1, nsP2, nsP3, and nsP4) (Rashad et al., 2014). The envelope glycoproteins are believed to be involved in the viral entry, and non-structural proteins are playing crucial role in viral replication and transcription (Pastorino et al., 2008). Hence, these proteins are considered potential targets of CHIKV (Keramagi and Skariyachan, 2018; Rashad et al., 2014). In the present study, a library of phytocompounds was virtually screened against the protein targets of CHIKV using computational docking, and anti-CHIKV activity of lead molecules was identified using in vitro studies.
2 Materials and Methods
2.1 Preparation of receptors
The 3D crystal structures of envelope glycoprotein (3N42) and nsP2 protease (3TRK) of CHIKV were downloaded from the protein data bank (PDB). The water molecules and sulfate ions were removed from the target proteins before performing the virtual screening.
2.2 Preparation of ligands
The phytocompounds were retrieved from different online databases such as PubChem, NPACT (Naturally Occurring Plant-based Anti-cancer Compound-Activity-Target) (Mangal et al., 2013), and KEGG (Kyoto Encyclopaedia for Gene and Genome). The structures of phytocompounds were converted into different structural file formats using the Open bable and were three-dimensionally optimized using ACD Chemsketch.
2.3 Virtual screening
The virtual screening was performed for the 6050 prepared ligand molecules obtained from different databases against the two important protein targets of CHIKV by rough docking, drug screening, and stable docking using iGEMDOCK software (Hsu et al., 2011).
2.4 Binding sites analysis
The binding sites of protein targets 3N42 (E protein) and 3TRK (nsP2 protein) of CHIKV were determined based on the high Dscore values provided by SiteMap module of Maestro 10.7 Schrödinger, LLC, New York, NY, 2016.
2.5 Molecular docking
Molecular docking was performed for the screened compounds against the binding sites of protein targets 3N42 and 3TRK using the glide module in Maestro 10.7. Schrödinger, LLC, New York, NY, 2016. This software assessed the quality of the interactions involved with the ligands and the respective receptors. The protein preparation was made using the protein preparation wizard, and ligand preparation by ligPrep using specific parameters. All the ligands were generated at a pH of 7 ± 2. Finally, the energy minimization was carried out using in-built constraint force field OPLS 2005 (Optimized Potentials for Liquid Simulations 2005). Since all the ligands are phytocompounds, the grid was generated by setting dimension of 40x40x40. The prepared ligands and receptors were further used for docking within the generated grid, and XP docking precision was used. The binding energy is represented in kcal/mol. The more negative binding energy denotes stronger interaction between ligand and protein.
2.6 ADMET analysis
The Adsorption, Distribution, Metabolism, Excretion, and Toxicity of the ligand molecules were estimated using the PreADMET and admetSAR analyses (Cheng et al., 2012; Sangeetha et al., 2017). Mutagenicity and carcinogenicity of the compounds were checked, and other properties like Blood-Brain Barrier, CaCo2 permeability, aqueous solubility rate, acute toxicity, and fish oral toxicity results were also recorded.
2.7 Cytotoxicity assay
The in vitro cytotoxicity assay for the screened compounds was performed using Vero cells (Serkedjieva and Ivancheva, 1998). Confluent monolayers of cells were trypsinized and suspended in 10% Minimum Essential Medium (MEM). 100 µL of cell suspension was then transferred into a 96 well plate followed by addition of 10% MEM, and the plates were incubated at 37 °C with 5% CO2. The compounds to be tested were serially diluted for various concentrations at 1000 µg, 500 µg, 250 µg, 125 µg, 62.5 µg, 31.25 µg, 15.62 µg, 7.81 µg, 3.9 µg, and 1.95 µg. The drug in the dilution plate was finally transferred into the 96 well plate, and 100 µL of 2% MEM was added. The morphological changes of the cells were monitored every 24 h for three consecutive days. After 72 h of monitoring, the changes were captured. The highest concentration of the phytocompound not showing cellular toxicity was considered as the Maximum Non-Toxic Concentration (MNTC).
2.8 MTT assay
After 72 h of incubation, 20 µL of MTT (3-(4, 5-dimethylthiazol-2-YL)-2, 5-diphenyltetrazolium bromide) solution (5 mg/mL) was added into all the wells, and the plate was incubated at 37 °C with 5% CO2 for 4 h. After 4 h of incubation, the medium in the plate was carefully aspirated without disturbing the cell monolayer. 120 µL of dimethyl sulfoxide (DMSO) was then added to the wells to dissolve the formazan crystals, and the reading was recorded at 510 nm and 650 nm. Each experiment was performed in triplicate (Müller et al., 2007). The cell viability percentage was calculated using the formula: cell viability = treated × 100 %/untreated.
2.9 In vitro antiviral assay
In this assay, Vero cells cultured in MEM supplemented with 10% FBS was used for antiviral studies (Gibco, NY, USA). The cells were regularly sub-cultured and maintained in 2% MEM. The standard strains of both the Asian and African Chikungunya viruses were obtained from the National Institute of Virology (NIV), Pune, India. The lyophilized stains were processed by infecting the Vero cells, and the virus in the supernatant was harvested and stored at −80 °C until use. Screened phytocompounds were purchased from Chengdu Biopurify Phytochemicals Ltd, China. As per manufacturer protocol, all the phytocompounds were dissolved in 50% ethanol and stored at −20 °C until use. It was used by adding the stock solution in 2% MEM and sterilized it with a syringe filter 0.2 µm pore size (Millipore, MA, USA). Antiviral assays of the phytocompounds and the ribavirin were carried out at the MNTC estimated. Ribavirin is a standard antiviral drug used against RNA viruses. Because of the unavailability of CHIKV specific antiviral drug, Ribavirin was used as standard drug in this study (Gallegos et al., 2016). About 100 μL of the estimated virus concentration was added to the Vero cells on 96 well plates and incubated for 1 h at 37 °C. After adsorption of the virus, the phytocompounds and Ribavirin were added and incubated at 37 °C. After 72 h, the minimum inhibitory concentrations (MICs) of the phytocompounds were estimated.
3 Results
In this study, a total of 6050 phytocompounds were virtually screened against two protein targets (3N42 and 3TRK) of CHIKV. Based on the high interaction energy value of ≥ -100 kcal/mol by iGEMDOCK, top 10 lead phytocompounds such as astragaloside II, astragaloside III, astragaloside IV, 7-methoxypraecansone B, 5-desmethylnobiletin, artobiloxanthone, butein, erythribyssins A, glycitein, and hispidulin were selected for precise docking against CHIKV protein targets using the glide module of Maestro 10.7, Schrodinger Software. Finally, three lead phytocompounds viz. astragaloside II, astragaloside III, and astragaloside IV were selected.
3.1 Interaction profile of astragaloside II, III, and IV with 3N42
While looking at the interaction profile of the three lead phytocompounds against the protein target 3N42, astragaloside II had the highest binding energy value of −10.603 kcal/mol, forming seven hydrogen bond (HB) interactions with amino acid residue positions Thr42, Pro152, Asp183, Tyr180, Trp320, and Lys149. The compound astragaloside IV had a −10.548 kcal/mol binding energy value, forming six HB interactions with amino acid residue positions Asp183, Ile261, Leu244, Tyr180, and Gly248 and the compound astragaloside III had the binding energy value of −9.539 kcal/mol, forming five HB interactions with amino acid residue positions Asp183, Gly182, Pro152, and Gly137 (Table 1 and Figs. 1-3 and 7).
| Target | Ligand | Glide G Score | Glide Energy | XP H-Bond | Hydrogen bond interacting amino acids | No. of Hydrogen bond | Other non-bounded interactions |
|---|---|---|---|---|---|---|---|
| 3TRK | Astragaloside II | −10.603 | −54.022 | −5.678 | 2 –OH group-Gln1241; OH-Tyr1079; OH-Asp1246; OH-Asn1082; Leu1205; | 6 | Lys1091, Thr1268, Glu1050, Val1051, Arg1267, Arg1271, Val1272, Val1275, Gly1245, Arg1249, Mse1242, Gly1206, Ala1046, Pro1049, Tyr1047, Trp1084, Ser1048, Mse1238, Lys1091, Thr1268, Glu1050, Val1051, Arg1267, Arg1271 |
| Astragaloside IV | −10.548 | −40.413 | −3.548 | O-Lys1091; OH-Arg1271; OH-Glu1050; OH-Val1077; OH-Tyr1079; OH-Gly1245 | 6 | Arg1249, Asp1246, Lys1279, Val1275, Leu1248, Gly1090, Trp1084, Tyr1078, Arg1267, Gln1241 | |
| Astragaloside III | −9.539 | −44.407 | −2.542 | OH-Asp1081; OH-Tyr1079; OH-Glu1050; OH-Ser1048; OH-Lys1091; 2 – OH group – Gln1241 | 7 | Val1051, Thr1268, Val1272, Arg1271, Ala1080, Gly1089, Tyr1078, Val1077, Gly1090 | |
| 3N42 | Astragaloside II | −10.726 | −73.103 | −5.025 | OH-Thr42; OH-Pro152; OH-Asp183; OH– Tyr180; OH– Trp320; OH– Lys149; O-Lys149 | 7 | Ile136, Leu244, Tyr185, Pro134, Lys245, Glu246, Lys140, Val135, Gly248, Arg247, Leu294, Arg138, Ala249, Hie253, Glu334, Gln252, Ser250, Ile261, Leu151, Lys245, Pro134, Tyr185, Ile136, Leu244, Cys153, Lys181, Asn33, Thr329, Asn332, Pro335, Gly331 |
| Astragaloside IV | −9.007 | −69.560 | −5.513 | OH-Asp183; 2 –OH group-Ile261, OH-Leu244;OH-Tyr180; OH-Gly248 | 6 | Thr42, Lys181, Pro134, Arg247, Tyr185, Glu246, Lys245, Lys140, Trp310, Leu294, Ala249, Tyr306, Val135, Glu308, Arg308, Arg138, Gly137, Gly182, Ala262, Gln252, Ser250, Hie253, Leu151, Pro152, Ile136 | |
| Astragaloside III | −6.197 | −53.818 | −3.746 | 2-OH-Asp183; OH-Gly182, OH-Pro152, OH-Gly137 | 5 | Hie253, Arg138, Ser250, Tyr185, Ala249, Leu294, Gly248, Glu308, Lys245, Lys140, Pro134,Val135, Trp310, Leu244, Arg247, Asn332, Thr329, Lys149, Gly331, Gln252, Leu151, Ile136, Tyr180, Lys181 | |
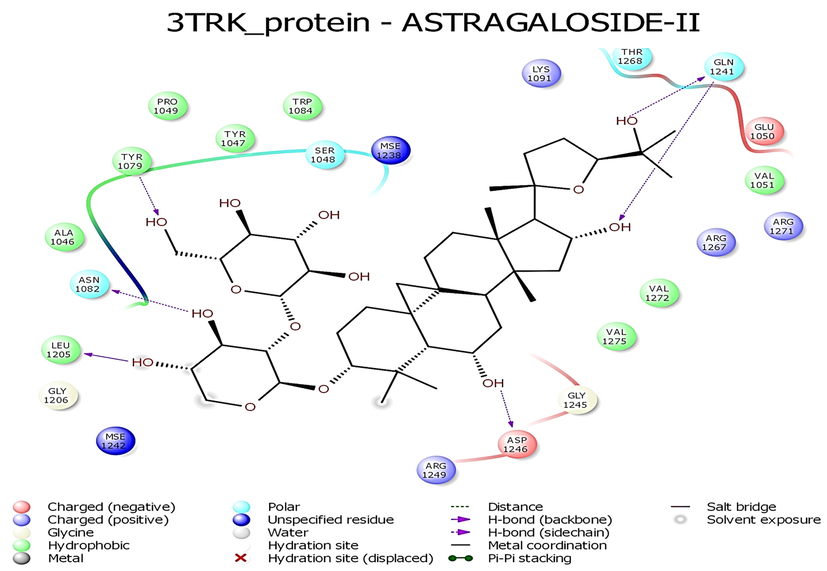
- Interaction profile of Astragaloside II with 3TRK protein of Chikungunya virus.
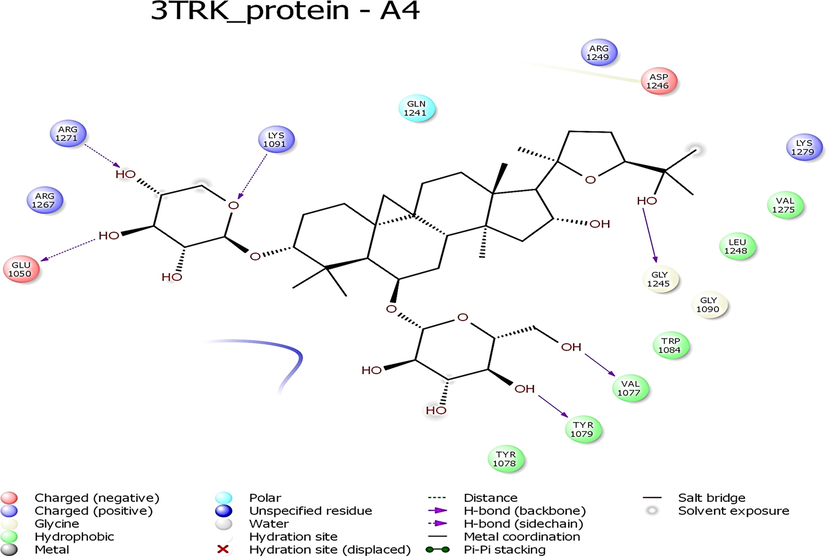
- Interaction profile of Astragaloside IV with 3TRK protein of Chikungunya virus.
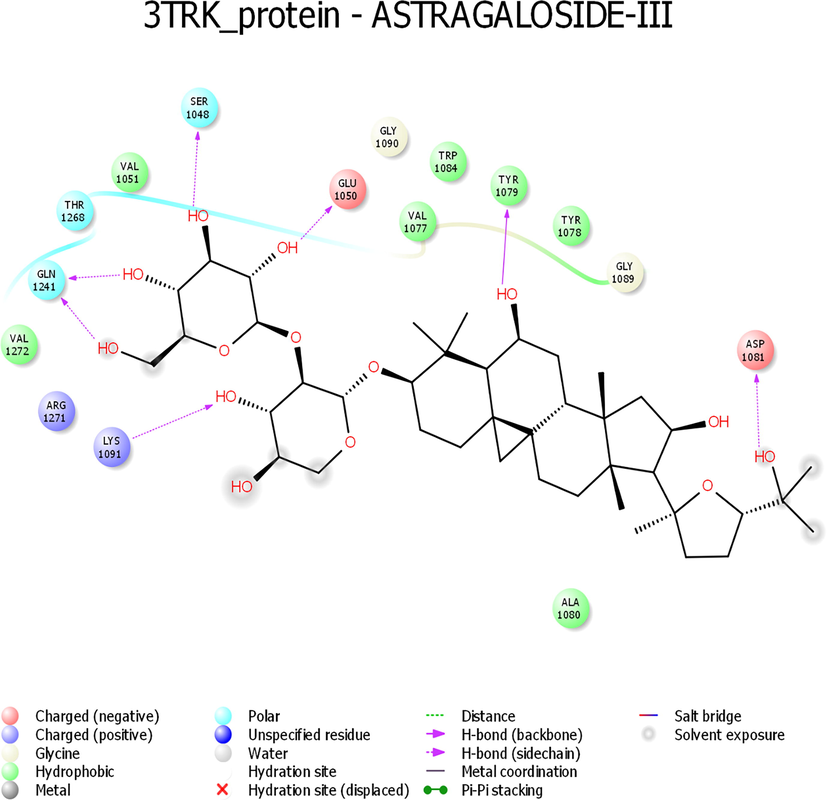
- Interaction profile of Astragaloside III with 3TRK protein of Chikungunya virus.
3.2 Interaction profile of astragaloside II, III, and IV with 3TRK
While looking at the interaction profile of the three lead phytocompounds against the protein target 3TRK, astragaloside II showed the highest binding energy of −10.726 kcal/mol, forming six HB interactions of which one interaction was from the backbone and five from the side chain amino acids. The amino acids which contributed to the HB interactions with astragaloside II were Gln1241, Tyr1079, Asp1246, Asn1082 and Leu1205. The compound astragaloside IV exhibited −9.007 kcal/mol binding energy value, forming six HB interactions. While analyzing the amino acids contributed to the HB interactions, it was noted that three interactions each were contributed by the backbone and side chain amino acids, respectively. The amino acids which contributed to the HB interactions with astragaloside IV were Lys1091, Arg1271, Glu1050, Val1077, Tyr1079, and Gly1245. The compound astragaloside III exhibited a docking score of −6.197 kcal/mol, forming seven HB interactions of which one interaction was contributed by the backbone and six by side-chain amino acids. The amino acids which contributed to the HB interactions with astragaloside III were Asp1081, Tyr1079, Glu1050, Ser1048, Lys1091, and Gln1241 (Table 1 and Figs. 4-6 and 8).
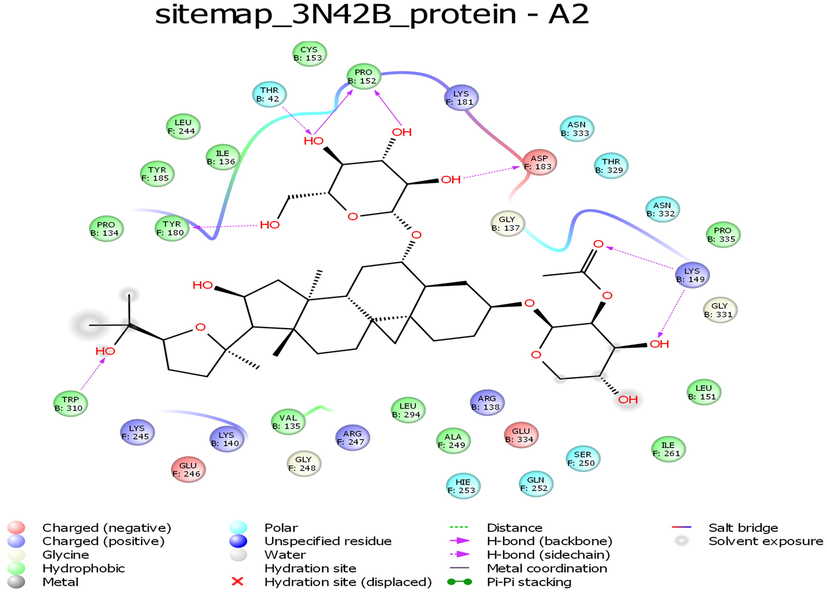
- Interaction profile of Astragaloside II with 3N42B protein target of Chikungunya virus.
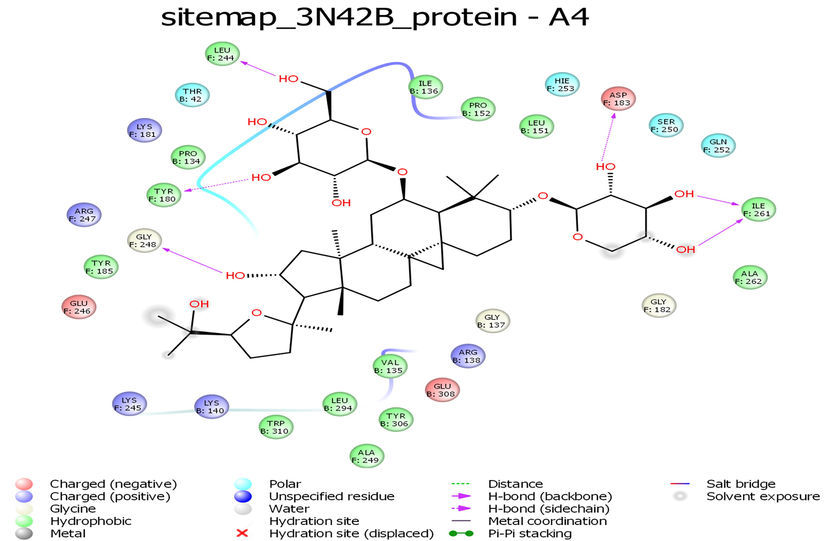
- Interaction profile of Astragaloside IV with 3N42B protein target of Chikungunya virus.
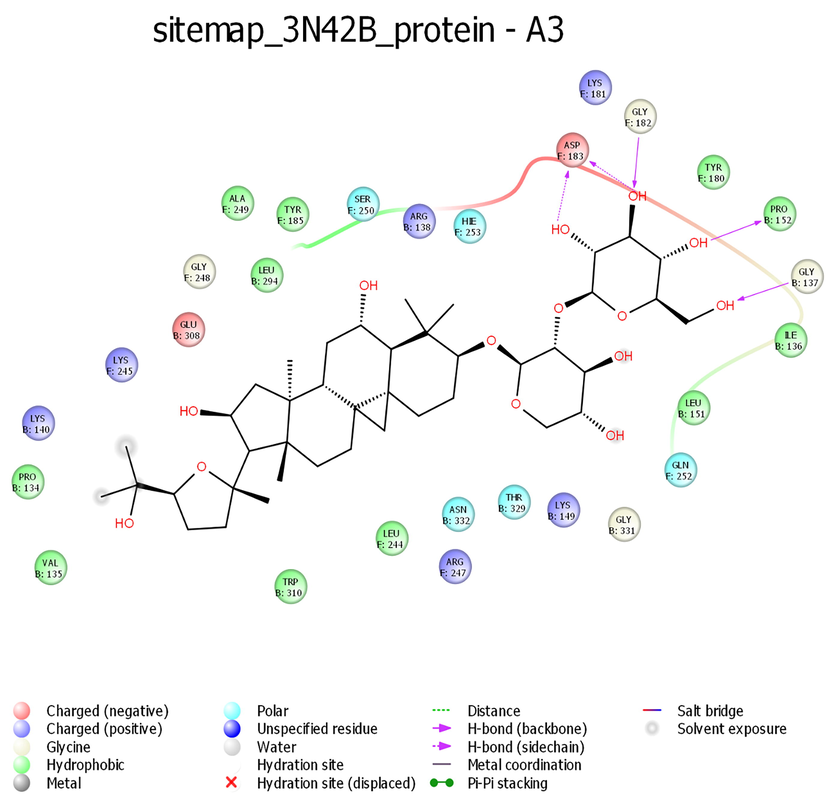
- Interaction profile of Astragaloside III with 3N42B protein target of Chikungunya virus.
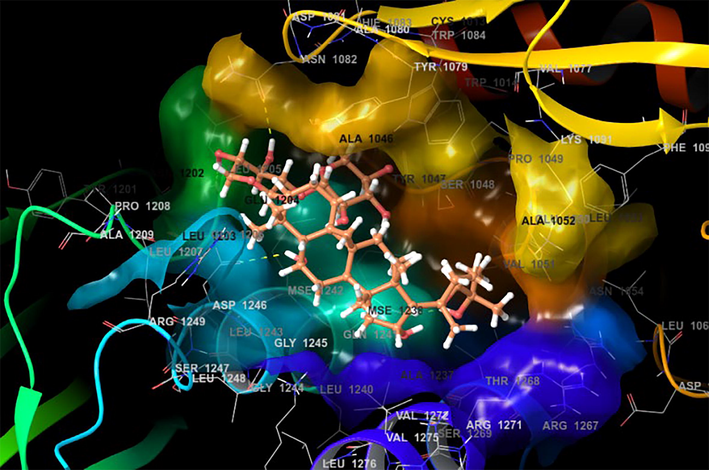
- Docking pose of Astragaloside with 3TRK protein target of Chikungunya virus.
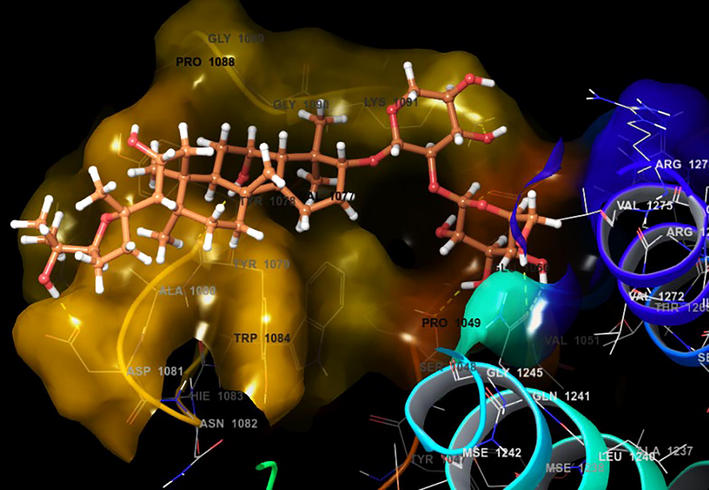
- Docking pose of Astragaloside with 3N42 protein target of Chikungunya virus.
3.3 ADMET analysis and in vitro cytotoxicity assay
The three lead compounds, astragaloside II, III, and IV were subjected to ADMET analysis, and it was found that all three phytocompounds were non-mutagenic and non-carcinogenic. The in-vitro cytotoxicity assay revealed that all the three compounds, astragaloside II, III, and IV were non-toxic up to 500 µg/mL on Vero cells, and more than 90% of the cells were viable at the highest concentration. The concentration of 250 µg/mL which was lower than the maximum non-toxic concentration was considered for the antiviral assay.
3.4 Antiviral activity of astragaloside II, III, and IV
All the three lead phytocompounds showed significant antiviral activity against both Asian and African strains of CHIKV. Astragaloside II showed significant antiviral activity against Asian strain of CHIKV at the concentration of 1.56 µg/mL and both astragaloside III and IV at 3.12 µg/mL. Against the African strain of CHIKV, astragaloside II also exhibited significant antiviral activity at 3.12 µg/mL, followed by astragaloside IV at 6.25 µg/mL and astragaloside III at 12.5 µg/mL (Fig. 9 and Table 2).
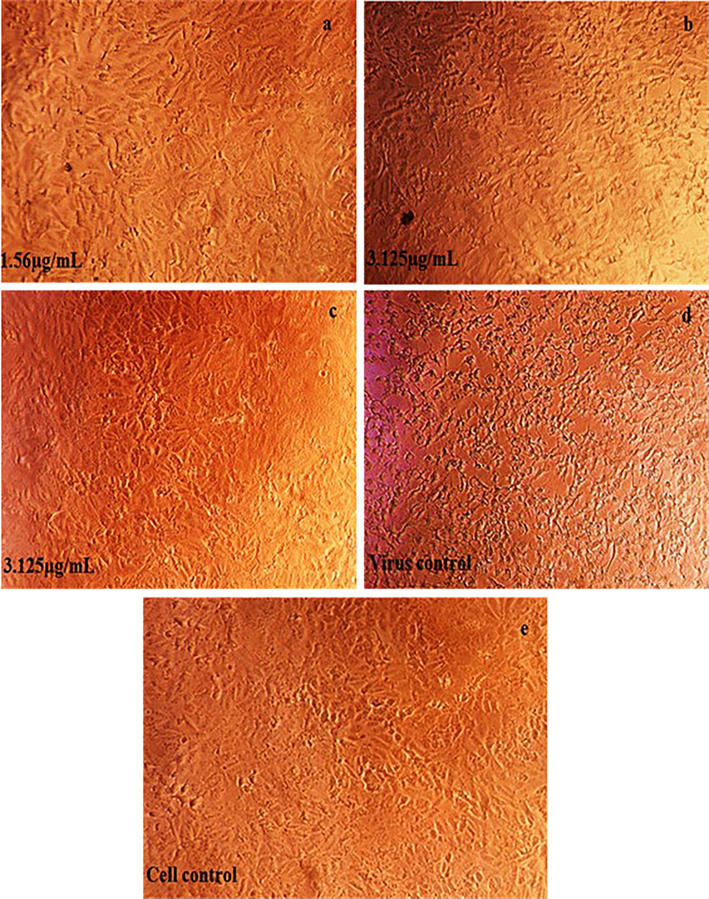
- Antiviral activity of screened phytocompounds Astragaloside II, III and IV against the Asian strain of Chikungunya virus on Vero cells.
| Phytocompounds | Antiviral Activity | |
|---|---|---|
| Asian Strain (µg/mL) | African Strain (µg/mL) | |
| Astragaloside II | 1.56 | 3.125 |
| Astragaloside III | 3.125 | 12.5 |
| Astragaloside IV | 3.125 | 6.25 |
4 Discussion
CHIKV is a re-emerging infectious agent causing chikungunya infection globally, and high mortality was observed among elderly patients, especially those who have hypertension and diabetes (Economopoulou et al., 2009). In the present study, a library of phytocompounds was virtually screened against E and nsP2 proteins of CHIKV. These two proteins are functionally important targets since they are involved in viral pathogenicity and virulence (Russo et al., 2010; Silva and Dermody, 2017). Since the predetermined active sites of these proteins were not already known the potential binding sites were predicted using a SiteMap followed by molecular docking studies. High throughput screening (HTS) of natural products and computer-aided drug designs against diseases are the major components of modern drug discovery approaches (Hughes et al., 2011; Indu et al., 2020).
The E protein has great importance since it is involved in the virion attachment to the host cell. Hence, E protein was considered as a vital target for drug discovery (Subudhi et al., 2018). In a study, various sites of the E proteins were used and it emphasized that the site 4 exhibited the highest volume of 31.3% containing amino acids Arg13, His18, Ser27, Val32, Leu241, and Ala246 (Rashad and Keller, 2013). In our study, among the five binding sites found, the site which had the highest volume and better D score was chosen. The binding site chosen in this study had amino acids that make contact with the B and F chains which form E1 and E2 envelope proteins. The amino acids that are involved in the B chain include Ile136, Pro134, Lys140, Val135, Leu294, Arg138, Glu334, Gln252, Leu151, Pro134, Ile136, Cys153, Asn33, Thr329, Asn332, Pro335, Gly331, Thr42, Pro152, Trp320, Lys149 and Lys149 and the amino acids involved in the F chain include Leu244, Tyr185, Lys245, Glu246, Gly248, Arg247, Ala249, His253, Ser250, Ile261, Lys245, Tyr185, Leu244, Lys181, Asp183 and Tyr180. While looking at the interactions formed by the three screened phytocompounds, it was interesting to note that all the lead compounds astragaloside II, astragaloside IV, and astragaloside III, exhibited high binding energy and shared two common hydrogen bonds in the amino acid positions Asp183 and Tyr180. The hydrogen bond interactions formed with the phytocompounds were not mostly in the solvent-exposed regions so that the interactions would remain stable even during simulation studies. No pi-pi interactions were observed in our study compounds. E2 protein of CHIKV was targeted and found compounds phenothiazine (-5.8 kcal/mol) and bafilomycin (-6.2 kcal/mol) as inhibitors (Deeba et al., 2017). The HB was contributed by the amino acids Pro176, Tyr15, His18, Pro240, and Leu241. In our study, the active site is contributed by both E1 and E2 proteins, and the amino acids Thr42, Pro152, Asp183, Tyr180, Trp320, Lys149, Ile261, Leu244, Gly248, Gly182, and Gly137 contributed towards HB interactions.
The nsP2 protein is another potential target against CHIKV (Bassetto et al., 2013). A study on molecular docking analysis by targeting nsP2 of CHIKV reported that andrographoside, deoxyandrographoside, neoandrographolide, and 14-Deoxy-11-oxoandrographolide had the best interaction values of −9.10 kcal/mol, −8.72 kcal/mol, −8.25 kcal/mol, and −7.38 kcal/mol, respectively with crucial amino acid residues Glu1043, Lys1045, Gly1176, Leu1203, His1222, and Lys1239 of nsP2 protein (Koushik Kumar et al., 2015). In this current study, we have used a new active site for the nsP2 with the amino acid residues Lys1091, Thr1268, Glu1050, Val1051, Arg1267, Arg1271, Val1272, Val1275, Gly1245, Arg1249, Gly1206, Ala1046, Pro1049, Tyr1047, Trp1084, and Ser1048.
The present study also targeted nsP2 protein for docking analysis and found that the amino acid residue Tyr1079 was mainly responsible for HB interaction with all the three lead phytocompounds. It is reported that the lead molecules had a docking score of −9.601 kcal/mol, and the amino acids involved in the interactions were Lys1045, Gly1176, His1222, and Lys1239 (Singh et al., 2012). In our study, astragaloside II exhibited the highest binding energy value of −10.726 kcal/mol, forming six HB interactions mainly contributed by amino acid residues Gln1241, Tyr1079, Asp1246, Asn1082, and Leu1205 and this amino acid profile was different from the study of Singh et al. (2012). Another study suggested that the molecules drodrenin and flinderole B are inhibitors of CHIKV nsP2 protein (Byler et al., 2016). Oo et al. (2016) reported that the amino acids such as Ala1046, Tyr1047, Ser1048, Tyr1079, Asn1082, Leu1206, Gln1241, and Gly1245 were found to be involved in the interaction of the inhibitor hesperidin with nsP2 and HB interactions were contributed by Asp1246, Trp1084, and Ser1048. A similar interaction profile was observed in our study and the amino acids Lys1091, Thr1268, Glu1050, Val1051, Arg1267, Arg1271, Val1272, Val1275, Gly1245, Arg1249, Gly1206, Ala1046, Pro1049, Tyr1047, Trp1084 and Ser1048 contributed for HB interactions.
Since the lead phytocompounds had a good interaction profile, astragaloside II, astragaloside III, and astragaloside IV were purchased and analyzed for their antiviral activity against Asian and African strains of CHIKV. Among the three lead phytocompounds, astragaloside II exhibited more antiviral activity against both the Asian and African strains of CHIKV compared to astragaloside III and IV. The variation in the antiviral activity against the two strains could be due to the difference in the multiplication rate of the virus in the Vero cells and virulence characteristics. A study reported that Asian/American CHIKV strains are less virulent than the strains found in the West, East, Central, and South African (ECSA) regions (Langsjoen et al., 2018). Astragaloside is already reported for various biological activities, including anti-inflammatory, immunostimulant, antioxidative, anticancer, antidiabetic, cardioprotective, hepatoprotective, and antiviral activities (Li et al., 2014). In vitro and in vivo antiviral activities of astragaloside IV against hepatitis B virus were reported in a study which suggested that this compound is very safe and has no significant toxicity (Wang et al., 2009). Therefore, it is inferred that the lead astragaloside compounds screened in this study would be the safe antiviral molecules against the CHIKV because of their non-toxic, non-mutagenic and non-carcinogenic properties.
5 Conclusion
The findings of this study revealed that all the three lead phytocompounds screened had high binding energy values against the envelope and nsP2 proteins of chikungunya virus with desirable pharmacokinetic properties. It is concluded that astragaloside II, astragaloside IV, and astragaloside III have substantial antiviral activity against Asian and African strains of chikungunya virus.
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project Number(RSP2022R418), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Computer-aided identification, design and synthesis of a novel series of compounds with selective antiviral activity against chikungunya virus. Antiviral Res.. 2013;98:12-18.
- [CrossRef] [Google Scholar]
- Pharmacologic management of pain in patients with Chikungunya: a guideline. Rev. Soc. Bras. Med. Trop.. 2016;49(6):668-679.
- [Google Scholar]
- Alphavirus protease inhibitors from natural sources: A homology modeling and molecular docking investigation. Comput. Biol. Chem.. 2016;64:163-184.
- [CrossRef] [Google Scholar]
- admetSAR: A Comprehensive Source and Free Tool for Assessment of Chemical ADMET Properties. J. Chem. Inf. Model.. 2012;52:3099-3105.
- [CrossRef] [Google Scholar]
- Chikungunya virus pathogenesis: From bedside to bench. Antiviral Res.. 2015;121:120-131.
- [CrossRef] [Google Scholar]
- Potential entry inhibitors of the envelope protein (E2) of Chikungunya virus: in silico structural modeling, docking and molecular dynamic studies. VirusDis.. 2017;28:39-49.
- [CrossRef] [Google Scholar]
- Atypical Chikungunya virus infections: clinical manifestations, mortality and risk factors for severe disease during the 2005–2006 outbreak on Réunion. Epidemiol. Infect.. 2009;137:534-541.
- [CrossRef] [Google Scholar]
- Arboviral Diseases Branch, National Center for Emerging and Zoonotic Infectious Diseases, CDC, Notes from the field: chikungunya virus spreads in the Americas – Caribbean and South America, 2013–2014 MMWR Morb. Mortal. Wkly. Rep.. 2014;63:500-501.
- [Google Scholar]
- Chikungunya Virus. In Vitro Response to Combination Therapy With Ribavirin and Interferon Alfa 2a. J. Infect. Dis.. 2016;214(8):1192-1197.
- [Google Scholar]
- Chikungunya Virus: Pathophysiology, Mechanism, and Modeling. Viruses. 2017;9:368.
- [CrossRef] [Google Scholar]
- iGEMDOCK: a graphical environment of enhancing GEMDOCK using pharmacological interactions and post-screening analysis. BMC Bioinform.. 2011;12:S33.
- [CrossRef] [Google Scholar]
- Principles of early drug discovery: Principles of early drug discovery. Br. J. Pharmacol.. 2011;162:1239-1249.
- [CrossRef] [Google Scholar]
- Raltegravir, Indinavir, Tipranavir, Dolutegravir, and Etravirine against main protease and RNA-dependent RNA polymerase of SARS-CoV-2: A molecular docking and drug repurposing approach. J. Infect. Public Health. 2020;13:1856-1861.
- [CrossRef] [Google Scholar]
- Prediction of binding potential of natural leads against the prioritized drug targets of chikungunya and dengue viruses by computational screening. 3. Biotech. 2018;8:274.
- [CrossRef] [Google Scholar]
- Structural basis for complementary and alternative medicine: Phytochemical interaction with non-structural protein 2 protease-a reverse engineering strategy. Chin. J. Integr. Med.. 2015;21:445-452.
- [CrossRef] [Google Scholar]
- Chikungunya Virus Strains Show Lineage-Specific Variations in Virulence and Cross-Protective Ability in Murine and Nonhuman Primate Models. mBio.. 2018;9(2)
- [CrossRef] [Google Scholar]
- A Review of Recent Research Progress on the Astragalus Genus. Molecules. 2014;19:18850-18880.
- [CrossRef] [Google Scholar]
- NPACT: Naturally Occurring Plant-based Anti-cancer Compound-Activity-Target database. Nucleic Acids Res.. 2013;41:D1124.
- [CrossRef] [Google Scholar]
- Epidemiology, clinical manifestations, and diagnosis of chikungunya fever: Lessons learned from the re-emerging epidemic. Indian J. Dermatol.. 2010;55:54.
- [CrossRef] [Google Scholar]
- Evaluation of antiviral activity of South American plant extracts against herpes simplex virus type 1 and rabies virus. Phytother. Res.. 2007;21:970-974.
- [CrossRef] [Google Scholar]
- In silico study on anti-Chikungunya virus activity of hesperetin. PeerJ. 2016;4:e2602.
- [Google Scholar]
- Expression and biochemical characterization of nsP2 cysteine protease of Chikungunya virus. Virus Res.. 2008;131:293-298.
- [CrossRef] [Google Scholar]
- Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis.. 2007;7:319-327.
- [CrossRef] [Google Scholar]
- Structure based design towards the identification of novel binding sites and inhibitors for the chikungunya virus envelope proteins. J. Mol. Graph. Model.. 2013;44:241-252.
- [CrossRef] [Google Scholar]
- Chikungunya Virus: Emerging Targets and New Opportunities for Medicinal Chemistry. J. Med. Chem.. 2014;57:1147-1166.
- [CrossRef] [Google Scholar]
- Chikungunya, a paradigm of neglected tropical disease that emerged to be a new health global risk. J. Clin. Virol.. 2015;64:144-152.
- [CrossRef] [Google Scholar]
- Structural basis for substrate specificity of alphavirus nsP2 proteases. J. Mol. Graph. Model.. 2010;29:46-53.
- [CrossRef] [Google Scholar]
- Spectral characterisation, antiviral activities, in silico ADMET and molecular docking of the compounds isolated from Tectona grandis to chikungunya virus. Biomed. Pharmacother.. 2017;87:302-310.
- [Google Scholar]
- Antiherpes virus activity of extracts from the medicinal plant Geranium sanguineum L. J. Ethnopharmacol.. 1998;64:59-68.
- [CrossRef] [Google Scholar]
- Chikungunya virus: epidemiology, replication, disease mechanisms, and prospective intervention strategies. J. Clin. Investig.. 2017;127(3):737-749.
- [Google Scholar]
- Homology modeling, molecular dynamics, e-pharmacophore mapping and docking study of Chikungunya virus nsP2 protease. J. Mol. Model.. 2012;18:39-51.
- [CrossRef] [Google Scholar]
- Current Strategies for Inhibition of Chikungunya Infection. Viruses. 2018;10:235.
- [CrossRef] [Google Scholar]
- Global expansion of chikungunya virus: mapping the 64-year history. Int. J. Infect. Dis.. 2017;58:69-76.
- [CrossRef] [Google Scholar]
- Anti-hepatitis B Virus Activities of Astragaloside IV Isolated from Radix Astragali. Biol. Pharm. Bull.. 2009;32:132-135.
- [CrossRef] [Google Scholar]







