Translate this page into:
Olea europaea mediated bioengineered biocompatible gold nanoparticles for antimicrobial, cytotoxic applications, and molecular docking study
⁎Corresponding authors. sellami_hanen@yahoo.fr (Hanen Sellami), shakeel.khan@polyu.edu.hk (Shakeel Ahmad Khan)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Abstract
The objective of this research work is to demonstrate the ecologically friendly fabrication of gold nanoparticles (OL-Au NAPs) using the biomolecules of Olea europaea leaf extract. Several spectroscopic approaches were utilized to analyze green fabricated OL-Au NAPs efficiently. OL-Au NAPs were investigated against two different bacteriological strains for antibacterial and biofilm inhibition efficacy. The MTT technique was used to determine the cytotoxic activity against MCF-7 cancerous cells, which was expressed as a percentage of viable cells. The biocompatibility of the synthesized NAPs was evaluated further by incubating them for 24 h with hMSC and 293T cell lines. The results indicated that synthesized OL-Au NAPs effectively suppressed proliferation and biofilm formation in all tested bacteria. Their antibacterial activity was statistically equivalent to that of standard antibiotics (p > 0.05). In silico molecular docking studies confirmed that OL-Au NAPs can also bind and inhibit important S. aureus proteins involved in the cell wall and fatty acid biosynthesis pathways. Moreover, they outperformed plant leaf extract and CH-Au NAPs in terms of cytotoxic effects on MCF-7 cancerous cells. Green fabricated OL-Au NAPs seemed more biocompatible with 293T and hMSC cells than CH-Au NAPs. The promising biological properties of the OL-Au NAPs may be a result of the NAPs' properties interacting with the adsorbed bioactive molecules from plant leaf extract. As a consequence of this study, synthesized OL-Au NAPs may be a potential choice for their numerous pharmacological and nutritional properties. This discovery will also open the road for the creation of nontoxic nanomaterials with extra biologic properties obtained from plants.
Keywords
Au NAPs
Olea europaea
Antibiofilm
Antibacterial
Anticancer
Molecular docking
1 Introduction
Carcinoma, a disease characterized by abnormal cell development, is one of the leading global causes of mortality. According to the World Health Organization (WHO), cancer was responsible for 8.8 million deaths in 2015, and 17 million cancer-related deaths are anticipated by the year 2030 (Thun et al., 2010). Current cancer therapies rely on chemotherapeutic medications that are designed to destroy cancer cells (Singh et al., 2017a). Unfortunately, these medicines frequently lead to adverse effects due to the harm they inflict on healthy cells in the surrounding area. Finding a natural therapeutic treatment for cancer cells is a difficult task. Consequently, a strategy that integrates technology and targeted medicine administration is necessary. By enabling simultaneous diagnosis and therapy, the nanoparticles-based drug delivery method plays an important role in addressing the limitations of traditional cancer therapies (Singh et al., 2017b).
In addition to cancer disorders, there has been significant growth in drug-resistant bacterial infections during the last several decades. This resistance to antibiotics has become a worldwide health concern and one of the leading causes of death and morbidity (Husain et al., 2019). Furthermore, the development of resistance diminishes the effectiveness of the drug, so making it more difficult to treat infections linked with chronic conditions like cancer and diabetes (Shakoor et al., 2019; Andleeb et al., 2020). However, a number of studies indicate that antibiotic resistance in bacterial infections is not attributable to free bacteria but instead to biofilm-dwelling bacteria (Liu et al., 2019). Therefore, biofilm-forming bacteria develop resistance to antibacterial drugs, either because the antibiotic cannot penetrate the biofilm or because they display complicated biofilm-drug resistance features (Elbourne et al., 2019). Nowadays, antibiotics based on nanoparticles are now being tested for their efficacy against drug-resistant bacteria and biofilm-forming bacteria.
Recently, nanobiotechnology is the convergence of nanotechnology and biology, leading to the creation of biogenic and biocompatible nanomaterials with potential biological applications (Singh et al., 2016). In recent years, noble metal nanoparticles, particularly gold nanoparticles (Au NAPs), have attracted a great deal of interest due to their exceptional optoelectronic and Surface Plasmon Resonance (SPR) properties and their promising biomedical applications, such as chronic disease diagnosis, drug delivery to selectively target, and biosensors (Peralta-Videa et al., 2016; Majdalawieh et al., 2014). Currently, Au NAPs are employed in anticancer treatment for numerous kinds of cancer cells, including hepatocellular carcinoma HepG-2 (Fadel et al., 2018), colon carcinoma HCT-116 and HT-29 cell lines (Rezaee et al., 2017), and breast adenocarcinoma MCF-7 (Hasanzadeh et al., 2018). Au NAPs have been manufactured utilizing multiple physical, chemical, and biological processes, as have all other metal nanoparticles. However, physical and chemical procedures include the use of toxic substances, which may result in serious human and environmental problems (Ijaz et al., 2017; Khan et al., 2020a,b,c; Sellami et al., 2021; Khan et al., 2021a,b).
Thus, in order to prevent the aforementioned concerns, it is necessary to synthesize nanoparticles using a process that is ecologically benign. Due to its utilization of natural materials and subsequent manufacture of biocompatible nanoparticles, the green synthesis technique has received substantial interest as an alternative (Kumari et al., 2020). As natural biological resources for the production of metallic nanoparticles, viruses, bacterium, fungus, algae, and vegetation have been widely used (Jayaramudu et al., 2013; Clarance et al., 2020; Syed et al., 2021). However, a green approach based on plant extracts has attracted considerable interest and has become popular over the years. In these biosynthesis approaches, plant extracts are cheap, stable, scalable, and readily available compared to other biomolecule resources (Ahmad et al., 2016; Baker et al., 2021; Kumari et al., 2020; Sellami et al., 2021; Khan et al., 2021b). In addition, extracts of plants have been employed as reducing and capping agents for nanoparticles biosynthesis owing to their numerous phytochemical contents, such as sugars, enzymes, polyphenols, flavonoids, alkaloids, and terpenoids, which stimulate the reduction of metal ions and the subsequent creation of metal nanoparticles (Elbagory et al., 2019; Gharehyakheh et al., 2020).
In addition, it is believed that the biogenic phytochemical elements of plants may enhance the inherent properties of nanoparticles, such as their antibacterial, antioxidant, and anticancer properties, by adsorbing onto the surface of nanoparticles (Chandran et al., 2006; Kumar and Yadav, 2009; Muthukumar et al., 2016). About 100 plant species have been researched for their propensity to produce metallic nanoparticles, and various plant components, including stem, root, seed, fruit, peel, callus, leaves, and flowers, have been used (Shkryl et al., 2021; Kuppusamy et al., 2016). In this regard, leaves of Olea europaea L., as a plant part, were used for the biosynthesis of Au NAPs. Olea europaea L. is a member of the Oleaceae family and an essential crop in Mediterranean nations, which produce around 98 percent of the world's olive oil (Gutiérrez et al., 2004). Multiple studies have shown that Olea europaea leaf extract contains significant secondary metabolites such as oleuropein and its derivatives with various biological properties, including antioxidants, anti-inflammatory, antiviral, anticancer, and antimicrobial properties (Bouaziz and Sayadi, 2005; Briante et al., 2002; Goulas et al., 2009; Visioli et al., 1998; Micol et al., 2005; Goulas et al., 2009; Pereira et al., 2007). In consideration of the health advantages of Olea europaea leaf extract in the field of biomedicine, biologically produced OL-Au NAPs were examined for their antibacterial, antibiofilm, cytotoxic, and biocompatibility properties.
2 Materials and methods
2.1 Chemicals
Chemicals and reagents of analytical quality and chemically synthesized gold nanoparticles were procured from Sigma-Aldrich. The staining agents such as Hoechst 33342 and Propidium Iodide (PI) were acquired from Thermo Fisher, USA. Experiments were conducted on leaves of the Olea europaea L. Chetoui cultivar, which is grown in Borj El Amri, Tunisia. We collected fresh green leaves till the conclusion of the olive morphology (February 2019). Professor Fathi Ben Amar of Tunisia's olive institution identified and validated Olea europaea leaves. The plant’s specimen voucher (MMC) was deposited at the herbarium of Oliviers de Tunisie as previously reported (Trigui and Msallem, 2002).
2.2 Plant extract preparation and OL-Au NAPs synthesis
Fresh Olea europaea leaves were washed in distilled water (DI) to remove contaminants, phytoplankton, and bugs, then dried (25–30 °C) in the shade. Using a commercial grinder (TSK-949, West point, France), the dried leaves were ground into powder. For future usage, the powdered plant leaves were preserved in glass bottles. The extraction of the plant and the synthesis of OL-Au NAPs were carried out according to the process described by (Khan et al., 2020a). To 100 mL of DI water, ten grams of powdered Olea europaea leaves were added. The aqueous mixture was then brought to a boil for 5 min while being vigorously stirred. The filter paper was used to filter the boiling mixture to get a yellowish-colored extract. The extract of the leaves was then utilized as a reducing and capping agent in order to convert gold salt to its metallic form. To 25 mL of Olea europaea leaf extract, one mM HAuCl4·3H2O was added. Following that, the mixture was then heated to 80 °C for 65 min while constantly stirring to acquire a ruby red dispersion of OL-Au NAPs. After 15 min at 15,000 rpm centrifugation, the resulting OL-Au NAPs were rinsed three times with DI water and then dried in a 70 °C oven.
2.3 Characterization of OL-Au NAPs
The green synthesis OL-Au NAPs were characterized using different spectroscopic techniques. X-Ray Diffraction (XRD) was performed using Bruker D2 PHASER with LYNXEYE XE-T detector diffractometer (Haidian, Beijing, China) operating at 40 kV and 30 mA with Cu Kα radiation (k = 1.54056 A°) to test the crystallinity and purity of OL-Au NAPs. Energy-dispersive X-ray spectroscopy using Thermo Fisher Scientific Ultradry (Madison, WI, USA) was performed to determine the elemental composition of the green-synthesized OL-Au NAPs. Morphology of OL-Au NAPs was identified using Transmission electron microscopy (TEM) (FEI/Philips Tecnai 12 BioTWIN, Baltimore, MD, USA). The LEICA TCS SP8 confocal laser scanning microscope (CLSM) was used for acquiring the LIVE/DEAD stained images of bacteria and cancer cells.
2.4 Cytotoxic activity
The cytotoxic activity of the OL-Au NAPs was determined using the MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) colourimetric assay against MCF-7 (breast cancer cells) (Sellami et al., 2021). MCF-7 carcinomatous cells were cultured in 100 µL Dulbecco's Modified Eagle's Medium (DMEM) at 37 °C in a humidified ambience composed of 5% CO2 and 95% air to achieve confluency (5 × 108 cells/well). Then, 200 µL from each sample solution (OL-Au NAPs, Olive leaves extract, and CH-Au NAPs) at 125 and 250 µg/mL concentrations were added separately to each well. The plate was then incubated at 37 °C for an additional 24 h. Following centrifugation to remove the supernatant, the plate was rinsed with phosphate buffer saline solution. Each well received 15 µL of MTT reagent at a concentration of 0.5 mg/mL, and the plate was then incubated at 37 °C for 4 h. Following incubation, 150 µL DMSO was added to each well to solubilize the formazan crystals and shaken for 10 to 15 min. The formazan product's absorbance at 570 nm was measured using a spectrophotometer. The following formula was used to determine the percentage of viable cells: % Cell viability = ODsample/ODcontrol × 100.
2.5 Antibacterial activity
The synthesized OL-Au NAPs were scrutinized for their antibacterial activity against Gram-negative bacterial strain (Klebsiella ATCC 13883) and Gram-positive bacterial strain (S. aureus ATCC 15564) using the standard well diffusion method as described by Shahid et al. (2018). Briefly, each bacterial strain (5 × 105 CFU/ml) was grown in Mueller-Hinton agar plates, and then holes with a diameter of 6 mm were bored on top of the bacterial lawn using a sterilized hollow iron rod. Then, 100 µL from each sample solution (OL-Au NAPs, Olive leaves extract, CH-Au NAPs, and standard drug (ciprofloxacin)) with a 500 µg/mL concentration were dropped separately on each hole. The plates were then incubated at 37 °C for 24 h, and the diameter of inhibition zones (ZOIs) was measured. The test was performed in triplicates, and the average was reported.
2.6 Biofilm inhibition activity
The biofilm inhibition performance of the green synthesized OL-Au NAPs was measured against the two bacterial strains following the polyvinyl chloride biofilm formation assay reported by (Hussain et al., 2019). In brief, both bacteria were cultured in nutrient agar and incubated at 37 °C for 24 h. These overnight cultures of both bacteria at 5 × 105 colony forming units (CFU) per well were resuspended in nutrient broth in a 96 microtiter well plate. After, 100 µL from each sample solution (OL-Au NAPs, Olive leaves extract, CH-Au NAPs, and standard drug (ciprofloxacin)) with a 500 µg/mL concentration were dropped separately in each well. The well plate was then incubated at 30 °C for 24 h. The biofilms in each well were stained with 90 µL crystal violet solution (1% w/v), and quantification was performed by solubilizing the dye in ethanol and measuring absorbance at 470 nm. The following formula was used for calculating the percentage of biofilm inhibition of each bacteria: Biofilm inhibition percentage = [(OD control – OD treated)/ OD control] × 100
Here OD is the optical density.
2.7 Cytobiocompatibility analysis
As described in Section 2.4, a similar MTT assay was carried out on normal cell lines (293T and human mesenchymal stem (hMSC) cell line) for cytobiocompatibility analysis (Sellami et al., 2021). Both cell lines were treated separately with 100 µL of each sample (plant extract, CH-Au NAPs, and OL-Au NAPs) at the concentration of 120 µg/mL. The optical density at 570 nm was measured using a spectrophotometer. The following formula was used to determine the percentage of viable cells: % Cell viability = OD sample/OD control × 100
Moreover, using a fluorescence staining technique, we determined the viability of the 293T cell line with a live/dead double staining kit. The same method (as described in Section 2.4) was repeated till the treatment of cells with OL-Au NAPs (100 µL at a concentration of 120 µg/mL). Following that, each well was incubated for 20 min at 37 °C with the staining solution as per the manufacturer’s recommendations. CLSM was used to acquire the images (535/617 nm and 361/497 nm excitation/emission wavelengths for PI and Hoechst 33342, respectively).
2.8 Molecular docking analysis of OL-Au NAPs against S. aureus
In recent years, in silico or computational studies have piqued researchers' attention since they have empowered them to understand biomolecule function at the atomic level better. With the antibacterial efficacy of OL-Au NAPs against S. aureus in mind, molecular docking studies were conducted against protein targets implicated in the cell wall and fatty acid production pathways, both of which are essential for bacterial survival. Molecular docking was performed on two enzyme targets from the cell wall biosynthesis pathway, β-lactamase and D-alanine-D-alanine ligase B. At the same time, enoyl-[acyl-carrier-protein] reductase was considered a likely target from the fatty acid biosynthesis pathway (Ikram et al., 2020). The crystal structures of β-lactamase, D-alanine-D-alanine ligase B, and enoyl-[acyl-carrier-protein] reductase were retrieved from the protein data bank with PDB IDs such as 1MWU, 2I80, and 4Z8D, respectively (RCSB PDB). The ligands and water molecules were retrieved from protein crystal structure complexes using the software BIOVIA Discovery Studio Visualizer 2021.
Moreover, it was utilized to design the grid and prepare the protein targets (Grosdidier et al., 2011). The PDB files were transferred to AutoDock Vina (version 1.2.0), and Kollman and Gasteiger partial charges were assigned to the receptor protein targets. The OL-Au NAPs were then uploaded to AutoDock Vina in PDB format (version 1.2.0.). Protein targets and OL-Au NAPs were both translated to pdbqt format using Au-to-Dock Vina (version 1.2.0). The pdbqt files were then used to generate scripts for molecular docking using AutoDock Vina (version 1.2.0), and the results of the docked complex were obtained (Trott and Olson, 2010). Using the software BIOVIA Discovery Studio Visualizer 2021, the docked complex results were then analyzed to determine the binding capacities of the ligands and targets. A score of 0 for binding energy means that a ligand may spontaneously bind to the receptor. It is generally understood that the lower the energy score of the ligand-receptor binding configuration, the greater the likelihood of binding (Trott and Olson, 2010).
2.9 Statistical analysis
The biological test was performed in triplicate, and the findings are expressed as mean, standard deviation. Furthermore, we used one-way and two-way ANOVA to assess the significance threshold at 0.05.
3 Results and discussion
3.1 Synthesis mechanism and characterization
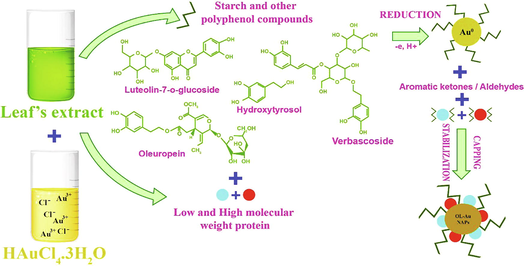
Olea europaea has a number of physiologically important natural compounds that may be used to produce pharmacologically important NAPs (Matteis et al., 2019). Therefore, we used this plant's leaf extract as a reductant and capping agent in the fabrication of bioactive gold nanoparticles. The possible mechanism of forming OL-Au NAPs utilizing plant leaf extract is shown in Fig. 1. Phenolics and flavonoids included in the leaf extract may contribute to the reduction of Au3+ ions to metallic form during the redox process. Concurrently, other natural compounds contained within the leaf extract, such as alkaloids, etc., may be readily used to cap and transform the produced Au0 into gold nanoparticles. Hussain et al. (2019) have also proposed a comparable mechanism for synthesis.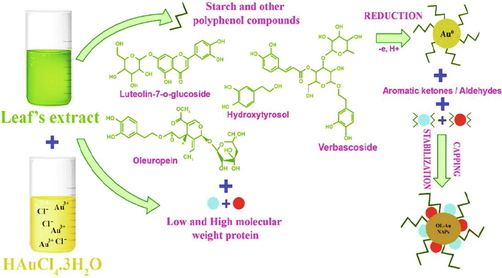
The possible mechanism for the green synthesis of OL-Au NAPs.
XRD was used to investigate the crystallinity and impurities of the green synthesized OL-Au NAPs. The intensity of the peak shows that the gold NAPs that were made are very crystalline. Also, the positions of the XRD peaks are very equivalent to those of metallic gold, and no peaks for impurities have been found (Fig. 2A). Krishnamurthy et al. earlier reported a similar XRD peak pattern for gold NAPs produced from yucca (Krishnamurthy et al., 2014). TEM was used to examine the morphology of the green synthesized OL-Au NAPs. TEM scans revealed that OL-Au NAPs had a spherical shape with a homogeneous dispersion and no aggregation (Fig. 2B). According to TEM measurements, the average particle size of synthesized OL-Au NAPs was 10.4 nm (Fig. 2C). EDX was used to conduct further elemental and compositional analyses. The sample consists mostly of gold, as shown by the EDX spectrum (Fig. 2D). Additional peaks of C and O in the EDX spectra may be attributable to the existence of phytomolecules on the surface of the produced OL-Au NAPs. Kumar et al. showed comparable EDX patterns for gold nanoparticles produced with Sansevieria roxburghiana leaf extract (Kumar et al., 2019).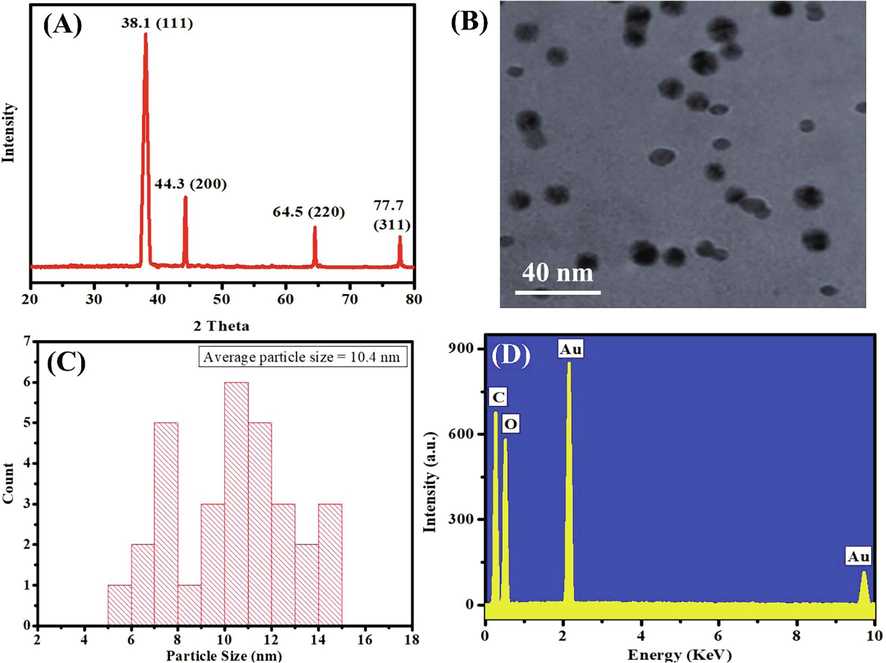
(A) XRD, (B) TEM, (C) size distribution, and (D) EDX pattern of the green synthesized OL-Au NAPs.
3.2 Cytotoxic potential of OL-Au NAPs
In contrast to CH-Au NAPs and plant leaf extract, the cytotoxicity of OL-Au NAPs on MCF-7 cancerous cells was determined using the MTT assay. As seen in Fig. 3, all of the samples tested exhibited a concentration-dependent lethal impact on MCF-7 cancerous cells. Additionally, the findings show that OL-Au NAPs were more cytotoxic than either plant leaf extract or CH-Au NAPs. In comparison to plant leaf extracts, CH-Au NAPs were more efficient in killing MCF-7 cancerous cells. Furthermore, the leaf extract of the plant demonstrated considerable lethal effectiveness on MCF-7 cells, which might have been attributable to the accumulation of cancer-killing bioactive molecules (saponins, terpenoids, alkaloids, flavonoids, polyphenols, etc.) (Essafi Rhouma et al., 2021; Antoniou and Hull, 2021). The higher cytotoxicity of OL-Au NAPs over CH-Au NPs, on the other hand, could be ascribed to the adsorption of therapeutically active bioactive molecules from a plant extract of leaves on their surfaces. Alqahtani et al. (2020) demonstrated that lichen-derived biogenic silver nanoparticles had a similar improvement in cytotoxicity.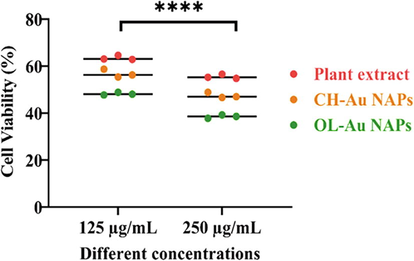
Cytotoxic effect of green synthesized OL-Au NAPs on MCF-7 cancer cells compared to plant leaf extract and CH-Au NAPs.
3.3 Antibacterial potential of OL-Au NAPs
The antibacterial properties of green-synthesized OL-Au NAPs against Gram-positive and Gram-negative bacterial species were investigated. In comparison to CH-Au NAPs and leaves extract, green synthesized OL-Au NAPs displayed the most efficient antibacterial activity and the highest ZOIs against all tested bacterial strains (Fig. 4). Surprisingly, the bactericidal efficacy of OL-Au NAPs was equivalent to that of commercially available antibiotics (p > 0.05). Moreover, the leaf extract showed considerable antibacterial activity against all tested microbes. A combination of the NAPs' physicochemical qualities and the phytochemical constituents from the plant extract of leaves that have been adsorbed on the surface of the NAPs may be responsible for the improved bioactivity of OL-Au NAPs against both bacteria (Fig. 2D).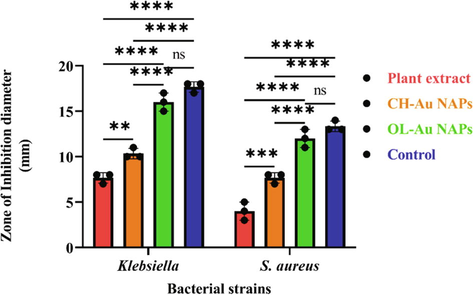
Antibacterial activity of OL-Au NAPs against bacterial strains in comparison to plant leaf extract, CH-Au NAPs, and standard drug. (** p = 0.0029, *** p = 0.0001, **** p < 0.0001, and ns p > 0.05).
It was observed that Gram-negative bacteria are more vulnerable to the effects of OL-Au NAPs manufactured using environmentally responsible methods than Gram-positive bacteria. NAPs have a stronger electrostatic attraction to surface-bound functionalities (such as sulfur proteins) on Gram-negative bacteria than on Gram-positive bacteria. Gram-negative bacteria have a much thinner cell wall as seen in Fig. 5; therefore NAPs may readily permeate them (Slavin et al., 2017; Yin et al., 2020; Pajerski et al., 2019). Gram-positive microorganisms have a multilayered cell wall comprised of wall teichoic acid, lipoteichoic acid, and thick peptidoglycan. Peptidoglycans and the cell membrane are also linked to lipoteichoic acid and teichoic wall acid. Gram-negative microorganisms, on the other hand, have a cell wall composed of outer/inner membrane layers, a thin peptidoglycan coating, lipopolysaccharides, and a gel-like periplasm. Lipopolysaccharides are negative charges macromolecules prevalent only in Gram-negative bacteria's outer membrane layers (Slavin et al., 2017; Pajerski et al., 2019; Yin et al., 2020). Consequently, it is shown that the synthesized OL-Au NAPs inhibit Gram-negative microbes more effectively than Gram-positive microbes. Bindhu et al. and Boomi et al. also showed that gold nanoparticles produced from diverse plants suppress Gram-negative bacteriological strains significantly more than Gram-positive bacteriological strains (Bindhu and Umadevi, 2014; Boomi et al., 2019).![The cell wall comparison between Gram-negative and Gram-positive bacteria [Matica et al., 2019].](/content/185/2022/34/6/img/10.1016_j.jksus.2022.102133-fig6.png)
The cell wall comparison between Gram-negative and Gram-positive bacteria [Matica et al., 2019].
Using the live and dead staining tests, the antibacterial mechanism of action of OL-Au NAPs was subsequently determined. Two bacteria (Klebsiella and S. aureus) were subjected to a live/dead staining assay using a double staining kit (Hoechst 33342/PI). Live/dead microbiological cells can be distinguished using these fluorescent dyes. Hoechst 33342 is a membrane-permanent dye that may stain live and nonliving cells when intercalated with DNA. However, PI is an impermeable membrane dye that can only enter bacteria when the cell membrane is breached and stained the dead cells (Ramalingam et al., 2016). Bacterial strains that have not been exposed to treatments stain only with Hoechst 33342, which indicates that they are viable and that their cell membranes have not been disrupted. Red fluorescence was observed in bacteria treated with OL-Au NAPs, suggesting that their cell membranes had been disrupted (Fig. 6 B and D). According to these findings, antibacterial activity may be attributed to OL-Au NAPs' ability to disrupt microbe membranes.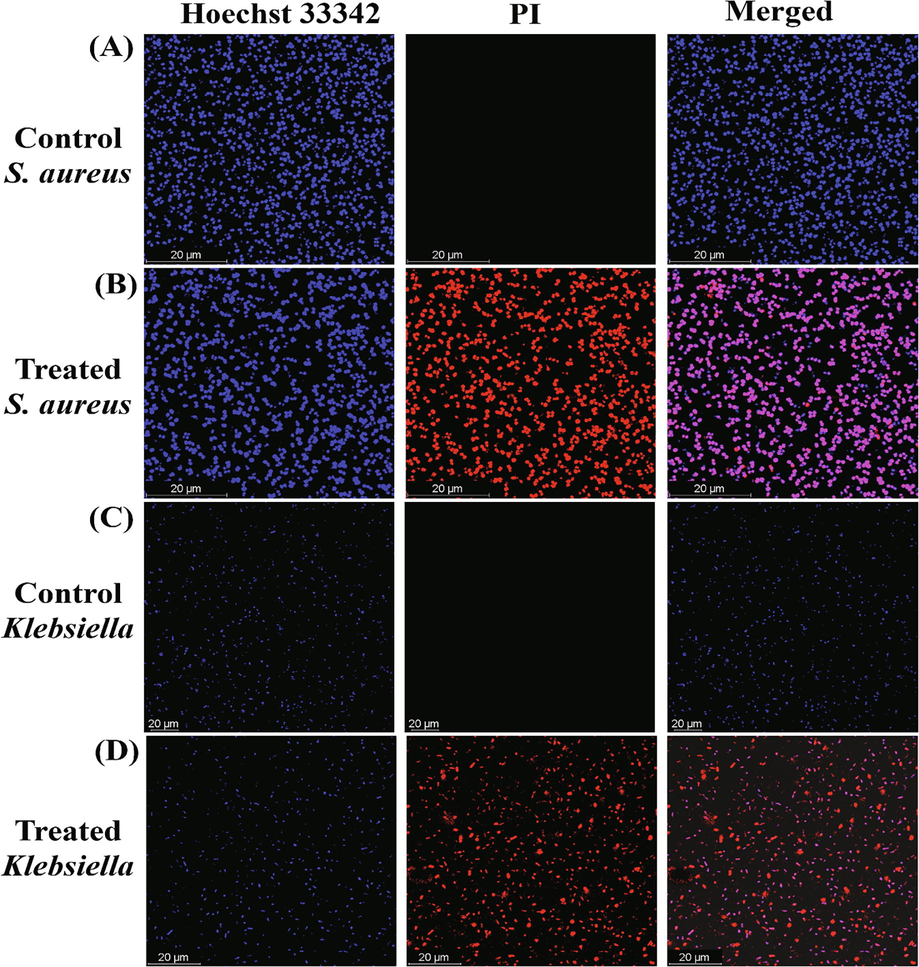
Live/dead stained images of Gram-positive and Gram-negative bacteria treated with green synthesized OL-Au NAPs.
Based on our live/dead assay results, we can deduce that antibacterial mechanisms of action of OL-Au NAPs might be due to the following reasons,
(i) direct contact with the bacterial cell wall; (ii) cell membrane annihilation; (iii) triggering of both innate and acquired host immune responses; (iv) production of reactive oxygen species (ROS); and (v) initiation of intracellular effects (e.g., interactions with DNA and/or proteins) (Fig. 7).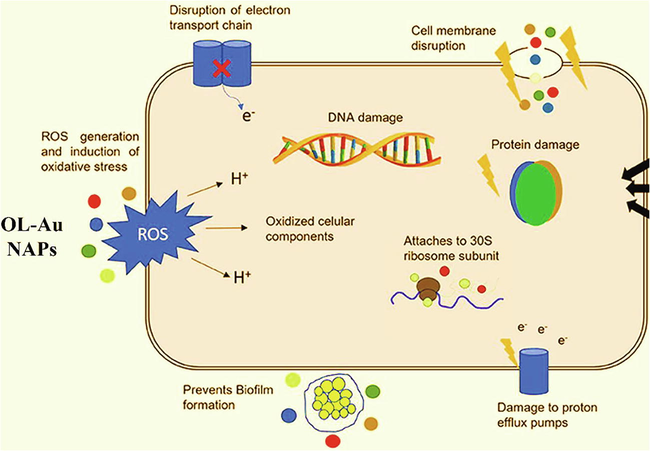
Antibacterial mechanism of action of OL-Au NAPs (Adapted from Baptista et al., 2018).
3.4 Biofilm inhibition performance
Biofilms play a critical role in the pathogenesis of several microorganisms. More significantly, biofilms are the primary biological entities contributing to bacteria gaining multidrug resistance. Bacteria in biofilms are resistant to many antibiotic dosages (Khan and Lee, 2020; Khan et al., 2021c). Therefore, we further assessed green synthesized OL-Au NAPs for their biofilm inhibition performance against different microbial species. Fig. 8 shows that green synthesized OL-Au NAPs demonstrated the maximum antibiofilm performance in comparison to other tested samples. Interestingly, the antibiofilm activity of green synthesized OL-Au NAPs was much better compared to used standard drug against both tested bacteria.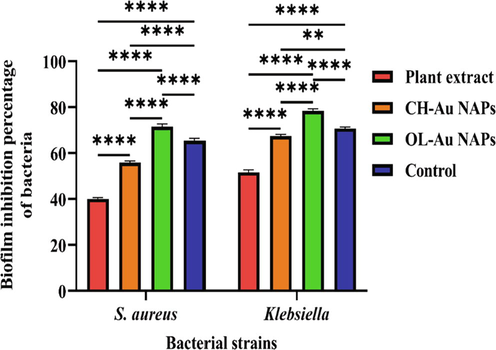
Biofilm inhibition performance of the green synthesized OL-Au NAPs against Gram-positive and Gram-negative bacteriological strains compared to plant leaves extract, CH-Au NAPs, and standard drug. (** p = 0.0022, and **** p < 0.0001).
Moreover, plant leaf extract also appeared to have antibiofilm activity against both microbial strains, which attributed to the presence of phytomolecules with biofilm inhibiting properties. CH-Au NAPs were found to have moderate antibiofilm activity. The OL-Au NAPs' outstanding antibiofilm efficiency may be due to a synergism of their physical–chemical properties and the adsorbed bioactive compounds from the leaf extract on their surfaces. Gram-negative bacterial biofilms were also inhibited more efficiently by OL-Au NAPs than Gram-positive bacterial biofilms. Similar antibiofilm performance of Gold NAPs toward Gram-negative than Gram-positive were also reported by Mu et al. (2016). Antibiofilm and antibacterial results are found in agreement with each other.
3.5 Molecular docking analysis
We performed molecular docking to investigate the mechanistic interactions of OL-Au NAPs with targeted enzymes, and the results are presented in Fig. 9 (A-F) and Table 1. Peptidoglycan synthase enzymes are well-characterized, appealing, and promising candidates for antibiotic development. Both β-lactamase and D-alanine-D-alanine ligase enzymes are members of the molecular machinery involved in peptidoglycan biosynthesis—their blockade results in bacterial cell wall rupture and eventual death (Ikram et al., 2020). The binding energies of OL-Au NAPs in the active pockets of D-alanine-D-alanine ligase and β-lactamase were −8.0 and −6.8 kcal/mol, respectively. Binding energy scores reveal that OL-Au NAPs have a high affinity for the proteins of targeted enzymes, disrupting and suppressing them.![Molecular docking analysis. (A and B) (2D and 3D) Binding of OL-Au NAPs with the active sites of D-alanine-D-alanine ligase. (C and D) (2D and 3D) Binding of OL-Au NAPs with the active sites of β-lactamase. (E and F) (2D and 3D) Binding of OL-Au NAPs with the active sites of enoyl-[acyl-carrier-protein] reductase.](/content/185/2022/34/6/img/10.1016_j.jksus.2022.102133-fig10.png)
Molecular docking analysis. (A and B) (2D and 3D) Binding of OL-Au NAPs with the active sites of D-alanine-D-alanine ligase. (C and D) (2D and 3D) Binding of OL-Au NAPs with the active sites of β-lactamase. (E and F) (2D and 3D) Binding of OL-Au NAPs with the active sites of enoyl-[acyl-carrier-protein] reductase.
Sr No.
Receptors
Nanoparticles
Binding affinity (Kcal/mol)
1
D-alanine-D-alanine ligase
OL-Au NAPs
−8.0
2
β-lactamase
OL-Au NAPs
−6.8
3
enoyl-[acyl-carrier-protein] reductase
OL-Au NAPs
−7.2
Since it has been shown that enoyl-[acyl-carrier-protein] reductase is a key target of broad-spectrum antibiotics such as biocide and triclosan, it has been verified as an effective target for antibacterial drug development (Heath et al., 1999). As a result, particular enoyl-[acyl-carrier-protein] reductase inhibitors may be promising lead molecules for the development of efficient antibacterial medicines. Molecular docking of OL-Au NAPs into an active pocket of enoyl-[acyl-carrier-protein] reductase revealed a docking score of −7.2 kcal/mol. We deduce from molecular docking studies that OL-Au NAPs are capable of binding to a variety of enzymes required for bacterial pathogenicity.
3.6 Cytobiocompatibility analysis of OL-Au NAPs
For nanoparticles to be used in biological systems, their cytobiocompatibility with different normal cells has significant importance. Therefore, we then employed the MTT test to determine the biocompatibility of synthesized OL-Au NAPs with the normal cell lines (hMSC and 293T) in comparison to CH-Au NPs and plant leaf extract. Fig. 10 illustrates the outcomes. The results indicated that the plant leaf extract was highly cytobiocompatible with both hMSC and 293T cell lines, exhibiting a high percentage of live cells compared to other tested samples. Whereas the green synthesized OL-Au NAPs demonstrated excellent cytobiocompatibility with hMSC and 293T cell lines. By contrast, the CH-Au NAPs showed the least cytocompatibility. Furthermore, all of the evaluated samples seemed to be highly biocompatible with hMSC than 293T cell lines, according to the findings.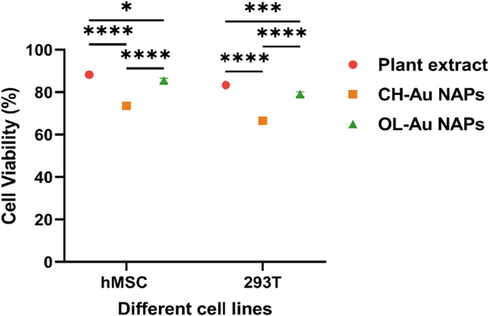
Cytobiocompatibility of OL-Au NAPs with hMSC and 293 T cell lines in comparison to plant leaf extract and CH-Au NAPs. (* p = 0.0187, *** p = 0.0008, and **** p < 0.0001).
Following that, we compared the OL-Au NAPs to the CH-Au NAPs and plant leaf extract in terms of cytobiocompatibility with the 293T cell line using the live/dead staining experiment. Fig. 11 (A–D) illustrates the findings. Plant leaf extract and OL-Au NAPs were shown to have the least harmful impact on 293T cells, resulting in minor cell damage. CH-Au NAPs, on the other hand, seemed to kill a significant number of cells (Red).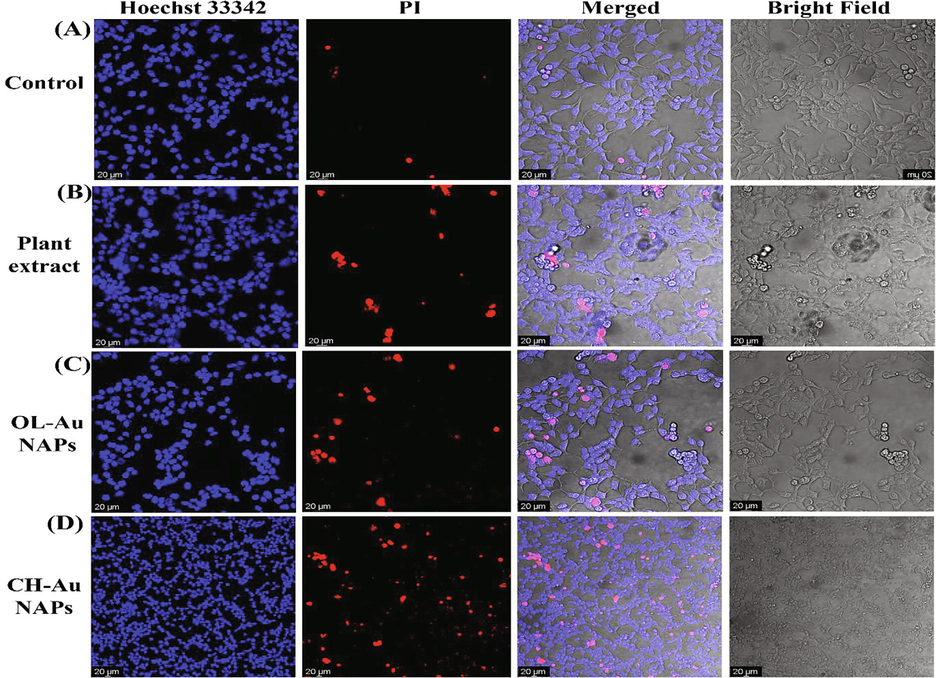
CLSM images of (A) untreated, and treated 293T cells with (B), plant extract (C), OL-Au NAPs, and (D) CH-Au NAPs.
The enhanced cytobiocompatibility of OL-Au NAPs with normal cell lines could be due to the surface immobilization of bioactive and biocompatible macromolecules from the plant leaf extract. Zaïri et al. (2020) have reported similar findings regarding the cytobiocompatibility of plant leaf extracts. Similarly, Rezaeian et al. (2021) demonstrated that silica NAPs had increased cytobiocompatibility when active biological phytomolecules from the plant extract were immobilized to their surface.
4 Conclusions
We conclusively achieved the green production of OL-Au NAPs utilizing biomolecules from Olea europaea leaf extract. Different spectroscopic approaches were used to effectively analyze green-produced OL-Au NAPs. The findings revealed that produced OL-Au NAPs significantly reduced the growth of all tested bacterial strains, which was statistically equal to the standard antibiotic medication (p > 0.05). The fabricated OL-Au NAPs very strongly inhibited the biofilm formation of both Gram-positive and Gram-negative bacterial strains. Furthermore, in silico molecular docking investigations verified the suppression of essential protein targets of S. aureus belonging to the cell wall and fatty acids biosynthetic pathways. In addition, they exhibited superior cytotoxicity against MCF-7 cancer cells when compared to plant lead extract and CH-Au NAPs. In comparison to CH-Au NAPs, green fabricated OL-Au NAPs seemed to be more biocompatible with 293T and hMSC cells. The excellent biological characteristics of the OL-Au NAPs may be a consequence of the synergistic impact of the NAPs' properties and the adsorbed secondary metabolites from plant leaf extract. As a result of this investigation, synthesized OL-Au NAPs may be a viable candidate for their many biomedical and pharmacological applications. Future research is needed to assess the dose-dependent cytocompatibility in vitro/in vivo. Furthermore, this research will pave the opportunities for sustainable development of biocompatible NAPs with additional and improved biological functions derived from important pharmacological plants.
Acknowledgements
This study was supported by the Tunisian Ministry of Higher Education and Scientific Research. The authors like to express their gratitude to Fathi Ben Amar of the olive institute in Sfax, Tunisia, for donating the Olea europaea leaves utilized in this study. The authors also want to express their gratitude to King Saud University's Deanship of Scientific Research for funding this research through the Research Group (RG-1441-362).
Funding
This research was funded by King Saud University, Riyadh, Saudi Arabia, grant number RG-1441-362.
Conflicts of interest
The authors declare no conflict of interest.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
References
- Biosynthesis of anti-proliferative gold nanoparticles using endophytic Fusarium oxysporum strain isolated from neem (A. indica) leaves. Curr. Top. Med. Chem.. 2016;16(18):2036-2042.
- [CrossRef] [Google Scholar]
- Bio fabrication of silver nanoparticles with antibacterial and cytotoxic abilities using lichens. Sci. Rep.. 2020;10:1-17.
- [CrossRef] [Google Scholar]
- Environmental and public health effects of antibiotics and AMR/ARGs. Antibiot. Antimicrob. Resist. Genes Environ.. 2020;1:269-291.
- [CrossRef] [Google Scholar]
- The anti-cancer effect of Olea europaea L. products: a review. Curr. Nutr. Rep.. 2021;10:99-124.
- [CrossRef] [Google Scholar]
- Fruit derived potentially bioactive bioengineered silver nanoparticles. Int. J. Nanomed.. 2021;16:7711.
- [CrossRef] [Google Scholar]
- Nano-strategies to fight multidrug resistant bacteria—“A Battle of the Titans”. Front. Microbiol.. 2018;9:1441.
- [CrossRef] [Google Scholar]
- Antibacterial activities of green synthesized gold nanoparticles. Mater. Lett.. 2014;120:122-125.
- [CrossRef] [Google Scholar]
- Biological synergy of greener gold nanoparticles by using Coleus aromaticus leaf extract. Mater. Sci. Eng. C. 2019;99:202-210.
- [CrossRef] [Google Scholar]
- Isolation and evaluation of antioxidants from leaves of a Tunisian cultivar olive tree. Eur. J. Lipid Sci. Technol.. 2005;107:497-504.
- [CrossRef] [Google Scholar]
- Olea europaea L. leaf extract and derivatives: antioxidant properties. J. Agric. Food Chem.. 2002;50:4934-4940.
- [CrossRef] [Google Scholar]
- Synthesis of gold nanotriangles and silver nanoparticles using Aloevera plant extract. Biotechnol. Prog.. 2006;22:577-583.
- [CrossRef] [Google Scholar]
- Green synthesis and characterization of gold nanoparticles using endophytic fungi Fusarium solani and its in-vitro anticancer and biomedical applications. Saudi J. Biol. Sci.. 2020;27(2):706-712.
- [CrossRef] [Google Scholar]
- The in vitro immunomodulatory effects of gold nanoparticles synthesized from Hypoxis hemerocallidea aqueous extract and hypoxoside on macrophage and natural killer cells. Int. J. Nanomed.. 2019;14:9007-9018.
- [CrossRef] [Google Scholar]
- The use of nanomaterials for the mitigation of pathogenic biofilm formation. Methods Microbiol.. 2019;46:61-92.
- [CrossRef] [Google Scholar]
- Olea europaea L. Flowers as a new promising anticancer natural product: phenolic composition, antiproliferative activity and apoptosis induction. Nat. Prod. Res.. 2021;35:1836-1839.
- [CrossRef] [Google Scholar]
- Comparative enhancement of curcumin cytotoxic photodynamic activity by nanoliposomes and gold nanoparticles with pharmacological appraisal in HepG2 cancer cells and Erlich solid tumor model. Drug Dev. Ind. Pharm.. 2018;44:1809-1816.
- [CrossRef] [Google Scholar]
- Effect of gold nanoparticles synthesized using the aqueous extract of Satureja hortensis leaf on enhancing the shelf life and removing Escherichia coli O157:H7 and Listeria monocytogenes in minced camel’s meat: the role of nanotechnology in the food indus. Appl. Organomet. Chem.. 2020;34:e5492.
- [Google Scholar]
- Phytochemicals in olive-leaf extracts and their antiproliferative activity against cancer and endothelial cells. Mol. Nutr. Food Res.. 2009;53:600-608.
- [CrossRef] [Google Scholar]
- Fast docking using the CHARMM force field with EADock DSS. J. Comput. Chem.. 2011;32:2149-2159.
- [CrossRef] [Google Scholar]
- Chemical characterization of lignin and lipid fractions in kenaf bast fibers used for manufacturing high-quality papers. J. Agric. Food Chem.. 2004;52:4764-4773.
- [CrossRef] [Google Scholar]
- Immunosensing of breast cancer prognostic marker in adenocarcinoma cell lysates and unprocessed human plasma samples using gold nanostructure coated on organic substrate. Int. J. Biol. Macromol.. 2018;118:1082-1089.
- [CrossRef] [Google Scholar]
- Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J. Biol. Chem.. 1999;274:11110-11114.
- [CrossRef] [Google Scholar]
- Nanoparticles as new emerging antibacterials: potentials and limitations. Antibact. Drug Discov. to Combat MDR. 2019;561–579
- [CrossRef] [Google Scholar]
- Biosynthesized silver nanoparticle (AgNP) from Pandanus odorifer leaf extract exhibits anti-metastasis and anti-biofilm potentials. Front. Microbiol.. 2019;10:8.
- [CrossRef] [Google Scholar]
- Green synthesis of copper oxide nanoparticles using abutilon indicum leaf extract: antimicrobial, antioxidant and photocatalytic dye degradation activities. Trop. J. Pharm Res.. 2017;16:743-753.
- [CrossRef] [Google Scholar]
- Dye degradation performance, bactericidal behavior and molecular docking analysis of Cu-doped TiO2 nanoparticles. RSC Adv.. 2020;10:24215-24233.
- [CrossRef] [Google Scholar]
- Development of novel biodegradable Au nanocomposite hydrogels based on wheat: for inactivation of bacteria. Carbohydr. Polym.. 2013;92:2193-2200.
- [CrossRef] [Google Scholar]
- Green synthesis of gold and silver nanoparticles using leaf extract of Clerodendrum inerme; characterization, antimicrobial, and antioxidant activities. Biomolecules. 2020;10:835.
- [CrossRef] [Google Scholar]
- Green synthesis of MnO nanoparticles using Abutilon indicum leaf extract for biological, photocatalytic, and adsorption activities. Biomolecules. 2020;10:785.
- [CrossRef] [Google Scholar]
- Green biological synthesis of nanoparticles and their biomedical applications. Appl. Nanotechnol. Green Synth.. 2020;10:247-280.
- [CrossRef] [Google Scholar]
- Phytomolecules-coated NiO nanoparticles synthesis using abutilon indicum leaf extract: antioxidant, antibacterial, and anticancer activities. Int. J. Nanomed.. 2021;16:1757-1773.
- [CrossRef] [Google Scholar]
- Green synthesis of chromium oxide nanoparticles for antibacterial, antioxidant anticancer, and biocompatibility activities. Int. J. Mol. Sci.. 2021;22:1-17.
- [CrossRef] [Google Scholar]
- Recent progress and strategies to develop antimicrobial contact lenses and lens cases for different types of microbial keratitis. Acta Biomater.. 2020;113:101-118.
- [CrossRef] [Google Scholar]
- Contact lenses coated with hybrid multifunctional ternary nanocoatings (Phytomolecule-coated ZnO nanoparticles: Gallic Acid:Tobramycin) for the treatment of bacterial and fungal keratitis. Acta Biomater.. 2021;128:262-276.
- [CrossRef] [Google Scholar]
- Yucca-derived synthesis of gold nanomaterial and their catalytic potential. Nanoscale Res. Lett.. 2014;9:1-9.
- [CrossRef] [Google Scholar]
- Plant-mediated synthesis of silver and gold nanoparticles and their applications. J. Chem. Technol. Biotechnol.. 2009;84:151-157.
- [CrossRef] [Google Scholar]
- Green one-pot synthesis of gold nanoparticles using Sansevieria roxburghiana leaf extract for the catalytic degradation of toxic organic pollutants. Mater. Res. Bull.. 2019;117:18-27.
- [CrossRef] [Google Scholar]
- Biocompatible biogenic silver nanoparticles interact with caspases on an atomic level to elicit apoptosis. Nanomedicine. 2020;15:2119-2132.
- [CrossRef] [Google Scholar]
- Biosynthesis of metallic nanoparticles using plant derivatives and their new avenues in pharmacological applications – an updated report. Saudi Pharm. J.. 2016;24:473-484.
- [CrossRef] [Google Scholar]
- Nanotechnology-based antimicrobials and delivery systems for biofilm-infection control. Chem. Soc. Rev.. 2019;48:428-446.
- [CrossRef] [Google Scholar]
- Recent advances in gold and silver nanoparticles: synthesis and applications. J. Nanosci. Nanotechnol.. 2014;14:4757-4780.
- [CrossRef] [Google Scholar]
- Chitosan as a wound dressing starting material: antimicrobial properties and mode of action. Int. J. Mol. Sci.. 2019;20:5889.
- [CrossRef] [Google Scholar]
- Cultivar-dependent anticancer and antibacterial properties of silver nanoparticles synthesized using leaves of different Olea Europaea trees. Nanomater.. 2019;9:1544.
- [CrossRef] [Google Scholar]
- The olive leaf extract exhibits antiviral activity against viral haemorrhagic septicaemia rhabdovirus (VHSV) Antiviral Res.. 2005;66:129-136.
- [CrossRef] [Google Scholar]
- Potent antibacterial nanoparticles against biofilm and intracellular bacteria. Sci. Rep.. 2016;6:1-9.
- [CrossRef] [Google Scholar]
- Green synthesis of gold nanoparticles and their enhanced synergistic antitumor activity using HepG2 and MCF7 cells and its antibacterial effects. Process Biochem.. 2016;51:384-391.
- [Google Scholar]
- Attachment efficiency of gold nanoparticles by Gram-positive and Gram-negative bacterial strains governed by surface charges. J. Nanoparticle Res.. 2019;21:186.
- [CrossRef] [Google Scholar]
- Plant-based green synthesis of metallic nanoparticles: scientific curiosity or a realistic alternative to chemical synthesis? Nanotechnol. Environ. Eng.. 2016;1:4.
- [CrossRef] [Google Scholar]
- Phenolic compounds and antimicrobial activity of olive (Olea europaea L. Cv. Cobrançosa) leaves. Molecules. 2007;12:1153-1162.
- [CrossRef] [Google Scholar]
- Ramalingam, B., Parandhaman, T., Das, S.K., 2016. Antibacterial effects of biosynthesized silver nanoparticles on surface ultrastructure and nanomechanical properties of gram-negative bacteria viz. Escherichia coli and Pseudomonas aeruginosa. ACS Appl. Mater. Interfaces. 8, 4963-4976. doi:10.1021/ACSAMI.6B00161.
- RCSB PDB: Accessed February 11, 2022.
- Gold nanoparticles and electroporation impose both separate and synergistic radiosensitizing effects in HT-29 tumor cells: an in vitro study. Int. J. Nanomed.. 2017;12:1431-1439.
- [CrossRef] [Google Scholar]
- Green synthesis of silica nanoparticles from olive residue and investigation of their anticancer potential. Nanomedicine. 2021;16:1581-1593.
- [CrossRef] [Google Scholar]
- Green synthesis of silver nanoparticles using Olea europaea leaf extract for their enhanced antibacterial, antioxidant, cytotoxic and biocompatibility applications. Int. J. Mol. Sci.. 2021;22:12562.
- [CrossRef] [Google Scholar]
- Size-dependent bacterial growth inhibition and antibacterial activity of Ag-doped ZnO nanoparticles under different atmospheric conditions. Indian J. Pharm Sci.. 2018;80:173-180.
- [CrossRef] [Google Scholar]
- Antibiotic-resistant enteric infections. Infect. Dis. Clin. North Am.. 2019;33:1105-1123.
- [CrossRef] [Google Scholar]
- Shkryl, Y., Rusapetova, T., Yugay, Y., Egorova, A., Silant’ev, V., Grigorchuk, V., Karabtsov, A., Timofeeva, Y., Vasyutkina, E., Kudinova, O., Ivanov, V., Kumeiko, V., Bulgakov, V., 2021. Biosynthesis and cytotoxic properties of Ag, Au, and bimetallic nanoparticles synthesized using Lithospermum erythrorhizon Callus culture extract. Int. J. Mol. Sci. 22, 9305. https://doi.org/10.3390/ijms22179305.
- In situ preparation of water-soluble ginsenoside Rh2-entrapped bovine serum albumin nanoparticles: in vitro cytocompatibility studies. Int. J. Nanomed.. 2017;12:4073-4084.
- [CrossRef] [Google Scholar]
- Engineering of mesoporous silica nanoparticles for release of ginsenoside CK and Rh2 to enhance their anticancer and anti-inflammatory efficacy: in vitro studies. J. Nanoparticle Res.. 2017;19
- [CrossRef] [Google Scholar]
- Biological synthesis of nanoparticles from plants and microorganisms. Trends Biotechnol.. 2016;34:588-599.
- [CrossRef] [Google Scholar]
- Metal nanoparticles: understanding the mechanisms behind antibacterial activity. J. Nanobiotechnol.. 2017;15:65.
- [CrossRef] [Google Scholar]
- Biological synthesis of α-Ag2S composite nanoparticles using the fungus Humicola sp. and its biomedical applications. J. Drug. Deliv Sci. Technol.. 2021;66(102770)
- [CrossRef] [Google Scholar]
- The global burden of cancer: priorities for prevention. Carcinogenesis. 2010;31:100-110.
- [CrossRef] [Google Scholar]
- Trigui, A., Msallem, M., 2002. Oliviers de Tunisie, Catalogue des Varietes Autochtones & Types Locaux, Identification Varietale & Caracterisation Morpho-Pomologique des Ressources Genetiques Oleicoles de Tunisie, Ministère de l’Agriculture, IRESA, Institut de l’Olivier, Tunisia. 1, 159.
- AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31:455-461.
- [CrossRef] [Google Scholar]
- Oleuropein, the bitter principle of olives, enhances nitric oxide production by mouse macrophages. Life Sci.. 1998;62:541-546.
- [CrossRef] [Google Scholar]
- The antibacterial mechanism of silver nanoparticles and its application in dentistry. Int. J. Nanomed.. 2020;15:2555.
- [CrossRef] [Google Scholar]
- Phytochemical profile, cytotoxic, antioxidant, and allelopathic potentials of aqueous leaf extracts of Olea europaea. Food Sci. Nutr.. 2020;8(9):4805-4813.
- [Google Scholar]







