Translate this page into:
The inhibitory effect of vitamins on O micron virus via targeting the ACE2 receptor. In silico analysis
⁎Corresponding author. khadidja.belkheir@univ-relizane.dz (Khadidja BELKHEIR)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objectives
The wide and rapid spread of epidemics like COVID 19, highlighted the need for the rapid development of effective solutions. In the present work, a therapeutic approach based on the use of vitamins against the Omicron variant spike was investigated through bioinformatics tools.
Methods
The inhibitory effect of vitamins A, B2, B3, C, E, D, K1 and K7 were tested in silico through a molecular docking and molecular dynamic simulation against four ACE2 grid boxes binding specifically the Omicron proteins 7T9K, 7T9L, 7WBL and 7WBP.
Results
The docking study showed that vitamins K1, A, D and E bind strongly and efficiently to ACE2 receptor. High docking scores with low binding energies and strong interactions with crucial amino acids residues were generally noted for these four vitamins. The dynamic simulation analysis for 100 ns of the best docked complexes using Gromacs tools at a temperature of 300 K allowed screening vitamin A-grid2, vitamin E- grid4, vitamin K1-grid2 and vitamin D-grid 2 as the most stable complexes compared to rest of systems.
Conclusion
These findings indicate that vitamins could interact directly with ACE2 receptor blocking in this way the Omicron binding to its host.
Keywords
ACE2
Vitamins
Docking
MD simulation
O micron
1 Introduction
The COVID 19 is a contagious respiratory disease caused by an enveloped coronavirus named SARS-CoV-2. Since the start of the COVID 19 pandemic for the first time in Wuhan, China at the end of 2019, several new variants of the SARS-CoV-2 have emerged in different regions of the globe (Solo and Doss, 2021). Some of these variants with multiple distinct mutations, increase COVID 19 cases and cause concern all over the world because most of the changes occur in the Spike protein located on the virion surface. This important protein mediates the viral attachment, membrane fusion, and intracellular entry through the angiotensin converting enzymes 2 receptor (ACE2r) (Verma and Subbarao, 2021). The viral spike protein is also the major antigenicity site and the target of many antibodies, or drugs which means the accumulation of mutations in this protein leads to decrease in vaccine and antiviral drug efficacies. The variant officially known as Omicron-B.1.1.529, shows about 50 genetic mutations with 37 of them distributed all over the three vital regions of the spike, the receptor binding domain (RDB), the N-terminal domain (NTD) and the furin cleavage site (FCS) (Bharathi et al., 2022). Some of the 15 mutations located in the RBD were reported to make this variant connection to the host cell stronger and its spread easier than all the other coronavirus. In addition, the vast remodeling in the NTD confers to the B.1.1.529 strain more extensive escape from antibodies and host's immune reaction than its predecessor (Dejnirattisai et al., 2022; Khan et al., 2022).
Omicron like all the other coronavirus variants, is relatively related to the renin-angiotensin system (RAS). After targeting cells that express ACE2r, the viral loading down regulates the ACE2 and increases Angiotensin II protein subsequently. In COVID19 infection, the accumulation of this protein induces inflammation, fibrosis and damage of critical cardiac, lung, liver and renal organ systems (Zwart and Smith, 2020; Xiao et al., 2021).
Some diet compounds like zinc, vitamins A, C, E and D have antiviral, antibacterial, anti-inflammatory and antioxidant characteristics besides their potential to enhance and boost immunity (Torres et al., 2022). For example, vitamin D facilitates the innate and adaptive immune activities by the Toll-like receptors (TLR) and the antimicrobial peptide stimulation (Mitra et al., 2022). Vitamins C and E seem to have direct antioxidant effects by suppressing free oxygen radicals liberated during the oxidative stress of the phagocytosis or the inflammatory reaction (Shao et al., 2021) and Zinc cations maintain the helper cells balance(Kumar et al., 2021). During the COVID 19 outbreak, these compounds have been associated to drugs as a preventive measure for disease (Khalid Omer, 2022) and recently it was indicated that Vitamin D could improve the COVID 19 inflammatory environment and the cytotoxic response against pseudotyped SARS-CoV-2 infected cells (Torres et al., 2022). Through a molecular docking it was noted also that vitamins C, B and D bind the virus RBD and ACE2 protein which makes them as potential inhibitors to this virus (Junaid et al., 2020; Shoemark et al., 2021). In general, a balanced diet rich of vitamins was usually reported as effective against various viruses such as influenza, hepatitis B virus (HBV), norovirus, cytomegalovirus, acute respiratory viruses including coronavirus, some HIV opportunist infections and MeV, either by a direct effect against them or by boosting the immunity of the body (Kumar et al., 2021;Thirumdas et al., 2021, Mitra et al., 2022).In this regard, a computational approach was used in the present study to investigate the docking of vitamins A, B2, B3,C, D, E,K1 and K7 to ACE2r with the objective to analyze their binding affinities to this site. Besides their positive effect on the immune system, vitamins could also block the ACE2r binding to the virus, avoiding in this way the viral loading entry and replication.
2 Materials and methods
2.1 Retrieval of protein sequences
The Cryo-EM structure of four SARS-CoV-2 Omicron spike protein in complex with ACE2r were downloaded from the RCSB Protein data bank, ID7T9K and ID 7T9L (Mannar et al., 2022), ID 7WBL and ID 7WBP (Han et al., 2022). The ligPlot + software was used to portray the main amino acids of the protein involved in the interaction between O micron and ACE2 receptor.
2.2 Grid boxes identification
At first Biovia Discovery Studio software (Biovia, 2015) was used to separate O micron from ACE2r then the Grid parameters were calculated using the same software and Arguslab software (Thompson,2004). The dimensions of the four Grid boxes used in the molecular docking were set (Table 1) (Feinstein and Brylinski, 2015).
ACE2 Grid boxes
Size ( Å)
Center( Å)
Grid 1
x = 11.288; y = 34.047; z = 20.098
X = 227.090744 Y = 174.155640 Z = 260.424504
Grid 2
x = 10.513; y = 34.047, z = 13.482
X = 228.163537 Y = 173.343683 Z = 259.522939
Grid 3
x = 13.821; y = 39.535; z = 11.119
X = 227.801773 Y = 170.168545 Z = 258.514114
Grid 4
x = 11.288; y = 39.535; z = 20.098
X = 226,736290 Y = 176,154391 Z = 260,548645
2.3 Protein-ligand docking
The molecular docking was performed using AutoDock Vina (Trott and Olson, 2010; Eberhardt et al., 2021) integrated into Galaxy web server. Vitamins used as ligands were downloaded from PubChem ( https://pubchem.ncbi.nlm.nih.gov) as SDF files, then were converted into Mol format by using the conversion tool on Open Babel software (O'Boyle et al., 2011). Proteins were next prepared for docking by ChimeraX ( https://www.cgl.ucsf.edu/chimerax/docs/credits.html) for hydrogen and charge adding and were converted to PDBqt files in Galaxy server. Finally, the docking result was downloaded for analysis by Biovia Discovery Studio.
2.4 Molecular dynamics simulation (MDS)
From the docking results, vitamin A-grid1, 2, and 3, vitamin K1-grid 1, 2 and 4, vitamin E-grid2 and 4 and vitamin D-grid 2, 3 complexes were selected for the MD simulation. Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), radius of gyration (Rg), Solvent accessible surface area (SASA) and the number of H‑bonds between vitamins and ACE2 grid boxes were calculated during 100 ns of simulation using the GROMACS package (Abraham et al., 2015) integrated in WebGRO for Macromolecular Simulations (https://simlab.uams.edu/). The ligand topology file was generated by PRODRG online server ( http://davapc1.bioch.dundee.ac.uk/cgi-bin/prodrg) and complexes were solvated in a simulation triclinic water box with SPC water model electro-neutralized by addition of 0.15 M NaCl. Solvent around proteins was equilibrated through NVT/ NPT ensembles at a constant temperature and pressure of 300 K and 1.0 bar. The approximate number of frames per simulation was 1000 and the final figures were prepared using Gnuplot 5.4 patch level3 packages ( http://www.gnuplot.info).
3 Results and discussion
3.1 Ligand receptor affinity
The novel highly contagious viral strain SARS-CoV-2 poses a serious threat to the worldwide population since its first apparition until now. This is the reason why the screening of effective treatment against coronavirus is still underway. Methods combining experimental and computational strategies allowed to test a large broad spectrum of natural compounds including vitamins (A, B, D and their derivates) against SARS-CoV-2 S-RBD and 3CLpro proteins (Essam et al., 2022; Eskandari, 2022). For this, we hypothesized in this study, that vitamins could also interact with crucial amino acids in ACE2r and reduce in this way the virus binding to its host receptor. So in the present work, a docking approach was employed to predict vitamins-ACE2 complexes binding. In first the binding interface for the four Omicron proteins 7T9K, 7T9L, 7WBL and 7WBP with ACE2r were identified (Fig. 1, a, b, c, d and e). A total of eight ACE2 residues, Lys353, Asp38, Tyr41, His34, Glu35, Ser19, Tyr83 and Gln24 were in close contact with the RBD of Omicron spike protein. These amino acids included in four Grid boxes within the binding site of ACE2 receptor (Table2), are generally reported to be involved in the binding of several variants spike protein likeN501Y, N501T, K417R, N501I, L455F, A475V, N501S, Y453F, Q493H, G446S, G446V, Q493L and Y495F. Some of these critical mutations in particular N501I, N501Y, Q493L, Q493H and K417R increased the binding affinity of the SARS-RBD with ACE2 which meansincreases in the viral infectivity and pathogenicity (Verma and Subbarao, 2021; Han et al., 2022). The binding affinities of the tested vitamins towards the four targeted Grid boxes are shown in Table 3. The docking results, identified in general the vitamin D as having the highest binding affinities of −6,01, −5,23 and − 5,28 kcal/mol for the three target Grid 1, 2 and 3, respectively and vitamin K1 for the target Grid 2 and 3 with highest binding affinities of −6,13 and −5,96 kcal/mol, respectively (Table 3). At the same time the vitamin C shows the least binding affinities for the three targets Grid 1, 2 and 4 in comparison with the rest of vitamins (Table3). Ligands with the highest binding scores (the most negative binding energies) were selected for further analysis.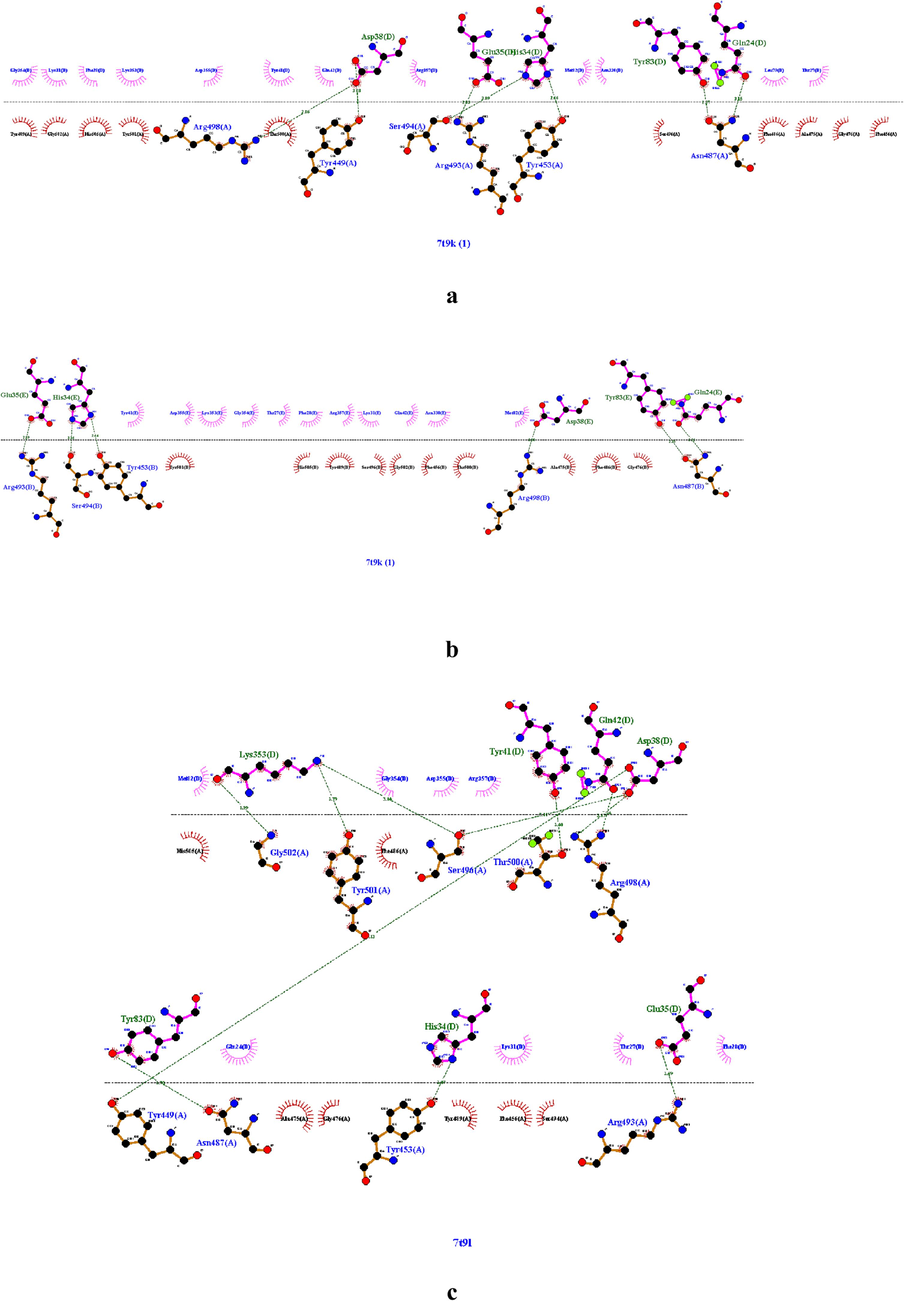
The intermolecular interaction of omicron protein’s with ace2r,(a) 7t9k chain a with chain d of ace2, (b) 7t9k chain b with chain e of ace2, (c) 7t9l chain a with chain d of ace2, (d) 7wbl chain b with chain a of ace2, (e) 7wbp chain b with a of ace2.
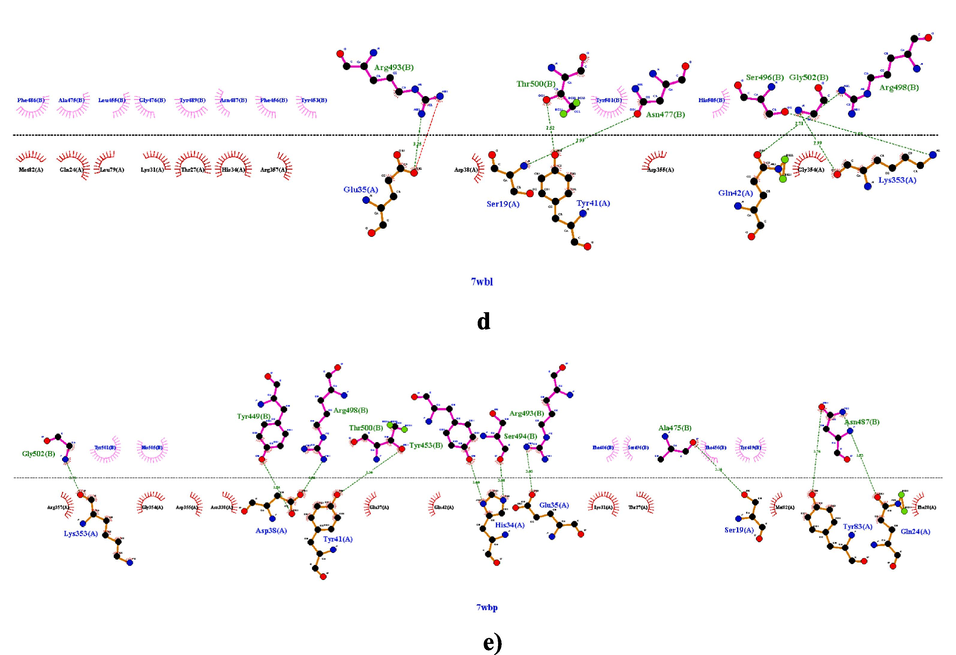
The intermolecular interaction of omicron protein’s with ace2r,(a) 7t9k chain a with chain d of ace2, (b) 7t9k chain b with chain e of ace2, (c) 7t9l chain a with chain d of ace2, (d) 7wbl chain b with chain a of ace2, (e) 7wbp chain b with a of ace2.
O micron protein
ACE2 residue
7t9k (A-D) (B-E) Grid 2
ASP 38
GLU 35
HIS 34
TYR 83
GLN 24
7t9l Grid 1
ASP 38
GLU 35
HIS 34
TYR 83
GLN 24
TYR 41
LYS 353
7wbl Grid 3
Glu 35
Ser19
Tyr41
Gln42
Lys353
7wbp Grid 4
Lys353
Asp38
Tyr41
His34
Glu35
Ser19
Tyr83
Gln24
vitamins
Grid 1
Grid 2
Grid 3
Grid 4
A
−5.22
−5.16
−5.22
−5.04
B2
−5.65
−4.88
−4.90
−5.41
B3
−4.56
−4.00
−3.64
−4.54
C
−4.02
−3.63
−4.00
−4.12
D
−6.01
−5.23
−5.28
−5.93
E
−4.45
−4.91
−4.64
−5.49
K1
−4.87
−6.13
−4.62
−5.96
K7
−4.62
−4.47
−3.81
−4.82
3.2 Ligand receptor interactions
The intermolecular interaction of vitamins with ACE2r showed that four amino acids namely Tyr83, Glu35, Gln 24 and His 34 were involved within crucial van der waals contacts (vdw), hydrophobic interactions or H-bonding with the tested vitamins. In general, these four amino acids play an important role in the wild type or Omicron RBD interactions with ACE2 (Table 4). For example, it was reported that Glu35 forms hydrogen bonds with Gln at position 493 giving stability to the central region of the RBD-ACE2 interface and hotspot-31(Lupala et al., 2022). In Q493H variant, this same amino acid makes a salt bridge and several non-bonded contacts when Gln is replaced by His at position 493 which favors stronger ACE2 and RBD interactions (Lupala et al., 2022). Furthermore His 34 and Tyr 83 are involved in key interactions with Asn 487 and Tyr 453 of wild type RBD or Omicron variant (Huang et al., 2020;Lupala et al., 2022). Previous studies showed also that Gln 24 interacts with Asn 377 in new favorable interactions enhanced by the Omicron substitutions (Wang et al., 2020; Han et al., 2022; Lupala et al., 2022). From our finding, it is clear that vitamins A, E, K1 and D cover a larger region in the essential subspace of ACE2 site indicating a possible reducing of ACE2-Omicron RBD binding affinities. Other amino acids like Asp30, Tyr31, Phe 28 and Thr 27, not included in the studied grid boxes but reported as playing key roles in Omicron or wild type RBD binding, increase interactions with some of the tested vitamins (Fig. 2). It is the special case of vitamins K1 and D which establish generally weak interactions with the four grid boxes of ACE2r although their highest binding affinities to this site. Effectively as shown in the Table 4, vitamin K1 evidences hydrogen bonding only in the Grid boxes1 and 2 with Tyr83 at distances of 2,35 Å and 2,60 Å respectively. Vitamin K1 also extends only one pi-alkyl bond at a distance of 4,90 Å with His 34 in the Grid 4 in addition to vdw contacts with Tyr83, Gln 24 and Glu 35 in the same grid box (Table 4, Fig. 2,a). However, vitamin K1 showed the ability to bind key residues of ACE2r like Lys 31 and Leu 79 residues with pi alkyl interactions, Thr 27 with H bonding, Phe 28 with pi-pi stacked or vdw contacts and Asp 30 with vdw interactions in the grid boxes 1, 2 and 4 which could explain the high binding score noted for this vitamin (Fig. 2,a).Similarly, vitamin D displays only vdw interactions with Tyr 83 and His 34 in the Grid box 4 and it exhibits one pi-alkyl bond with His 34 in the Grid boxes 2 and 3 at distances of 4,61 and 4,93 Å respectively, or only vdw contacts with Tyr 83 in the Grid box 1 (Table 4, Fig. 2,b) but it forms several pi alkyl with Lys 31 and Leu 79 in addition to vdw contacts with Thr27, Phe 28 and Asp 30 (Fig. 2,b).
vitamins
Grid 1
Grid 2
Grid 3
Grid 4
A
Tyr83
Gln24
His34: 3.71 pi-sigmaTyr83
Gln24: 2.61H bond
His34: 3.68 pi-sigmaTyr83: 2.56 Hbond
Gln24: 2.65 Hbond
His34: 3.69 pi-sigmaTyr83: 2.64 Hbond
Gln24: 2.87 unfavorable A/A
His34: 4.32 pi-alkyl
B2
Tyr83: 1.29 unfavorable D/D
Gln24Tyr83: 2.03/2.02 Hbond
Gln24Glu35
His34Tyr83
Gln24
B3
/
Tyr83
Gln24: 2.62H bondTyr83
Gln24: 2.74 Hbond/
C
/
Tyr83: 2.42 Hbond
Gln24: 2.29 Hbond/
/
D
Tyr83
His34: 4.61 pi-alkyl
Tyr83
His34: 4.93 pi-alkylTyr83
His34
E
Tyr83: 4.93 pi-alkyl
His34Tyr83
Gln24
His34: 4.92 pi-alkyl;
3.93 pi-sigma
Glu35Tyr83
His34
Glu35Tyr83: 5.39 pi-pi stacked
Gln24: 2.49 Hbond
K1
Tyr83: 2.60H bond
Gln24Tyr83: 2.35 Hbond
Gln24Tyr83
Gln24Tyr83
Gln24
His34: 4.90 pi-alkyl
Glu35
K7
/
Tyr83: 2.36 Hbond
Gln24His34
/
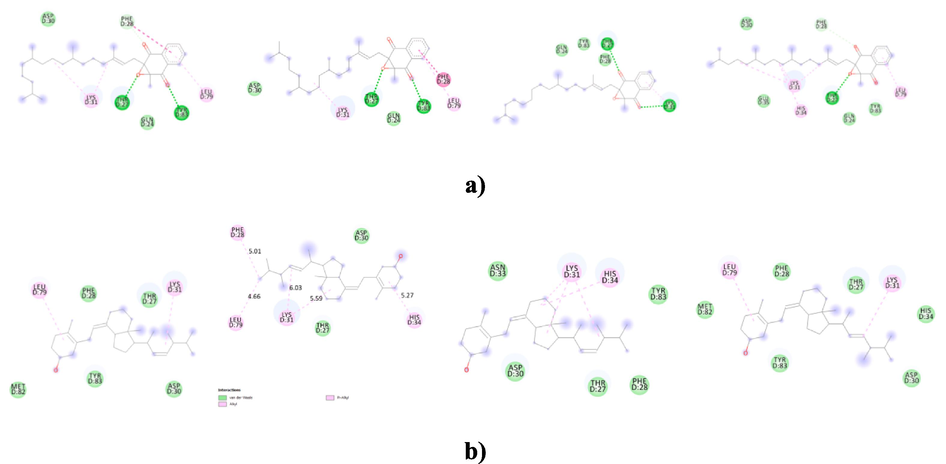
The intermolecular interaction of the 2D structure of vitamins with from left to right Grid 1, Grid 2,Grid 3,and Grid 4 of ACE2r. a) vitamin K1, b)vitamin D, c)vitamin A and d) vitamin E.
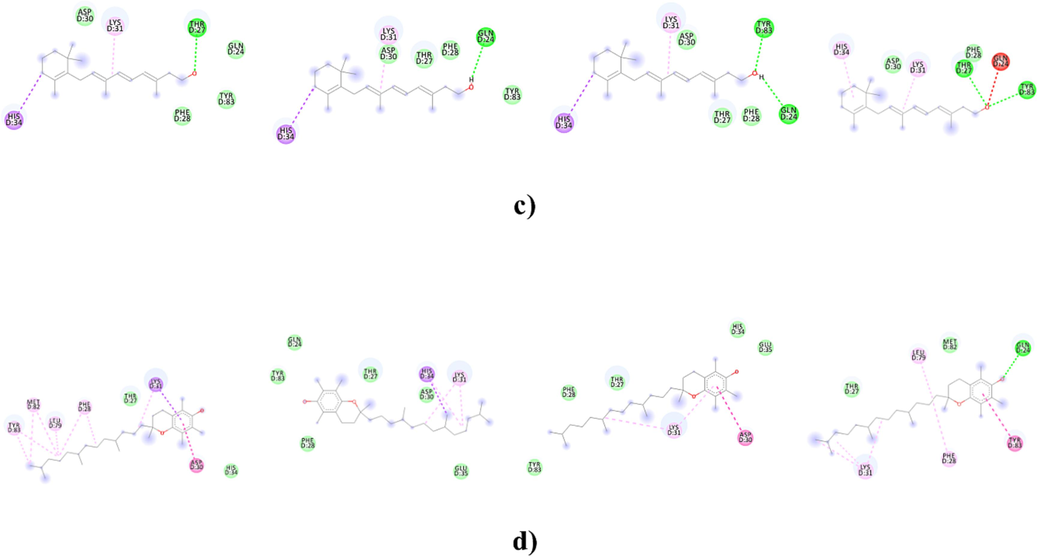
The intermolecular interaction of the 2D structure of vitamins with from left to right Grid 1, Grid 2,Grid 3,and Grid 4 of ACE2r. a) vitamin K1, b)vitamin D, c)vitamin A and d) vitamin E.
Notably, vitamin A forms strong H bonds interactions with both Tyr83 and Gln 24 and it forms pi-sigma bonding with His 34 in the Grid3 of ACE2. Vitamin A also constitutes strong interactions with both the Grid 1 and Grid 2 of ACE2 through vdw contacts with Tyr 83, pi-sigma interaction with His 34 and H bonding with Gln 24 (in the case of Grid 2)(Table 4, Fig. 2,c). In the Grid 4, vitamin A contacts Tyr 83, and His 34 from the ACE2r with H bond and pi- Alkyl interactions respectively. However unfavorable acceptor/acceptor interactions with repulsive forces reducing the stability of the other bonds (Ali et al., 2014) were shown between the same vitamin and Gln 24 at a distance of 2,87 Å in this box(Table 4, Fig. 2,c).
Other strong interactions with the ACE2r were noted for the vitamin E which shows to be in close proximity with Tyr83, Gln 24, His 34, and Glu 35 amino acids of the Grid boxes 2 and 4(Table 4, Fig. 2,d). Indeed, it links these two Grid boxes residues by several pi-, H and vdw interactions with binding energies of −4,91 and −5,49 respectively.
3.3 Complex stability
Next, to analyze the stability of each complex vitamins-ACE2 showing high affinities, statistical tools like RMSD, RMSF, H-bonds, SASA and Rg were calculated during 100 ns of simulation at a constant temperature of 300 K. RMSD analysis provides information about the stability of the ligand-receptor complex. RMSF informs about the residues fluctuations into the ligand-receptor complex. High deviations in these two parameters indicate low stability of the complex because the ligand may not fit very well into the binding site.
As seen in Fig. 3,a, vitamin A in complex with grid 2 and grid 3 of ACE2r, exhibited stable RMSDs of 3 and 3,5 A˚ respectively, with no significant differences between the initial and the final RMSD values during the whole simulation period (2,5–3 A˚ for both systems). The residue fluctuations range from 0,5 to 5 A˚ in both vitamin A-grid 2 and –grid 3 systems with less fluctuations in the case of vitamin A- grid 2 system suggesting more stability for this complex compared to the other (Fig. 3,b). Vitamin A-grid 2 system also showed highest in the number of hydrogen bonds formation (520 vs 500 for vitamin A-grid 3 complex) (Fig. 3,c). The plots of SASA total versus run time (ns) analysis gives information about the surface area (nm2) accessed by solvents or water molecules inside the active site. An increase or decrease in SASA value indicates a change in the protein’s structural conformation. The Vitamin A-grid 2 complex displays a stable SASA total value of about 250 nm2 after 20 ns of simulation but the vitamin A-grid 3 system indicates decreasing in the SASA value until 40 ns of simulation thereby more loss of solvent accessibility and more changes in the protein’s structural conformation are observed in this complex during the simulation period (Fig. 3, d). Finally, the Rg value which is closely related to the compactness of the protein structure remains almost constant (24,42 A˚ for vitamin A-grid2 and 24,5 A˚ for vitamin A-grid 3) from 20 ns to 100 ns with some marginal fluctuations for these two complexes (Fig. 3,e).
The MD simulation of vitamin A-Grid1,-Grid2 and -Grid 3 complexes. a) RMSD, b) RMSF, c) H-bonds, d) Rg and e) SASA analysis.
The molecular dynamics simulation shows also good stability for vitamin E-grid4 system which exhibits a steady course in the RMSD value of 4 A˚(Fig. 4,a). However, a gradual increase in RMSD value, stabilizing at an average of 5 A˚ was noted for vitamin E-grid 2 complex(Fig. 4,a). The RMSF graph analysis indicates higher flexibility of amino acids residues in vitamin E-grid 2 complex than in vitamin E-grid 4 system (Fig. 4,b). The high flexibility (RMSF value of 8 A˚) was noted for residue 50 in vitamin E-grid 2 complex suggesting better accommodation of vitamin E in the grid4 than the grid 2 of ACE2r (Fig. 4,b). The result of RMSD and RMSF analysis agree with the docking data as shown in Fig. 2,d, the total number of interactions involved in vitamin E-grid 4 complex was obviously more than those involved in vitamin E-grid2 system. The dynamic simulation has indicated also the occurrence of about 520H-bonds (versus 510 in vitamin E-grid 2 complex)between the vitamin E and the grid 4 of the ACE2 site confirming their consistent interactions (Fig. 4,c). Additionally to understand the solvent and proteins behaviors of these two complexes, Rg and SASA analysis were also performed (Fig. 4,d and e). Unstable Rg and SASA profiles were noted over the simulation time for both complexes indicating clearly that the ligand-receptor binding induces important changes in the protein’s tertiary structure (Fig. 4,d and e).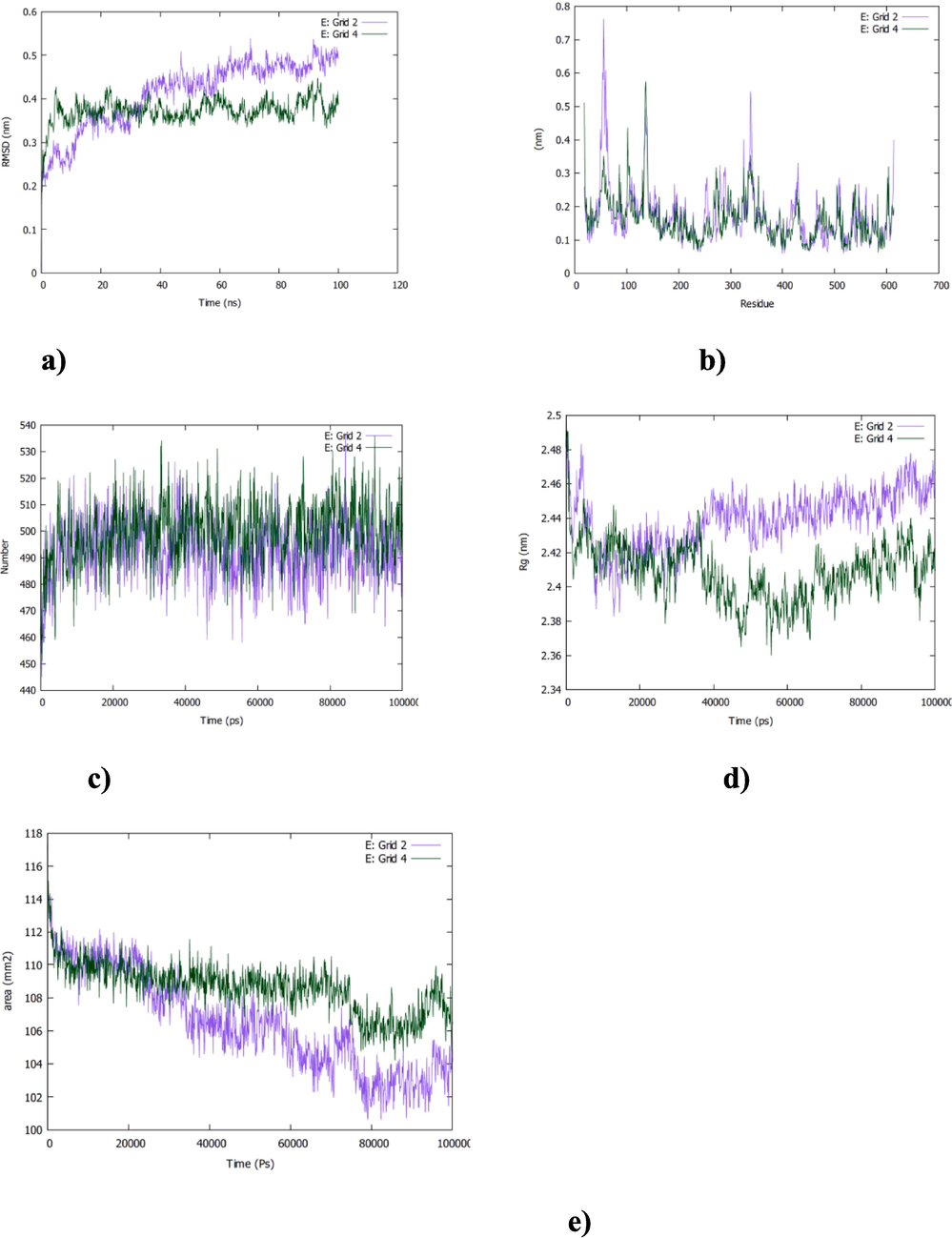
The MD simulation of vitamin E-Grid2 and -Grid4 complexes. a) RMSD, b) RMSF, c) H-bonds, d) Rg and e) SASA analysis.
Furthermore, different RMSD behaviors were noted for vitamin K1 in complex to ACE2r (Fig. 5,a). For example, the RMSD of the vitamin K1-grid1 complex has been found ranging from 3 to 4 A˚ with a general average of 3,5 A˚ and marginal deviations at 50 ns (Fig. 5,a) but it revealed constant RMSD values of about 3 and 3,5 A˚ respectively for vitamin K1-grid 4 and-grid 2 complexes suggesting a better stability in the binding of this vitamin to these two grid boxes than the first one (Fig. 5,a). Moreover, residues fluctuations were also analyzed. The residues fluctuation range is 1 A˚to 6 A˚ in the three complexes with no too deviations in vitamin K1-grid 2 complex (Fig. 5,b). As shown in Fig. 5,c the total number of hydrogen bonds formed between the vitamin K1 and the grid 2 was about 510 H-bonds during 100 ns of simulation time but it ranged from 500 to 520 H-bonds for the K1-grid 1 and grid 4 complexes which confirms that the vitamin K1 fit well in the grid2 than the grid boxes 1 and 4. Rg values of about 42,5 A˚ were noted for the three complexes with a long range of variations over the time for vitamin K1-grid1 complex indicating a misfolding of the ligand in the receptor site in this case (Fig. 5,d). SASA total for vitamin K1-grid 1 and grid 2 complexes are relatively constant (250 nm2) but it decreased from 255 nm2 to 240 nm2 in vitamin K1-grid 4 complex indicating a change in protein’s structural conformation during the simulation period (Fig. 5,e).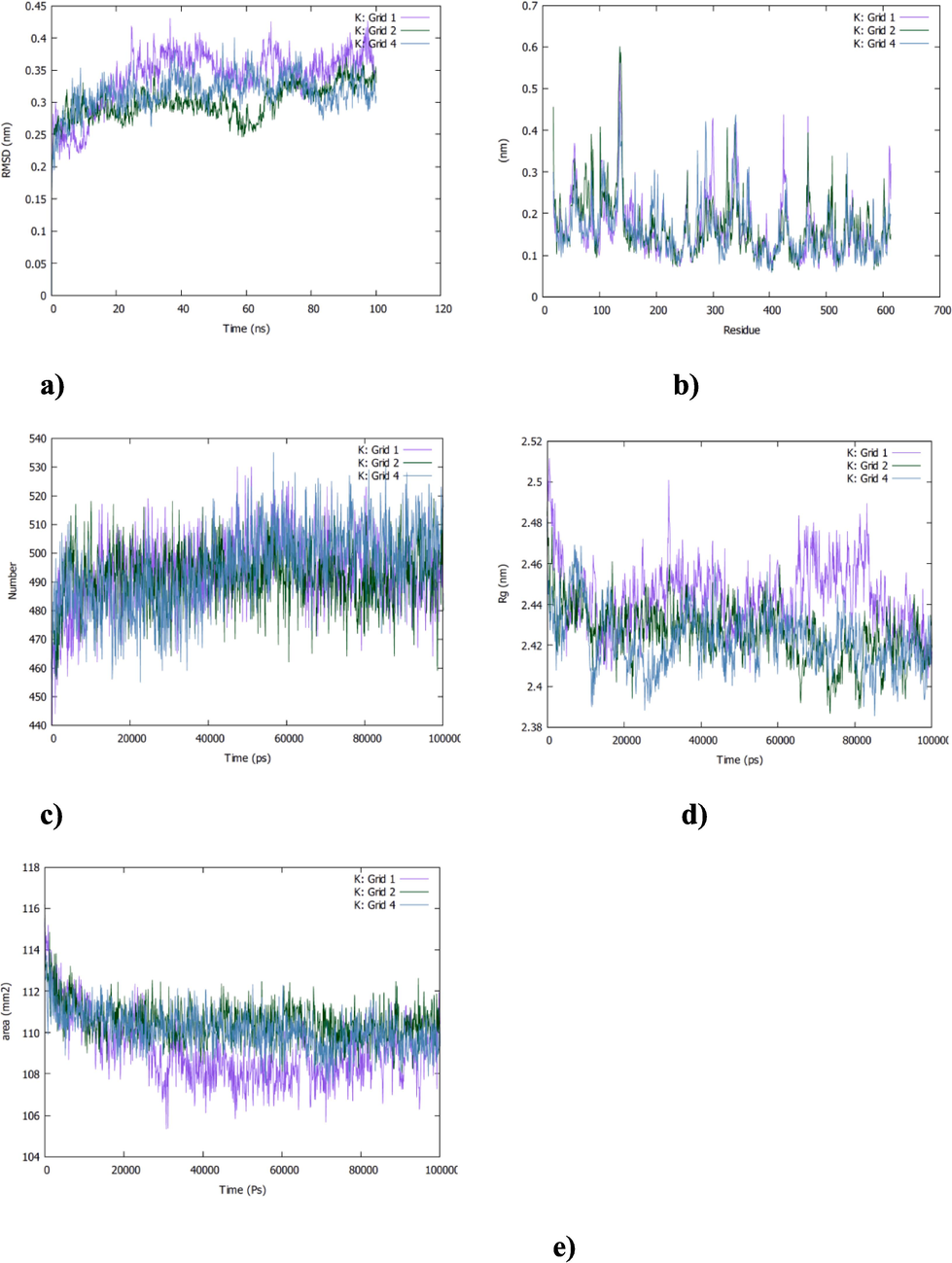
The MD simulation of vitamin K1-Grid1,-Grid2 and -Grid 4 complexes. a) RMSD, b) RMSF, c) H-bonds, d) Rg and e) SASA analysis.
As reported above vitamin D showed the highest affinities for the grid 1, 2 and 3. RMSD analysis for these three complexes shows more stability for vitamin D-grid 2 than vitamin D-grid 1 and -grid 3. Comparatively vitamin D-grid 2 system exhibits the lowest RMSD value (3 A˚ vs 4 A˚ for the vitamin D-grid 1 and 3) and a plateau curve from the beginning of the simulation (Fig. 6,a). The curve becomes in the equilibrated state after 25 ns of simulation for the vitamin D-grid1 and -grid3 systems (Fig. 6,a). As can be expected vitamin D-grid 1 and -grid 3 complexes show more fluctuations in RMSF values indicating more mobility of residues in these two systems than the other one (Fig. 6,b). Additionally, the number of H-bonds, the Rg and the SASA analysis confirmed the overall stability of vitamin D-grid 2 complex. Lower number of H–bonds and greater deviations are noted for Rg and SASA parameters curves in the case of vitamin D-grid 1 and -grid 3 systems (Fig. 6, c, d and e).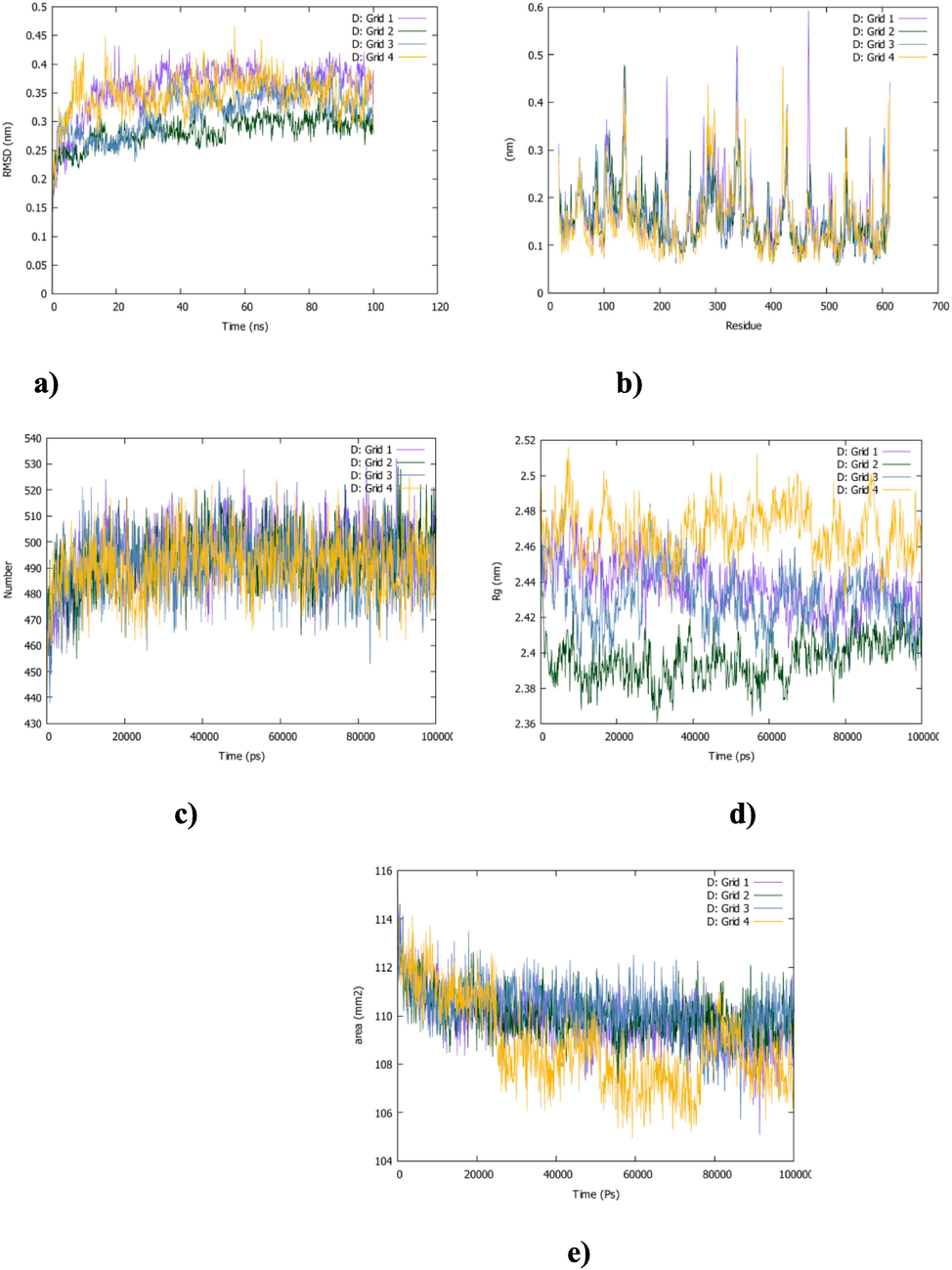
The MD simulation of vitamin D-Grid1,-Grid2,-Grid3 and -Grid 4 complexes. a) RMSD, b) RMSF, c)H-bonds, d) Rg and e) SASA analysis.
4 Conclusion
In the present work, computational approaches were used to study vitamins interactions with ACE2r. Firstly the virtual screening based on molecular docking protocol allowed selecting vitamins K1, A, D and E as the best docked to four ACE2 grid boxes binding Omicron proteins. Furthermore, the molecular dynamics simulation approach was used to infer with the stability, the flexibility, and the compactness of complexes. During the molecular simulation RMSD, RMSF, SASA, Rg and the number of H-bonds analysis indicate that these vitamins are in more stable binding with ACE2r. All the results suggest that vitamins could be considered as potential natural inhibitors compounds to the Omicron virus.
Author contribution statement
Belkheir K conceived and designed research. Laref N conducted computational experiments. Belkheir K analyzed data and wrote the manuscript. Belkheir K and Laref N authors read and approved the manuscript.
Acknowledgements
Authors would like to thank simlab.uams.edu on providing a free license to use GROMACS simulation package for performing the molecular dynamic simulation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19-25.
- [Google Scholar]
- A review of methods available to estimate solvent-accessible surface areas of soluble proteins in the folded and unfolded states. Curr. Protein Pept. Sci.. 2014;15(3):1-21.
- [Google Scholar]
- In silico screening of bioactive compounds of representative seaweeds to inhibit SARS-CoV-2 ACE2-bound Omicron B.1.1.529 spike protein trimer. Mar. Drugs. 2022;20(148)
- [CrossRef] [Google Scholar]
- Biovia D S (2015) Discovery Studio Modeling Environment. Dassault Syst. Release, San Diego, 4.
- SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185:467-484.
- [Google Scholar]
- AutoDock Vina 1.2.0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model.. 2021;61(8):3891-3898.
- [Google Scholar]
- Repurposing the natural compounds as potential therapeutic agents for COVID–19 based on the molecular docking study of the main protease and the receptor–binding domain of spike protein. J. Mol. Model.. 2022;28(153)
- [CrossRef] [Google Scholar]
- Molecular docking and dynamics studies of Nicotinamide Riboside as a potential multi-target nutraceutical against SARS-CoV-2 entry, replication, and transcription: A new insight. J. Mol. Struct.. 2022;1247(131394)
- [Google Scholar]
- Calculating an optimal box size for ligand docking and virtual screening against experimental and predicted binding pockets. J. Cheminform.. 2015;15(7):18. PMID: 26082804; PMCID: PMC4468813
- [CrossRef] [Google Scholar]
- Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARSCoV-2. Cell. 2022;185:630-640.
- [Google Scholar]
- Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin.. 2020;41:1141-1149.
- [CrossRef] [Google Scholar]
- Potential inhibitory effect of vitamins against COVID-19. Comput. Mater. Continua. 2020;66(1):707-721.
- [CrossRef] [Google Scholar]
- A review on the antiviral activity of functional foods against COVID-19 and viral respiratory tract infections. Int. J. Gen. Med.. 2022;15:4817-4835.
- [Google Scholar]
- The Omicron (B.1.1.529) variant of SARS-CoV-2 binds to the hACE2 receptor more strongly and escapes the antibody response: Insights from structural and simulation data. Int. J. Biol. Macromol.. 2022;200:438-448.
- [Google Scholar]
- Role of vitamins and minerals as immunity boosters in COVID–19. Infammopharmacology. 2021;29:1001-1016.
- [CrossRef] [Google Scholar]
- Mutations on RBD of SARS-CoV-2 Omicron variant result in stronger binding to human ACE2 receptor. Biochem. Biophys. Res. Commun.. 2022;590:34-41.
- [Google Scholar]
- SARS-CoV-2 Omicron variant: Antibody evasion and cryo-EM structure of spike protein–ACE2 complex. Science. 2022;375:760-764.
- [Google Scholar]
- Exploring the immune-boosting functions of vitamins and minerals as nutritional food bioactive compounds: A comprehensive review. Molecules. 2022;27:555.
- [CrossRef] [Google Scholar]
- Physical activity and nutritional influence on immune function: An important strategy to improve immunity and health status. Front. Physiol.. 2021;12:751374
- [CrossRef] [Google Scholar]
- Shoemark D K, Colenso C K, Toelzer C, Gupta K et al (2021) Molecular Simulations suggest Vitamins, Retinoids and Steroids as Ligands of the Free Fatty Acid Pocket of the SARS-CoV-2 Spike Protein. Angew. Chem. Int. Ed 60: 7098– 7110, International Ed doi.org/10.1002/anie.202015639, German Ed doi.org/10.1002/ange.202015639.
- Potential inhibitors of SARS-CoV-2 (COVID 19) spike protein of the delta and delta plus variant: In silico studies of medicinal plants of North-East India. Curr. Res. Pharmacol. Drug Discovery. 2021;2(100065)
- [Google Scholar]
- Role of food nutrients and supplementation in fighting against viral infections and boosting immunity: A review. Trends Food Sci. Technol.. 2021;110:66-77.
- [Google Scholar]
- Thompson M A (2004). Molecular docking using ArgusLab, an efficient shape-based search algorithm and the AScore scoring function. ACS meeting, Philadelphia, 172, CINF 42, PA.
- Changes in the immune response against SARS-CoV-2 in individuals with severe COVID-19 treated with high dose of vitamin D. Biomed. Pharmacother.. 2022;150(112965)
- [Google Scholar]
- AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem.. 2010;31(2):455-461.
- [Google Scholar]
- In silico study on the effect of SARS-CoV-2 RBD hotspot mutants’ interaction with ACE2 to understand the binding affinity and stability. Virology. 2021;561:107-116.
- [Google Scholar]
- Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894-904.
- [Google Scholar]
- Network pharmacology reveals that resveratrol can alleviate COVID-19-related hyperinflammation. Hindawi Disease Markers 2021:4129993.
- [CrossRef] [Google Scholar]
Appendix A
Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2023.103082.
Appendix A
Supplementary material
The following are the Supplementary data to this article:







