Translate this page into:
In-silico analysis of phylogenetic relationship and potentially damaging nsSNPs in human SLC2A2 gene
⁎Corresponding author. mamoona.noreen@gmail.com (Mamoona Noreen)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Transport of glucose across the eukaryotic cell membranes is carried out by members of the glucose transporter (GLUT) family which is mainly divided into three classes (I, II and III) on the basis of phylogenetic relationship. In humans, one member of Class I called GLUT2 is encoded by solute carrier family 2, facilitated glucose transporter member 2 (SLC2A2) gene located on third chromosome. Protein mediated glucose movement across cell membranes is made possible through GLUT2 that is a trans-membrane carrier protein. It regulates the entry of glucose and secretion of insulin in the pancreatic cell. It has three isoforms and the longest isoform consists of 524 amino acid. There are 13 extracellular, 12 transmembrane and 5 cytoplasmic domains in human GLUT2. The risk of Fanconi-Bickel syndrome (FBS), diabetes, breast cancer (BC) and Alzheimer disease (AD) is associated with improper functioning of GLUT2. The most frequent form of genetic changes is single nucleotide polymorphism (SNPs). Non synonymous SNPs (nsSNPs) can result in alterations of amino acids and subsequent changes in phenotype. In this study, in-silico analysis was done to find phylogenetic relationship of human GLUT2 protein and possible deleterious effect of nsSNPs of its coding region. Clustal Omega was used to make phylogenetic tree. SIFT, PolyPhen, PROVEAN and SNPeffect were used to predict deleterious or tolerated SNPs. 167 nsSNPs were predicted to be damaging by SIFT, 65 to be possibly damaging and 77 to be probably damaging by PolyPhen. PROVEAN predicted 162 nsSNPs to be neutral and 138 to be deleterious. 101 SNPs were found to be damaging by three algorithms; SIFT, PolyPhen and PROVEAN.
Keywords
GLUT2
SLC2A2 gene
In silico analysis
nsSNP
Phylogenetic analysis
1 Introduction
Glucose is a major energy source and is an important substrate for both protein and lipid synthesis in mammalian cells. Through glycolysis and the citric acid cycle it supplies energy in the form of adenosine tri phosphate (ATP). In the form of nicotinamide adenine dinucleotide phosphate (NADPH), it provides reducing power through the pentose phosphate shunt. Glucose plays a vital role in retaining cellular homeostasis and metabolic functions. For generating ATP molecules, every vertebrate cell depends on the continuous delivery of glucose (Cant et al., 2002). Glucose transport across the plasma membranes is done by two distinct processes depending on the type of cells and tissues. First is facilitative transport that is interceded by a group of facilitative glucose transporters (GLUT) (Mueckler, 1994; Joost and Thorens, 2001) and the other is sodium dependent transport that is interceded by the Na+/glucose linked transporters (SGLT) (Wright, 2001).
GLUTs are proteins of ∼500 amino acids and are estimated to have 12 transmembrane-spanning alpha helices and a single N-linked oligosaccharide. Based on the phylogenetic analysis and sequence similarities 14 members of GLUT family can be divided into three different classes; Class I, II and III (Thorens and Mueckler, 2010; Joost et al., 2002). Class I includes GLUT 1, 2, 3, 4 and 14, class II includes GLUT 5, 7, 9 and 11 while GLUT 6, 8, 10, 12 and 13 are classified as class III (Manolescu et al., 2007). In humans the solute carrier family 2 (facilitated glucose transporter), member 2 (SLC2A2) gene is located on q26.2 of chromosome 3 (Fig. 1) which encodes GLUT2 protein. There are three isoforms of GLUT2; NP_000331.1, NP_001265587.1 and NP_001265588.1.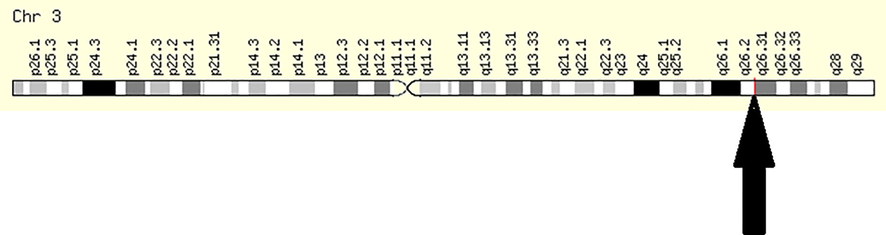
Location of GLUT2 on human chromosome. Arrow head shows that GLUT2 occupies 3q26.2 on third human chromosome. (https://www.genecards.org/cgi-bin/carddisp.pl?gene=SLC2A2).
The GLUT2 is a transmembrane carrier protein and it permits protein facilitated glucose movement across cell membranes. GLUT2 is involved in discharge of absorbed or reabsorbed glucose and is mainly expressed in the kidney and intestinal absorptive epithelial cells of basolateral membrane (Wright et al., 2003; Kellett and Brot-Laroche, 2005). GLUT2 is also present in the liver, brain and pancreas. It serves as major glucose transporter for the islet cells. As GLUT2 is a high capacity transporter, the concentration of glucose in the cell is directly proportional to the extracellular level of glucose. Enhanced level of intracellular glucose is supposed to increase the ATP/ADP ratio which will shut ATP sensitive potassium channels, depolarize the cells and secrete insulin (Schuit et al., 2001).
In hepatocytes, as a result of gluconeogenesis, GLUT2 supports the uptake of glucose into the blood. It also yields insulin secretion as glucose-sensing functions in the pancreatic β-cell and hypothalamus as low level of glucose stimulated insulin was described in GLUT2 knockout mice (Guillam et al., 1997; Burcelin et al., 2001; Bady et al., 2006). In β-cells of pancreatic islets and hepatocytes, the major glucose transporter is GLUT2. In both cell types, GLUT2 helps the facilitated diffusion of glucose across the cell membranes, and then intracellular glucose metabolism is initiated by the glucose-phosphorylating enzyme, hexokinase IV or glucokinase. In the β-cells, the rate of glucose metabolism controls insulin secretion, whereas in the liver, glucose metabolism and transport are essential to subsequent glycogen synthesis and gluconeogenesis (Meglasson et al., 1986; Thorens et al., 1988).
Fanconi Bickel Syndrome (FBS) is an autosomal recessive disease of glucose metabolism characterized by accumulation of hepatorenal glycogen, Fanconi nephropathy, and impaired use of glucose and galactose (Fanconi, 1949; Santer et al., 1998). The risk of diabetes has been found to be associated with SNPs of SLC2A2 gene in some (Barroso et al., 2003; Alcolado and Alcolado, 1991) but not in other studies (Matsutani et al.,1990). Globally, breast cancer (BC) is the most frequent cancer among women (Ilahi et al., 2016; Noreen et al., 2015a, 2015b). One way to stimulate growth of cancer cells may be fructose metabolism and the expression of GLUT5 and GLUT2 in BC is elevated and helpful in fructose metabolism (Godoy et al., 2006). The increased level of GLUT2 was observed in patients of Alzheimer disease (AD) (Liu et al., 2008).
Diseases susceptibility and development is affected by presence of genetic variations, one very common type of which is single nucleotide polymorphism or SNPs. In DNA, SNPs are found in both exonic and intronic regions (Noreen et al. 2012). Non synonymous SNPs (nsSNPs) residing in exonic region of protein involve amino acid substitution which can result in alteration of structure and functioning of the protein. Thus nsSNPs can play more significant role in diseases development. In this study we predicted the structural and functional effect of potential genetic variations of human SLC2A2 gene by using some sequence and structural homology based algorithms.
2 Materials and methods
2.1 Dataset compilation
National centre for biological information; NCBI (http://www.ncbi. nlm.nih.gov) was accessed. Protein and nucleotide sequences were acquired in FASTA format. Public SNP database named as dbSNP (http://www. ncbi.nlm.nih.gov/SNP), Gene Cards (http://www.genecards.org/) and UniProt (http://www.uniprot.org) were searched to assemble information related to longest isoform of GLUT2 regarding protein sequence. (GLUT2 gene ID: 6514 SNPs NCBI Reference Sequence: NP_000331). The coding region nsSNPs were screened and were subjected to further computational analysis.
2.2 Phylogenetic analysis
To carry out phylogenetic analysis, protein sequence of human GLUT2 (GenBank Accession Number: NP 000331.1) along other 9 species of Hominidae family; Gorilla gorillagorilla (XP_004038053.1), Pongo abelli (XP_002814322.1), Rhinopithecus roxellana (XP_010359570.1), Microcebus murinus (XP_012591846.1), Bos taurus (DAA33241.1), Myotis brandtii (XP_005886638.1), Neomonachus schauinsland (XP_021558267.1), Galeopterus variegates (XP_008586350.1) and Ailuropoda melanoleuca (XP_002920886.1) were retrieved from the NCBI. Protein sequence in FASTA format of all 10 species was obtained and saved. Protein sequence alignment was performed using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). In Clustal Omega, output format “ClustalW with charater counts” was chosen and option of phylogenetic tree was clicked.
2.3 Membrane topology
The number of machine learning algorithms wre used to align the set of protein sequences and clustering of topological data. These approaches or algorithms are transmembrane helices prediction (PHDhtm) (Rost et al. 1996), statistical algorithm TMAP (Persson and Argos 1994) and Hidden Markov models (HMMs) (HMMTOP (Tusnady and Simon, 1998, 2001); TMHMM (Sonnhammer et al., 1998; Krogh et al., 2001). The membrane topology of GLUT2 protein was predicted with the TMHMM program, server 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
2.4 Analysis of nsSNPs by SIFT
Sorting Intolerant from Tolerant (SIFT) is an algorithm that forecasts whether substitution of an amino acid influences protein function or not. It is based on the principle that essential amino acids in a protein family remain conserved, and any replacement at these positions is deleterious, which influences function/activity of protein. Protein sequence in FASTA format along substitution of amino acids was used as input at (http://www.blocks.fhcrc.org/sift/SIFT.html). It then considered the normalized probability for the provided substitution at the related position in the alignment. Substitutions with the normalized probability i.e. Tolerance Index (TI) less than 0.05 were forecasted to be intolerant or harmful whereas substitutions with TI higher than 0.05 were predicted to be tolerant (Adzhubei et al., 2010).
2.5 Analysis of nsSNPs by using PolyPhen
PolyPhen-2 is a tool for forecasting of the feasible impact of an amino acid substitution on the protein structure and function (Adzhubei et al., 2010). Automated divinations of this type are vital for interpreting rare genetic variants, which may have potential approach in recent research of human genetics. Its importance in modern research involves identification of rare alleles that generate Mendelian disease (Bamshad et al., 2011), scanning for medically important alleles in an individual’s genome (Ashley et al., 2010), and profiling the spectrum of rare variation uncovered by deep sequencing of large populations (Tennessen et al., 2012).
PolyPhen (http://genetics.bwh.harvard.edu/pph2) utilizes UniProtKB database which is used as source of reference. Protein sequence of GLUT2 in FASTA format and position of protein sequence was entered. AA1 wild type (query sequence) as well as substitution reside AA2 were selected. Two pair’s datasets were used. First pair of HumDiv and the second pair; HumVar were compiled in UniProt database.
2.6 Analysis of nsSNPs by using PROVEAN
PROVEAN is not only effective to predict single amino acid change but it can also perform task for other changes in the protein sequence including indels (deletion and insertion) (Choi et al., 2012). The PROVEAN web server (http://provean.jcvi.org) gives online approach for the functioning of distribution for the software package of stand-alone. Its major role is to give divination from any of organism’s protein sequence. It takes the sequence of protein and variation of amino acid as input. BLAST search was done to recognize the homologous sequence and produce the PROVEAN result. Generally the BLAST search took 10–20 min to produce the divination for a provided protein query. Sequence identifiers list for supporting sequences and clustering data was stored in database. Focused on sequence of query protein consecutive prediction data for supporting sequences were indexed.
2.7 Analysis of nsSNPs by using SNPeffect
SNPeffect (De Baets et al., 2012) was used to predict the molecular phenotypic influence of nsSNPs lying in coding region of GLUT2 protein. It works beyond the scores got on conservational basis and mainly emphasizes to map the effect of SNPs on the capability of cells to uphold suitable concentration of the properly folded proteins in appropriate cellular region i.e. protein homeostasis landscape (Powers et al., 2009). For this assessment, wild type protein sequence in FASTA format along with each of its variants was given to the SNPeffect server (http://snpeffect.switchlab.org) for analysis. Homology threshold was fixed to 90%. It uses TANGO (Fernandez-Escamilla et al., 2004) which forecasts the regions in given protein sequence that are prone to aggregation and calculated TANGO score with wild and variant amino acid. On the basis of TANGO score difference (dTANGO), the server assesses effect of these variants on protein aggregation. Aggregate morphology is more distinctively evaluated by WALTZ server (Maurer-Stroh et al., 2010) that forecasts amyloid-forming regions in given protein sequence with accuracy and specificity on basis of dWALTZ score. Chaperone binding propensity is forecasted by LIMBO (Van Durme et al., 2009) for the Hsp70 chaperones and effect of variant is determined by dLIMBO score. SNPeffect also uses high resolution crystal structure of proteins from Protein Data Bank (PDB) (Deshpande et al., 2005) and models the variants using the empirical force field FoldX (version 2.5) (Schymkowitz et al., 2005) to determine possible effects on stability and binding properties of given protein.
3 Results
3.1 Phylogenetic analysis
Evolutionary relationship of GLUT2 protein among 10 different species of primates was carried out using human GLUT2 protein as a reference sequence (Fig. 2). The phylogenetic tree generated from Clustal Omega helps to understand the evolutionary relationship of GLUT2 among different species. Evolutionary tree demonstrates that GLUT2 of human and Pongo abelli lie close to each other whereas that of Bos taurusis most distant from human.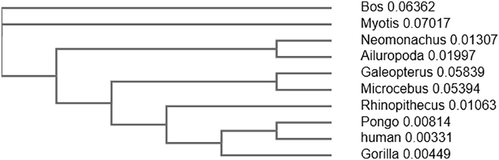
Phylogenetic analysis of human GLUT2 protein. A phylogenetic tree of the amino acid sequences of GLUT2 protein was constructed using the Clustal Omega.
3.2 Membrane topology
A significant server TMHMM was used to predict the significant features of helices in transmembrane protein and GLUT2 was found to comprise of 12 predicted transmembrane helices with intracellular amino (N-terminal) and carboxyl (C-termini) in cytosol. It also contains extracellular and intracellular loops which are located between the segments of 1st and 2nd transmem brane domains and between 6th and 7th transmembrane segments (Fig. 3). While, the remaining transmembrane segments from seven to twelve have ability to perform transportation of fructose and glucose to the membrane.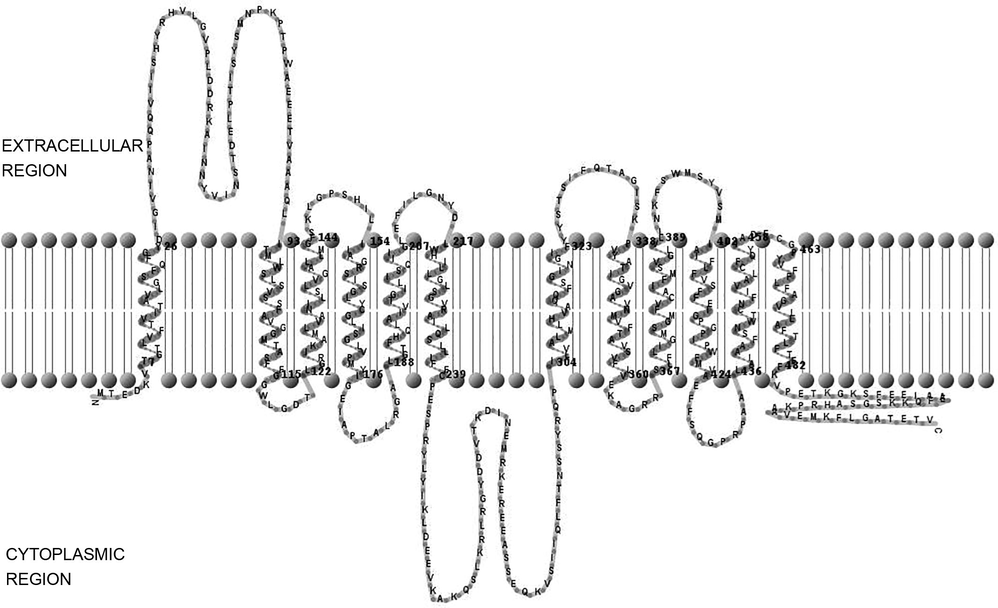
Toplogy prediction of human GLUT2 protein. The human GLUT2 protein comprises of 12 predicted transmembrane helices with intracellular amino (N-terminal) and carboxyl (C-termini) in cytosol. It also contains extracellular and intracellular loops which are located between the segments of 1st and 2nd transmembrane domains and between 6th and 7th transmembrane segments.
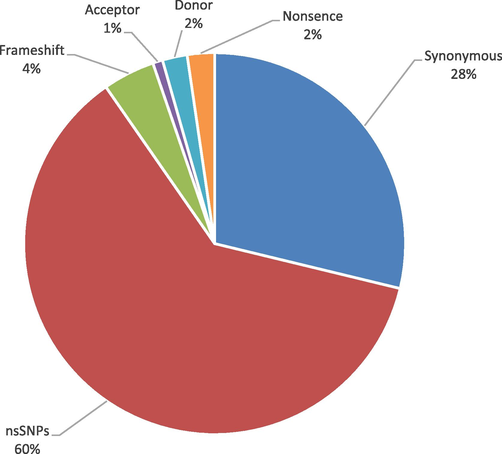
3.3 Distribution of SNPs in GLUT2 protein
In the human SLC2A2 gene the non coding region of gene contains 6,628 SNPs in which 83 SNPs were near 3′gene, 340 in 3′UTR, 427 SNPs in 5′ near gene, 62 in 5′UTR and in the intron 5716 SNPs were found (Fig. 4). While the coding region contained 484 SNPs in which 137 SNPs were synonymous, 293 were nsSNPs, 21 were frameshifts; (rs766082034), (rs1447936042), (rs769888108), (rs1255595607), (rs776307487), (rs1162215911), (rs771361095), (rs1326032349), (rs1350704340), (rs1290975016), (rs765132996), (rs1181030797), (rs771799491), (rs746178753), (rs1316522125), (rs748296868), (rs34066960), (rs1174349159), (rs1384674663),(rs1386374799) and (rs1290412048). 11 were nonsense; (rs774841662), (rs771477447), (rs1475086161), (rs1114167428), (rs753629940), (rs773581866), (rs121909746), (rs121909743), (rs121909742), (rs121909745) and (rs1379944645). Out of 7 SNPs, 4 were inframe deletion; (rs772999215), (rs1169887677), (rs763620441), (rs774721090) and 3 were inframe insertion; (rs1161394690), (rs1463507753) and (rs749789723). 10 (rs1294679246), (rs756163471), (rs371977235), (rs1240053337), (rs756874949), (rs1303795800), (rs1281471314), (rs985090030), (rs757587931), (rs867530965) were donor and 4 (rs754220999), (rs1318756243), (rs776248984), (rs1230247311) were acceptor shown (Fig. 5). (rs749789723) SNPs also contain stop gained. 1 SNPs was stop lost (rs750463981). 10 additional SNPs were detected by dbSNP in which 4 SNPs; (rs766082034), (rs1255595607), (rs1162215911), (rs746178753) were frameshifts and 6; (rs766762468), (rs1800572), (rs777020657), (rs867996396), (rs121909747), (rs1309197020) were nsSNPs (Table 1)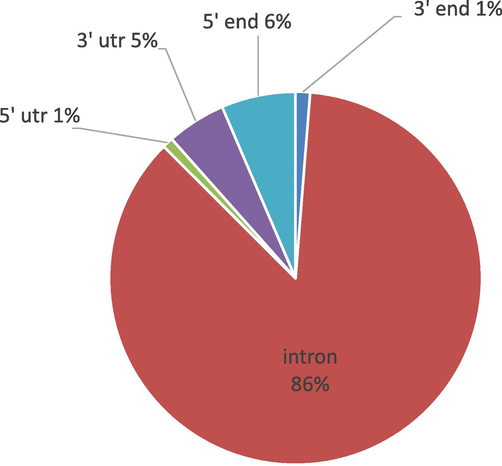
Distribution of SNPs in human GLUT2 Protein.
Distribution of nsSNPs in coding region of GLUT2 protein.
dbSNP id
Function
dbSNPallele
Codonpos.
mRNApos.
Proteinresidue
Amino acidpos
rs759495940
nsSNPs
G /A
3
312
Met [M]/Ile [I]
1
rs773205789
nsSNPs
A /C
1
313
Thr [T]/Pro [P]
2
rs200073044
nsSNPs
G /C
1
319
Asp [D]/His [H]
4
rs1372689853
nsSNPs
A /G
1
322
Lys [K]/Glu [E]
5
rs767313610
nsSNPs
G /A
1
325
Val [V]/Ile [I]
6
rs369700669
nsSNPs
C /A
2
329
Thr [T]/Asn [N]
7
rs766082034
frame shift
G /-
3
333
Thr [T]/Pro [P]
9
rs766082034
frameshift
G/-
3
333
(Gly)G/(Gly)G
9
rs1181030797
frame shift
G /-
1
340
Val [V]/Phe [F]
11
rs1481905618
nsSNPs
C /A
3
345
Phe [F]/Leu [L]
12
rs1247912820
nsSNPs
C /A
2
347
Thr [T]/Asn [N]
13
rs1247912820
nsSNPs
C /G
2
347
Thr [T]/Ser [S]
13
rs372441014
nsSNPs
G /C
1
349
Val [V]/Leu [L]
14
rs766364438
nsSNPs
T /C
2
353
Ile [I]/Thr [T]
15
rs1463507753
INFRAME INSERTION
TCA
1
355
Ile[I] /IleIle[II]
15
rs867315854
nsSNPs
G /A
1
358
Ala [A]/Thr [T]
17
rs761992056
nsSNPs
G /A
2
368
Gly [G]/Asp [D]
20
rs369781481
nsSNPs
C /A
3
375
Phe [F]/Leu [L]
22
rs1409436045
nsSNPs
T /A
2
380
Phe [F]/Tyr [Y]
24
rs1049223265
nsSNPs
T /C
2
392
Ile [I]/Thr [T]
28
rs1437312005
nsSNPs
G /A
1
394
Gly [G]/Ser [S]
29
rs1437312005
nsSNPs
G /C
1
394
Gly [G]/Arg [R]
29
rs775531825
nsSNPs
A /G
2
404
Asn [N]/Ser [S]
32
rs143528640
nsSNPs
G /A
1
406
Ala [A]/Thr [T]
33
rs746158263
nsSNPs
C /A
1
409
Pro [P]/Thr [T]
34
rs774841662
nonsense
C/ T
1
412
Gln [Q]/
35
rs1158195535
nsSNPs
C /A
1
415
Gln [Q]/Lys [K]
36
rs1158195535
nsSNPs
C /G
1
415
Gln [Q]/Glu [E]
36
rs1451394300
nsSNPs
G /A
1
418
Val [V]/Ile [I]
37
rs1477523180
nsSNPs
A /C
1
421
Ile [I]/Leu [L]
38
rs1360464436
nsSNPs
T /A
2
422
Ile [I]/Lys [K]
38
rs1176350402
nsSNPs
T /C
2
425
Ile [I]/Thr [T]
39
rs772999215
Inframe Deletion
II [ATA] > I []
1
424
IleIle[II] / Ile [I]
39
rs758670698
nsSNPs
G /T
2
437
Arg [R]/Ile [I]
43
rs760618624
nsSNPs
C /T
1
439
His [H]/Tyr [Y]
44
rs775791143
nsSNPs
G /A
1
442
Val [V]/Ile [I]
45
rs149108283
nsSNPs
T /A
2
443
Val [V]/Asp [D]
45
rs149108283
nsSNPs
T /C
2
443
Val [V]/Ala [A]
45
rs1159338702
nsSNPs
G /A
1
448
Gly [G]/Ser [S]
47
rs561765982
nsSNPs
G /A
1
451
Val [V]/Ile [I]
48
rs983907950
nsSNPs
C /T
1
454
Pro [P]/Ser [S]
49
rs1211075508
nsSNPs
T /C
2
458
Leu [L]/Pro [P]
50
rs1311902495
nsSNPs
G /A
1
463
Asp [D]/Asn [N]
52
rs1311902495
nsSNPs
G /T
1
463
Asp [D]/Tyr [Y]
52
rs771477447
nonsense
C /T
1
466
Arg [R]/
53
rs771477447
nsSNPs
C /G
1
466
Arg [R]/Gly [G]
53
rs145210664
nsSNPs
G /A
2
467
Arg [R]/Gln [Q]
53
rs546539032
nsSNPs
A /C
3
471
Lys [K]/Asn [N]
54
rs747555903
nsSNPs
G /A
1
487
Val [V]/Ile [I]
60
rs1447936042
frame shift
- /(26 bp)
1
490
Ile [I]/Thr [T]
61
rs977284195
nsSNPs
T /C
2
491
Ile [I]/Thr [T]
61
rs780903829
nsSNPs
C /G
3
492
Ile [I]/Met [M]
61
rs1373290524
nsSNPs
G /C
2
497
Ser [S]/Thr [T]
63
rs1310901426
nsSNPs
A /G
1
499
Thr [T]/Ala [A]
64
rs1169887677
Inframe Deletion
INS [ATCA] / [ACA]
2
491
INS (IleAsnSer) / ()
61
rs1354126805
nsSNPs
C /G
2
500
Thr [T]/Arg [R]
64
rs754585542
nsSNPs
G /A
1
502
Asp [D]/Asn [N]
65
rs1217666649
nsSNPs
T /A
3
504
Asp [D]/Glu [E]
65
rs1391257598
nsSNPs
C /A
1
511
Pro [P]/Thr [T]
68
rs7637863
nsSNPs
C /T
2
512
Pro [P]/Leu [L]
68
rs779977931
nsSNPs
C /A
2
515
Thr [T]/Lys [K]
69
rs1182852354
nsSNPs
A /G
1
517
Ile [I]/Val [V]
70
rs750405382
nsSNPs
A /G
1
529
Met [M]/Val [V]
74
rs1207297111
nsSNPs
A /G
1
532
Asn [N]/Asp [D]
75
rs1342979475
nsSNPs
A /C
2
533
Asn [N]/Thr [T]
75
rs1274084408
nsSNPs
C /G
1
535
Pro [P]/Ala [A]
76
rs778655073
nsSNPs
C /T
2
542
Pro [P]/Leu [L]
78
rs769888108
frame shift
C /-
2
548
Pro [P]/Leu [L]
80
rs549263048
nsSNPs
T /C
1
550
Trp [W]/Arg [R]
81
rs531049536
nsSNPs
G /T
3
552
Trp [W]/Cys [C]
81
rs150851401
nsSNPs
G /A
1
556
Glu [E]/Lys [K]
83
rs766762468
nsSNPs
C /G
2
566
Thr [T]/Ser [S]
86
rs766762468
nsSNPs
C/T
2
566
T (Thr) > I (Ile)
86
rs763255363
nsSNPs
G /T
1
571
Ala [A]/Ser [S]
88
rs144715667
nsSNPs
A /G
1
586
Ile [I]/Val [V]
93
rs1415169647
nsSNPs
T /G
2
593
Met [M]/Arg [R]
95
rs1407375423
nsSNPs
G /T
3
600
Trp [W]/Cys [C]
97
rs1800572
nsSNPs
G /A
1
610
Val [V]/Ile [I]
101
rs1800572
nsSNPs
G/C
1
610
Val [V] / Leu [L]
101
rs770135219
nsSNPs
T /C
2
611
Val [V]/Ala [A]
101
rs1399091893
nsSNPs
C /T
2
623
Ala [A]/Val [V]
105
rs1332764085
nsSNPs
G /A
1
625
Val [V]/Ile [I]
106
rs5400
nsSNPs
C /T
2
638
Thr [T]/Ile [I]
110
rs377238940
nsSNPs
G /C
1
640
Ala [A]/Pro [P]
111
rs1475086161
nonsense
C /G
2
644
Ser [S]/
112
rs1255595607
frame shift
C /-
3
648
Phe [F]/Leu [L]
114
rs1255595607
frame shift
C /-
3
648
Phe [F]/ Phe [F]
114
rs768407637
nsSNPs
G /T
2
656
Gly [G]/Val [V]
116
rs753980727
nsSNPs
G /T
3
660
Trp [W]/Cys [C]
117
rs746632604
nsSNPs
G /C
2
665
Gly [G]/Ala [A]
119
rs772002572
nsSNPs
C /T
2
671
Thr [T]/Ile [I]
121
rs1476520648
nsSNPs
T /A
2
683
Ile [I]/Asn [N]
125
rs760201098
nsSNPs
C /A
2
689
Ala [A]/Asp [D]
127
rs1267904495
nsSNPs
T /C
2
692
Met [M]/Thr [T]
128
rs1212980167
nsSNPs
G /C
3
693
Met [M]/Ile [I]
128
rs367856967
nsSNPs
T /C
2
698
Val [V]/Ala [A]
130
rs970550665
nsSNPs
C /G
2
701
Ala [A]/Gly [G]
131
rs775283150
nsSNPs
A /T
1
706
Ile [I]/Phe [F]
133
rs913419413
nsSNPs
T /G
2
710
Leu [L]/Arg [R]
134
rs771843187
nsSNPs
T /C
1
712
Ser [S]/Pro [P]
135
rs144125084
nsSNPs
G /C
1
718
Val [V]/Leu [L]
137
rs993833041
nsSNPs
T /C
2
719
Val [V]/Ala [A]
137
rs1300072764
nsSNPs
G /A
2
722
Gly [G]/Glu [E]
138
rs778548964
nsSNPs
C /T
1
727
Leu [L]/Phe [F]
140
rs776307487
frame shift
T /-
2
728
Leu [L]/Pro [P]
140
rs1016384738
nsSNPs
T /A
2
734
Met [M]/Lys [K]
142
rs770941010
nsSNPs
G /C
1
736
Gly [G]/Arg [R]
143
rs777718289
nsSNPs
G /A
2
752
Gly [G]/Glu [E]
148
rs777718289
nsSNPs
G /T
2
752
Gly [G]/Val [V]
148
rs372621339
nsSNPs
C /G
2
755
Pro [P]/Arg [R]
149
rs747025551
nsSNPs
T /C
2
764
Ile [I]/Thr [T]
152
rs376064965
nsSNPs
C /T
1
766
Leu [L]/Phe [F]
153
rs1188886679
nsSNPs
A /C
1
769
Ile [I]/Leu [L]
154
rs1188886679
nsSNPs
A /G
1
769
Ile [I]/Val [V]
154
rs192720796
nsSNPs
T /A
2
770
Ile [I]/Lys [K]
154
rs763620441
INFRAME DELETION
ILI [CTTATA] / I []
1
766
IleLeuIle [ILI] / Ile [I]
153
rs910976682
nsSNPs
C /T
2
776
Ala [A]/Val [V]
156
rs750836049
nsSNPs
G /C
1
778
Gly [G]/Arg [R]
157
rs1278964539
nsSNPs
G /A
2
782
Arg [R]/Lys [K]
158
rs1231468128
nsSNPs
T /A
2
797
Leu [L]/Gln [Q]
163
rs1445887606
nsSNPs
T /C
1
799
Tyr [Y]/His [H]
164
rs1271546287
nsSNPs
T /C
2
812
Ile [I]/Thr [T]
168
rs760095835
nsSNPs
G /A
2
818
Gly [G]/Asp [D]
170
rs1335888503
nsSNPs
G /C
1
823
Val [V]/Leu [L]
172
rs1293130515
nsSNPs
T /C
2
830
Met [M]/Thr [T]
174
rs1019696977
nsSNPs
A /G
1
835
Ile [I]/Val [V]
176
rs144822218
nsSNPs
G /A
1
838
Gly [G]/Ser [S]
177
rs759047405
nsSNPs
G /A
2
839
Gly [G]/Asp [D]
177
rs1441606652
nsSNPs
C /T
2
848
Ala [A]/Val [V]
180
rs368626129
nsSNPs
G /A
1
856
Ala [A]/Thr [T]
183
rs368626129
nsSNPs
G /T
1
856
Ala [A]/Ser [S]
183
rs771799491
frame shift
G /-
2
866
Gly [G]/Glu [E]
186
rs200213178
nsSNPs
G /A
1
868
Ala [A]/Thr [T]
187
rs200213178
nsSNPs
G /C
1
868
Ala [A]/Pro [P]
187
rs748052042
nsSNPs
C /T
1
871
Leu [L]/Phe [F]
188
rs1412289847
nsSNPs
G /A
1
874
Gly [G]/Ser [S]
189
rs776498787
nsSNPs
G /A
2
875
Gly [G]/Asp [D]
189
rs779065938
nsSNPs
C /G
1
889
Leu [L]/Val [V]
194
rs1469335096
nsSNPs
G /A
1
892
Ala [A]/Thr [T]
195
rs771182536
nsSNPs
T /A
2
896
Ile [I]/Asn [N]
196
rs771182536
nsSNPs
T /C
2
896
Ile [I]/Thr [T]
196
rs121909741
nsSNPs
G /A
1
898
Val [V]/Ile [I]
197
rs149460434
nsSNPs
C /A
2
902
Thr [T]/Lys [K]
198
rs149460434
nsSNPs
C /T
2
902
Thr [T]/Met [M]
198
rs1276756236
nsSNPs
C /A
1
910
Leu [L]/Ile [I]
201
rs779591826
nsSNPs
T /G
3
924
Ile [I]/Met [M]
205
rs1262860274
nsSNPs
T /C
2
926
Ile [I]/Thr [T]
206
rs1186359171
nsSNPs
G /A
1
928
Gly [G]/Ser [S]
207
rs1114167428
nonsense
G /T
1
934
Glu [E]/
209
rs1215469128
nsSNPs
T /C
2
944
Leu [L]/Ser [S]
212
rs1347267249
nsSNPs
A /C
1
949
Asn [N]/His [H]
214
rs573292685
nsSNPs
A /G
2
950
Asn [N]/Ser [S]
214
rs764799427
nsSNPs
A /T
2
956
Asp [D]/Val [V]
216
rs753629940
nonsense
G /A
3
963
Trp [W]/
218
rs1380319602
nsSNPs
T /A
2
968
Ile [I]/Asn [N]
220
rs760641937
nsSNPs
G /C
2
977
Gly [G]/Ala [A]
223
rs1413841367
nsSNPs
T /C
2
980
Leu [L]/Pro [P]
224
rs771075989
nsSNPs
G /A
1
988
Val [V]/Met [M]
227
rs773581866
nonsense
C /T
1
991
Arg [R]/
228
rs773581866
nsSNPs
C /G
1
991
Arg [R]/Gly [G]
228
rs770126214
nsSNPs
C /A
1
1000
Leu [L]/Ile [I]
231
rs1374154306
nsSNPs
T /A
2
1013
Leu [L]/Gln [Q]
235
rs748588515
nsSNPs
C /T
1
1015
Leu [L]/Phe [F]
236
rs1162215911
frame shift
CT /-
3
1017
Phe [F]/Leu [L]
238
rs1162215911
frame shift
CT /-
3
1017
Leu [L] /Leu[L]
238
rs757087261
nsSNPs
T /A
1
1021
Phe [F]/Ile [I]
238
rs777020657
nsSNPs
T /G
2
1022
Phe [F]/Cys [C]
238
rs777020657
nsSNPs
T/C
2
1022
Phe [F] /Ser[S]
238
rs769089021
nsSNPs
G /A
2
1034
Ser [S]/Asn [N]
242
rs780381836
nsSNPs
A /G
1
1039
Arg [R]/Gly [G]
244
rs1480881050
nsSNPs
T /C
1
1042
Tyr [Y]/His [H]
245
rs1250722271
nsSNPs
T /C
1
1048
Tyr [Y]/His [H]
247
rs1158317020
nsSNPs
A /T
2
1049
Tyr [Y]/Phe [F]
247
rs867996396
nsSNPs
T /A
2
1052
Ile [I]/Asn [N]
248
rs867996396
nsSNPs
T/C
2
1052
Ile [I] / Thr [T]
248
rs536261161
nsSNPs
G /C
3
1056
Lys [K]/Asn [N]
249
rs745373269
nsSNPs
G /A
1
1060
Asp [D]/Asn [N]
251
rs745373269
nsSNPs
G /C
1
1060
Asp [D]/His [H]
251
rs1367431424
nsSNPs
A /C
2
1064
Glu [E]/Ala [A]
252
rs865881030
nsSNPs
G /A
1
1066
Glu [E]/Lys [K]
253
rs778607566
nsSNPs
A /G
2
1067
Glu [E]/Gly [G]
253
rs1309254226
nsSNPs
A /C
3
1068
Glu [E]/Asp [D]
253
rs777225980
nsSNPs
C /A
3
1086
Ser [S]/Arg [R]
259
rs76026576
nsSNPs
G /T
3
1089
Leu [L]/Phe [F]
260
rs746178753
frame shift
AG /-
3
1101
Gly [G]/Ile [I]
265
rs746178753
frame shift
AG /-
3
1101
Arg [R]/ Arg[R]
265
rs1490504926
nsSNPs
G /A
1
1108
Asp [D]/Asn [N]
267
rs935009475
nsSNPs
T /A
3
1113
Asp [D]/Glu [E]
268
rs774721090
Inframe Deletion
DD [GATG] / D [GTC]
2
1112
AspAsp[DD]/ Asp [D]
268
rs140285191
nsSNPs
G /A
1
1123
Asp [D]/Asn [N]
272
rs140285191
nsSNPs
G /T
1
1123
Asp [D]/Tyr [Y]
272
rs754932741
nsSNPs
G /A
3
1137
Met [M]/Ile [I]
276
rs750579210
nsSNPs
A /G
1
1138
Arg [R]/Gly [G]
277
rs1304842107
nsSNPs
A /C
1
1141
Lys [K]/Gln [Q]
278
rs765426962
nsSNPs
A /C
2
1142
Lys [K]/Thr [T]
278
rs761756532
nsSNPs
A /C
3
1143
Lys [K]/Asn [N]
278
rs771361095
frame shift
-/A
1
1147
Arg [R]/Lys [K]
280
rs1329237779
nsSNPs
G /A
1
1153
Glu [E]/Lys [K]
282
rs1384542256
nsSNPs
A /C
2
1154
Glu [E]/Ala [A]
282
rs776912318
nsSNPs
A /C
1
1162
Ser [S]/Arg [R]
285
rs776912318
nsSNPs
A /T
1
1162
Ser [S]/Cys [C]
285
rs121909746
nonsense
C /T
1
1168
Gln [Q]/
287
rs764161243
nsSNPs
A /C
2
1172
Lys [K]/Thr [T]
288
rs780873643
nsSNPs
T /G
2
1175
Val [V]/Gly [G]
289
rs775691314
nsSNPs
C /G
2
1178
Ser [S]/Cys [C]
290
rs772619265
nsSNPs
A /G
1
1180
Ile [I]/Val [V]
291
rs760061096
nsSNPs
T /A
2
1181
Ile [I]/Lys [K]
291
rs749789723
inframe insertion/stop gained
TTT
2
1181
Leu [L]
291
rs1205719797
nsSNPs
T /A
2
1190
Leu [L]/His [H]
294
rs1364855365
nsSNPs
C /G
2
1196
Thr [T]/Ser [S]
296
rs368432491
nsSNPs
A /G
2
1199
Asn [N]/Ser [S]
297
rs182778895
nsSNPs
T /C
1
1201
Ser [S]/Pro [P]
298
rs777540740
nsSNPs
A /T
1
1204
Ser [S]/Cys [C]
299
rs769325995
nsSNPs
T /C
1
1207
Tyr [Y]/His [H]
300
rs121909743
nonsense
C /T
1
1210
Arg [R]/
301
rs374492763
nsSNPs
G /A
2
1211
Arg [R]/Gln [Q]
301
rs374492763
nsSNPs
G /T
2
1211
Arg [R]/Leu [L]
301
rs1326032349
frame shift
-/TTGG
3
1212
Gln [Q]/Leu [L]
302
rs1316522125
frame shift
C /-
2
1217
Pro [P]/Leu [L]
303
rs938526894
nsSNPs
T /C
2
1223
Leu [L]/Pro [P]
305
rs1312418962
nsSNPs
T /C
2
1235
Met [M]/Thr [T]
309
rs1421580115
nsSNPs
G /A
1
1243
Val [V]/Met [M]
312
rs1324205444
nsSNPs
G /A
1
1246
Ala [A]/Thr [T]
313
rs1324205444
nsSNPs
G /T
1
1246
Ala [A]/Ser [S]
313
rs780067980
nsSNPs
G /A
1
1261
Gly [G]/Arg [R]
318
rs369101584
nsSNPs
A /G
2
1268
Asn [N]/Ser [S]
320
rs757366672
nsSNPs
G /T
1
1270
Gly [G]/Cys [C]
321
rs767670296
nsSNPs
T /A
2
1274
Ile [I]/Asn [N]
322
rs1272816101
nsSNPs
T /G
3
1275
Ile [I]/Met [M]
322
rs759952425
nsSNPs
T /C
1
1282
Tyr [Y]/His [H]
325
rs751917665
nsSNPs
C /T
2
1304
Thr [T]/Met [M]
332
rs1441375275
nsSNPs
G /A
1
1306
Ala [A]/Thr [T]
333
rs763345848
nsSNPs
G /A
2
1310
Gly [G]/Asp [D]
334
rs748296868
frame shift
G /-
2
1310
Gly [G]/Val [V]
334
rs1461795294
nsSNPs
A /G
1
1315
Ser [S]/Gly [G]
336
rs773717998
nsSNPs
C /A
2
1334
Thr [T]/Asn [N]
342
rs1162318193
nsSNPs
T /C
2
1337
Ile [I]/Thr [T]
343
rs1050103029
nsSNPs
T /C
2
1343
Val [V]/Ala [A]
345
rs764683908
nsSNPs
G /A
2
1346
Gly [G]/Asp [D]
346
rs776435170
nsSNPs
G /A
1
1348
Ala [A]/Thr [T]
347
rs1236921754
nsSNPs
C /T
2
1349
Ala [A]/Val [V]
347
rs746863503
nsSNPs
T /G
2
1358
Met [M]/Arg [R]
350
rs775407568
nsSNPs
G /C
3
1359
Met [M]/Ile [I]
350
rs771855037
nsSNPs
G /A
1
1369
Ala [A]/Thr [T]
354
rs140815551
nsSNPs
G /A
1
1372
Val [V]/Ile [I]
355
rs1348497054
nsSNPs
C /G
2
1376
Ser [S]/Cys [C]
356
rs1469035471
nsSNPs
G /C
1
1378
Val [V]/Leu [L]
357
rs372845210
nsSNPs
C /A
1
1384
Leu [L]/Ile [I]
359
rs372845210
nsSNPs
C /T
1
1384
Leu [L]/Phe [F]
359
rs1380054283
nsSNPs
G /C
1
1390
Glu [E]/Gln [Q]
361
rs999185720
nsSNPs
G /T
3
1392
Glu [E]/Asp [D]
361
rs745619267
nsSNPs
G /C
3
1395
Lys [K]/Asn [N]
362
rs76362149
nsSNPs
G /T
1
1396
Ala [A]/Ser [S]
363
rs121909742
nonsense
C /T
1
1402
Arg [R]/
365
rs34066960
frame shift
-/C
3
1401
Arg [R]/Pro [P]
365
rs781225543
nsSNPs
G /A
2
1403
Arg [R]/Gln [Q]
365
rs1321655963
nsSNPs
C /T
1
1405
Arg [R]/Cys [C]
366
rs755000812
nsSNPs
G /A
2
1406
Arg [R]/His [H]
366
rs1174349159
frame shift
CT /-
3
1413
Phe [F]/Ser [S]
369
rs1223071449
nsSNPs
T /A
3
1416
Phe [F]/Leu [L]
369
rs1430684701
nsSNPs
T /C
2
1418
Leu [L]/Pro [P]
370
rs747262541
nsSNPs
G /A
2
1430
Ser [S]/Asn [N]
374
rs868182136
nsSNPs
G /A
2
1433
Gly [G]/Glu [E]
375
rs780255530
nsSNPs
A /G
1
1435
Met [M]/Val [V]
376
rs758699271
nsSNPs
G /A
3
1437
Met [M]/Ile [I]
376
rs946622803
nsSNPs
T /C
1
1438
Phe [F]/Leu [L]
377
rs1381085405
nsSNPs
T /A
2
1439
Phe [F]/Tyr [Y]
377
rs1381085405
nsSNPs
T /C
2
1439
Phe [F]/Ser [S]
377
rs1384674663
frame shift
TTGT /-
3
1443
Cys [C]/Pro [P]
379
rs750782646
nsSNPs
T /A
2
1451
Ile [I]/Asn [N]
381
rs765728439
nsSNPs
T /C
2
1457
Met [M]/Thr [T]
383
rs757805176
nsSNPs
G /A
1
1465
Gly [G]/Arg [R]
386
rs757805176
nsSNPs
G /C
1
1465
Gly [G]/Arg [R]
386
rs1199637811
nsSNPs
T /G
2
1472
Val [V]/Gly [G]
388
rs121909747
nsSNPs
T /G
2
1475
Leu [L]/Arg [R]
389
rs 121,909,747
nsSNPs
T/C
2
1475
L (Leu) > P (Pro)
389
rs760200790
nsSNPs
T /G
2
1478
Leu [L]/Arg [R]
390
rs766191732
nsSNPs
C /A
3
1488
Phe [F]/Leu [L]
393
rs1464417991
nsSNPs
T /C
1
1489
Ser [S]/Pro [P]
394
rs762668792
nsSNPs
T /A
2
1505
Val [V]/Glu [E]
399
rs1457657980
nsSNPs
A /G
1
1510
Met [M]/Val [V]
401
rs374702599
nsSNPs
T /C
2
1514
Ile [I]/Thr [T]
402
rs1161394690
inframe insertion
GAT
2
1514
Met [M]/ MetMet [MM]
401
rs1419532672
nsSNPs
A /G
1
1519
Ile [I]/Val [V]
404
rs2229608
nsSNPs
T /C
2
1520
Ile [I]/Thr [T]
404
rs760729620
nsSNPs
T /C
2
1529
Phe [F]/Ser [S]
407
rs140791627
nsSNPs
A /T
1
1534
Ser [S]/Cys [C]
409
rs746136121
nsSNPs
T /C
1
1537
Phe [F]/Leu [L]
410
rs1353890919
nsSNPs
T /G
2
1541
Phe [F]/Cys [C]
411
rs966424064
nsSNPs
T/ C
2
1547
Ile [I]/Thr [T]
413
rs779212294
nsSNPs
T /G
3
1548
Ile [I]/Met [M]
413
rs121909744
nsSNPs
C /G
2
1559
Pro [P]/Arg [R]
417
rs121909744
nsSNPs
C /T
2
1559
Pro [P]/Leu [L]
417
rs1309197020
nsSNPs
C /G
3
1563
Ile [I]/Met [M]
418
rs1309197020
nsSNPs
C/A
3
1563
Ile [I] / Ile [I]
418
rs121909745
nonsense
G /A
2
1568
Trp [W]/
420
rs749661374
nsSNPs
A /G
1
1573
Met [M]/Val [V]
422
rs778490867
nsSNPs
T /C
2
1574
Met [M]/Thr [T]
422
rs28928874
nsSNPs
T /A
2
1577
Val [V]/Glu [E]
423
rs1386374799
frame shift
T /-
2
1589
Phe [F]/Ser [S]
427
rs367980651
nsSNPs
C /T
1
1603
Arg [R]/Cys [C]
432
rs75144723
nsSNPs
G /A
2
1604
Arg [R]/His [H]
432
rs754405476
nsSNPs
G /A
1
1612
Ala [A]/Thr [T]
435
rs1379813904
nsSNPs
C /A
2
1619
Ala [A]/Glu [E]
437
rs751226875
nsSNPs
T /C
2
1622
Ile [I]/Thr [T]
438
rs762675284
nsSNPs
G /T
1
1624
Ala [A]/Ser [S]
439
rs1262058831
nsSNPs
C /T
2
1625
Ala [A]/Val [V]
439
rs758246412
nsSNPs
C /A
2
1628
Ala [A]/Glu [E]
440
rs1203908311
nsSNPs
A /G
2
1637
Asn [N]/Ser [S]
443
rs765196886
nsSNPs
A /C
1
1654
Ile [I]/Leu [L]
449
rs761784655
nsSNPs
T /C
2
1655
Ile [I]/Thr [T]
449
rs776395971
nsSNPs
T /C
2
1658
Val [V]/Ala [A]
450
rs1238600269
nsSNPs
G /A
1
1660
Ala [A]/Thr [T]
451
rs1418589512
nsSNPs
T /C
1
1666
Cys [C]/Arg [R]
453
rs1350704340
frame shift
-/G
3
1665
Cys [C]/Val [V]
453
rs759480075
nsSNPs
T /C
1
1675
Tyr [Y]/His [H]
456
rs771274850
nsSNPs
T /C
2
1679
Ile [I]/Thr [T]
457
rs749710583
nsSNPs
C /A
2
1682
Ala [A]/Glu [E]
458
rs749710583
nsSNPs
C /T
2
1682
Ala [A]/Val [V]
458
rs1290975016
frame shift
-/T
2
1688
Cys [C]/Leu [L]
461
rs774542648
nsSNPs
G /A
1
1693
Gly [G]/Arg [R]
462
rs765132996
frame shift
G /-
2
1694
Gly [G]/Asp [D]
462
rs1272353608
nsSNPs
C /A
2
1697
Pro [P]/His [H]
463
rs1381049817
nsSNPs
A /G
2
1700
Tyr [Y]/Cys [C]
464
rs1336322605
nsSNPs
T /C
1
1708
Phe [F]/Leu [L]
467
rs140138702
nsSNPs
C /G
1
1711
Leu [L]/Val [V]
468
rs770197371
nsSNPs
T /G
2
1712
Leu [L]/Arg [R]
468
rs748401954
nsSNPs
T /C
2
1715
Phe [F]/Ser [S]
469
rs1290412048
frame shift
TT /-
2
1715
Phe [F]/Cys [C]
469
rs374342938
nsSNPs
G /C
2
1721
Gly [G]/Ala [A]
471
rs769037887
nsSNPs
G /A
1
1723
Val [V]/Met [M]
472
rs556023421
nsSNPs
T /C
1
1735
Phe [F]/Leu [L]
476
rs5397
nsSNPs
C /G
1
1741
Leu [L]/Val [V]
478
rs5398
nsSNPs
C /A
3
1746
Phe [F]/Leu [L]
479
rs757137603
nsSNPs
C /T
2
1748
Thr [T]/Ile [I]
480
rs1441249949
nsSNPs
T /A
1
1750
Phe [F]/ Ile [I]
481
rs1195253424
nsSNPs
G /A
1
1759
Val [V]/Ile [I]
484
rs753575081
nsSNPs
C /A
1
1762
Pro [P]/Thr [T]
485
rs777806589
nsSNPs
A /G
2
1772
Lys [K]/Arg [R]
488
rs766600474
nsSNPs
T /A
1
1780
Ser [S]/Thr [T]
491
rs766600474
nsSNPs
T /G
1
1780
Ser [S]/Ala [A]
491
rs1379944645
nonsense
G /T
1
1786
Glu [E]/
493
rs1379944645
nsSNPs
G /A
1
1786
Glu [E]/Lys [K]
493
rs1353603250
nsSNPs
G /C
1
1789
Glu [E]/Gln [Q]
494
rs1446857276
nsSNPs
A /T
3
1791
Glu [E]/Asp [D]
494
rs1283734332
nsSNPs
T /C
2
1793
Ile [I]/Thr [T]
495
rs1445660295
nsSNPs
C /T
2
1796
Ala [A]/Val [V]
496
rs201797691
nsSNPs
G /A
1
1798
Ala [A]/Thr [T]
497
rs201797691
nsSNPs
G /C
1
1798
Ala [A]/Pro [P]
497
rs200160167
nsSNPs
C /A
2
1799
Ala [A]/Glu [E]
497
rs200160167
nsSNPs
C /T
2
1799
Ala [A]/Val [V]
497
rs762305192
nsSNPs
T /C
1
1804
Phe [F]/Leu [L]
499
rs776826621
nsSNPs
G /C
3
1812
Lys [K]/Asn [N]
501
rs5399
nsSNPs
G /C
3
1815
Lys [K]/Asn [N]
502
rs1412427073
nsSNPs
G /A
1
1819
Gly [G]/Ser [S]
504
rs966895511
nsSNPs
G /T
1
1825
Ala [A]/Ser [S]
506
rs1199349184
nsSNPs
C /T
2
1826
Ala [A]/Val [V]
506
rs374630641
nsSNPs
G /C
3
1833
Arg [R]/Ser [S]
508
rs771586150
nsSNPs
C /A
2
1835
Pro [P]/Gln [Q]
509
rs776597156
nsSNPs
A /C
3
1839
Lys [K]/Asn[N]
510
rs770462591
nsSNPs
C /A
2
1841
Ala [A]/Asp [D]
511
rs770462591
nsSNPs
C /T
2
1841
Ala [A]/Val [V]
511
rs141574520
nsSNPs
T /A
2
1859
Phe [F]/Tyr [Y]
517
rs147959014
nsSNPs
G /A
2
1865
Gly [G]/Glu [E]
519
rs752687355
nsSNPs
A /G
2
1874
Glu [E]/Gly[G]
522
rs781215842
nsSNPs
A /T
1
1876
Thr [T]/Ser [S]
523
rs758607933
nsSNPs
G/ A
1
1879
Val[V]/Met[M]
524
rs750463981
stop lost
T /C
1
1882
(Ter) /Gln[Q]
525
3.4 Analysis of nsSNPS using SIFT
SIFT forecasts whether amino acid substitution in protein has phenotypic effect or not. Alignment also gave forecasted changes of all amino acid locations. The SIFT intolerance index starting point was 0.05. The color code of non polar, basic, uncharged polar and acidic amino acids were shown by black, red, green and blue color respectively. On the other hand, the amino acid alignment is shown by capital letter while the results of prediction are shown by small letter. 'Seq Rep' was the proportion of sequences that has one of the important amino acids. The position either severely gapped or unalignable was determined by low fraction and it has little information which has poor prediction. Its prediction score ranges from 0 to 1. On phenotypic substitution by aligning orthologous and paralogous protein sequence, this study mainly considers the physical properties of amino acids, effect of natural nsSNPs and alignment of homologous sequences (Noreen et al., 2015a, 2015b). For positions 1–524 of amino acids, the prediction of possible substitutions was made (Figure 6). It was used to predict damaging and tolerated effect of 293 nsSNPs that occurs in coding region of GLUT2 protein. As a consequence, 167 were predicted to be damaging (Table 2). Variants with tolerance index ≤ 0.05 score of SIFT was considered as deleterious while others are taken to be tolerant. By PolyPhen, the variations with probabilistic score above 0.85 and 0.15 were considered to be “Probably damaging” and “possibly damaging” respectively while all the resting were categorized to be “Benign”. Provean score was equal to or below −2.5 it may be considered as deleterious and if the score was above −2.5, it was considered as neutral.
dbSNP id
Proteinresidue
SIFT score
SIFTprediction
PolyPhenscore
PolyPhenprediction
PROVEANscore
PROVEAN prediction
rs759495940
Met [M]/Ile [I]
0.02
Intolerant
0.016
Benign
−1.576
Neutral
rs773205789
Thr [T]/Pro [P]
0.08
Tolerant
0.003
Benign
−0.251
Neutral
rs200073044
Asp [D]/His [H]
0.16
Tolerant
0.001
Benign
−1.256
Neutral
rs1372689853
Lys [K]/Glu [E]
0.27
Tolerant
0.009
Benign
−1.503
Neutral
rs767313610
Val [V]/Ile [I]
0.24
Tolerant
0.003
Benign
0.189
Neutral
rs369700669
Thr [T]/Asn [N]
0.05
Tolerant
0.505
Possibly damaging
−4.345
Deleterious
rs1481905618
Phe [F]/Leu [L]
1
Tolerant
0.002
Benign
−1.974
Neutral
rs1247912820
Thr [T]/Asn [N]
0
Intolerant
0.255
Benign
−1.769
Neutral
rs1247912820
Thr [T]/Ser [S]
0.37
Tolerant
0.007
Benign
0.439
Neutral
rs372441014
Val [V]/Leu [L]
0.04
Intolerant
0.534
Possibly damaging
−2.404
Neutral
rs766364438
Ile [I]/Thr [T]
0.48
Tolerant
0.007
Benign
−0.108
Neutral
rs1463507753
I (Ile) > II (IleIle)
–
–
–
–
–
–
rs867315854
Ala [A]/Thr [T]
0
Intolerant
0.951
Probably damaging
−3.332
Deleterious
rs761992056
Gly [G]/Asp [D]
0
Intolerant
0.967
Probably damaging
−5.211
Deleterious
rs369781481
Phe [F]/Leu [L]
1
Tolerant
0.253
Benign
−0.832
Neutral
rs1409436045
Phe [F]/Tyr [Y]
0.08
Tolerant
0.582
Possibly damaging
−1.774
Neutral
rs1049223265
Ile [I]/Thr [T]
0.61
Tolerant
0.041
Benign
−0.47
Neutral
rs1437312005
Gly [G]/Ser [S]
0
Intolerant
0.996
Probably damaging
−5.242
Deleterious
rs1437312005
Gly [G]/Arg [R]
0
Intolerant
1
Probably damaging
−6.99
Deleterious
rs775531825
Asn [N]/Ser [S]
0
Intolerant
0.979
Probably damaging
−4.384
Deleterious
rs143528640
Ala [A]/Thr [T]
0.05
Tolerant
0.934
Probably damaging
−3.505
Deleterious
rs746158263
Pro [P]/Thr [T]
0
Intolerant
0.999
Probably damaging
−7.021
Deleterious
rs1158195535
Gln [Q]/Lys [K]
1
Tolerant
0.004
Benign
0.622
Neutral
rs1158195535
Gln [Q]/Glu [E]
0.14
Tolerant
0.002
Benign
−0.245
Neutral
rs1451394300
Val [V]/Ile [I]
0.59
Tolerant
0.006
Benign
−0.067
Neutral
rs1477523180
Ile [I]/Leu [L]
0.16
Tolerant
0.437
Benign
−1.338
Neutral
rs1360464436
Ile [I]/Lys [K]
0
Intolerant
0.977
Probably damaging
−4.893
Deleterious
rs1176350402
Ile [I]/Thr [T]
0.02
Intolerant
0.012
Benign
0.3
Neutral
rs772999215
II (IleIle) > I (Ile)
–
–
–
–
−4.083
Deleterious
rs758670698
Arg [R]/Ile [I]
0.14
Tolerant
0.016
Benign
−2.042
Neutral
rs760618624
His [H]/Tyr [Y]
0.04
Intolerant
0
Benign
−1.705
Neutral
rs775791143
Val [V]/Ile [I]
0.09
Tolerant
0.098
Benign
−0.283
Neutral
rs149108283
Val [V]/Asp [D]
0.02
Intolerant
0.689
Possibly damaging
−2.294
Neutral
rs149108283
Val [V]/Ala [A]
0.14
Tolerant
0.034
Benign
−1.116
Neutral
rs1159338702
Gly [G]/Ser [S]
0.01
Intolerant
0.653
Possibly damaging
−1.878
Neutral
rs561765982
Val [V]/Ile [I]
0.11
Tolerant
0.006
Benign
−0.204
Neutral
rs983907950
Pro [P]/Ser [S]
0.41
Tolerant
0.004
Benign
−1.397
Neutral
rs1211075508
Leu [L]/Pro [P]
0.32
Tolerant
0.01
Benign
−1.458
Neutral
rs1311902495
Asp [D]/Asn[N]
0.34
Tolerant
0.493
Possibly damaging
−0.762
Neutral
rs1311902495
Asp [D]/Tyr [Y]
0.01
Intolerant
0.873
Possibly damaging
−2.194
Neutral
rs771477447
Arg [R]/Gly [G]
0.05
Tolerant
0.013
Benign
−1.477
Neutral
rs145210664
Arg [R]/Gln [Q]
0.18
Tolerant
0.039
Benign
−0.58
Neutral
rs546539032
Lys [K]/Asn [N]
0.48
Tolerant
0.038
Benign
−0.733
Neutral
rs747555903
Val [V]/Ile [I]
0.43
Tolerant
0.006
Benign
−0.048
Neutral
rs977284195
Ile [I]/Thr [T]
0.65
Tolerant
0.003
Benign
−0.17
Neutral
rs780903829
Ile [I]/Met [M]
0.17
Tolerant
0.006
Benign
−0.304
Neutral
rs1169887677
INS (IleAsnSer) > ()
–
–
–
–
–
–
rs1373290524
Ser [S]/Thr [T]
0.61
Tolerant
0.005
Benign
−0.656
Neutral
rs1310901426
Thr [T]/Ala [A]
0.78
Tolerant
0.484
Possibly damaging
−1.49
Neutral
rs1354126805
Thr [T]/Arg [R]
0.53
Tolerant
0.93
Probably damaging
−1.61
Neutral
rs754585542
Asp [D]/Asn[N]
0.6
Tolerant
0.003
Benign
−0.514
Neutral
rs1217666649
Asp [D]/Glu [E]
1
Tolerant
0.001
Benign
−0.257
Neutral
rs1391257598
Pro [P]/Thr [T]
0.01
Intolerant
0.005
Benign
−0.821
Neutral
rs7637863
Pro [P]/Leu [L]
0.05
Intolerant
0.005
Benign
−0.881
Neutral
rs779977931
Thr [T]/Lys [K]
0.16
Tolerant
0.037
Benign
−0.951
Neutral
rs1182852354
Ile [I]/Val [V]
0.67
Tolerant
0.001
Benign
0.053
Neutral
rs750405382
Met [M]/Val[V]
0.6
Tolerant
0.001
Benign
0.191
Neutral
rs1207297111
Asn [N]/Asp[D]
0.84
Tolerant
0.001
Benign
−0.029
Neutral
rs1342979475
Asn [N]/Thr [T]
0.69
Tolerant
0
Benign
0.053
Neutral
rs1274084408
Pro [P]/Ala [A]
0.73
Tolerant
0.01
Benign
−0.158
Neutral
rs778655073
Pro [P]/Leu [L]
0.01
Intolerant
0.258
Benign
−1.466
Neutral
rs549263048
Trp [W]/Arg [R]
0.07
Tolerant
0
Benign
−1.382
Neutral
rs531049536
Trp [W]/Cys [C]
0.03
Intolerant
0.069
Benign
−1.624
Neutral
rs150851401
Glu [E]/Lys [K]
0.03
Intolerant
0.004
Benign
−0.777
Neutral
rs766762468
Thr [T]/Ser [S]
0.71
Tolerant
0.004
Benign
−0.271
Neutral
rs766762468
Thr (T)/ Ile (I)
0.18
Tolerant
0.039
Benign
−0.862
Neutral
rs763255363
Ala [A]/Ser [S]
0.74
Tolerant
0.016
Benign
0.281
Neutral
rs144715667
Ile [I]/Val [V]
0.53
Tolerant
0.002
Benign
−0.043
Neutral
rs1415169647
Met [M]/Arg[R]
0.07
Tolerant
0.179
Benign
−2.363
Neutral
rs1407375423
Trp [W]/Cys [C]
0
Intolerant
0.999
Probably damaging
−11.435
Deleterious
rs1800572
Val [V]/Ile [I]
0
Intolerant
0.997
Probably damaging
−0.887
Neutral
rs1800572
Val [V] / Leu[L]
0
Intolerant
0.996
Probably damaging
−2.66
Deleterious
rs770135219
Val [V]/Ala [A]
0
Intolerant
0.991
Probably damaging
−3.541
Deleterious
rs1399091893
Ala [A]/Val [V]
0
Intolerant
0.542
Possibly damaging
−2.559
Deleterious
rs1332764085
Val [V]/Ile [I]
0.39
Tolerant
0.026
Benign
−0.536
Neutral
rs5400
Thr [T]/Ile [I]
1
Tolerant
0
Benign
3.394
Neutral
rs377238940
Ala [A]/Pro [P]
0.01
Intolerant
0.808
Possibly damaging
−2.114
Neutral
rs768407637
Gly [G]/Val [V]
0
Intolerant
0.996
Probably damaging
−6.815
Deleterious
rs753980727
Trp [W]/Cys [C]
0.01
Intolerant
0.704
Possibly damaging
−2.72
Deleterious
rs746632604
Gly [G]/Ala [A]
1
Tolerant
0.011
Benign
−0.025
Neutral
rs772002572
Thr [T]/Ile [I]
0.02
Intolerant
0.003
Benign
−1.408
Neutral
rs1476520648
Ile [I]/Asn [N]
0.09
Tolerant
0.18
Benign
−0.471
Neutral
rs760201098
Ala [A]/Asp [D]
0.03
Intolerant
0.616
Possibly damaging
−2.467
Neutral
rs1267904495
Met [M]/Thr [T]
0
Intolerant
0.799
Possibly damaging
−5.134
Deleterious
rs1212980167
Met [M]/Ile [I]
0
Intolerant
0.175
Benign
−3.41
Deleterious
rs367856967
Val [V]/Ala [A]
0.27
Tolerant
0.004
Benign
−1.196
Neutral
rs970550665
Ala [A]/Gly [G]
0.53
Tolerant
0.526
Possibly damaging
−1.704
Neutral
rs775283150
Ile [I]/Phe [F]
0.05
Tolerant
0.082
Benign
−2.379
Neutral
rs913419413
Leu [L]/Arg [R]
0
Intolerant
0.954
Probably damaging
−4.778
Deleterious
rs771843187
Ser [S]/Pro [P]
0
Intolerant
0.82
Possibly damaging
−2.268
Neutral
rs144125084
Val [V]/Leu [L]
0.75
Tolerant
0.006
Benign
−0.509
Neutral
rs993833041
Val [V]/Ala [A]
0.6
Tolerant
0.004
Benign
−1.391
Neutral
rs1300072764
Gly [G]/Glu [E]
0
Intolerant
0.991
Probably damaging
−5.45
Deleterious
rs778548964
Leu [L]/Phe [F]
0.65
Tolerant
0.125
Benign
−1.799
Neutral
rs1016384738
Met [M]/Lys[K]
0
Intolerant
0.979
Probably damaging
−5.027
Deleterious
rs770941010
Gly [G]/Arg [R]
0
Intolerant
0.89
Possibly damaging
−5.634
Deleterious
rs777718289
Gly [G]/Glu [E]
0.02
Intolerant
0.588
Possibly damaging
−3.269
Deleterious
rs777718289
Gly [G]/Val [V]
0.04
Intolerant
0.065
Benign
−3.177
Deleterious
rs372621339
Pro [P]/Arg [R]
0.54
Tolerant
0.017
Benign
−0.696
Neutral
rs747025551
Ile [I]/Thr [T]
0.14
Tolerant
0.158
Benign
0.37
Neutral
rs376064965
Leu [L]/Phe [F]
0
Intolerant
0.114
Benign
−2.421
Neutral
rs763620441
ILI (IleLeuIle) /I(Ile)
–
–
–
–
–
–
rs1188886679
Ile [I]/Leu [L]
1
Tolerant
0.006
Benign
−0.029
Neutral
rs1188886679
Ile [I]/Val [V]
0.28
Tolerant
0.02
Benign
−0.518
Neutral
rs192720796
Ile [I]/Lys [K]
0
Intolerant
0.616
Possibly damaging
−4.296
Deleterious
rs910976682
Ala [A]/Val [V]
0.24
Tolerant
0.007
Benign
0.447
Neutral
rs750836049
Gly [G]/Arg [R]
0
Intolerant
0.996
Probably damaging
−7.236
Deleterious
rs1278964539
Arg [R]/Lys [K]
0
Intolerant
0.999
Probably damaging
−2.714
Deleterious
rs1231468128
Leu [L]/Gln [Q]
0
Intolerant
0.951
Probably damaging
−4.187
Deleterious
rs1445887606
Tyr [Y]/His [H]
0.08
Tolerant
0.319
Benign
−3.492
Deleterious
rs1271546287
Ile [I]/Thr [T]
1
Tolerant
0.007
Benign
0.249
Neutral
rs760095835
Gly [G]/Asp [D]
0
Intolerant
0.998
Probably damaging
−6.71
Deleterious
rs1335888503
Val [V]/Leu [L]
0
Intolerant
0.48
Possibly damaging
−2.654
Deleterious
rs1293130515
Met [M]/Thr [T]
0
Intolerant
0.792
Possibly damaging
−5.465
Deleterious
rs1019696977
Ile [I]/Val [V]
1
Tolerant
0.014
Benign
−0.016
Neutral
rs144822218
Gly [G]/Ser [S]
0.19
Tolerant
0.153
Benign
−4.8
Deleterious
rs759047405
Gly [G]/Asp [D]
0
Intolerant
0.275
Benign
−6.064
Deleterious
rs1441606652
Ala [A]/Val [V]
0
Intolerant
0.93
Probably damaging
−3.261
Deleterious
rs368626129
Ala [A]/Thr [T]
0.8
Tolerant
0.015
Benign
−0.903
Neutral
rs368626129
Ala [A]/Ser [S]
0.86
Tolerant
0.012
Benign
−0.098
Neutral
rs200213178
Ala [A]/Thr [T]
0
Intolerant
0.838
Possibly damaging
−3.928
Deleterious
rs200213178
Ala [A]/Pro [P]
0
Intolerant
0.996
Probably damaging
−4.91
Deleterious
rs748052042
Leu [L]/Phe [F]
0.31
Tolerant
0.044
Benign
−1.051
Neutral
rs1412289847
Gly [G]/Ser [S]
0
Intolerant
0.994
Probably damaging
−5.883
Deleterious
rs776498787
Gly [G]/Asp [D]
0
Intolerant
0.999
Probably damaging
−6.867
Deleterious
rs779065938
Leu [L]/Val [V]
0.03
Intolerant
0.951
Probably damaging
−2.948
Deleterious
rs1469335096
Ala [A]/Thr [T]
0.01
Intolerant
0.688
Possibly damaging
−2.844
Deleterious
rs771182536
Ile [I]/Asn [N]
0
Intolerant
0.961
Probably damaging
−6.383
Deleterious
rs771182536
Ile [I]/Thr [T]
0.02
Intolerant
0.517
Possibly damaging
−4.26
Deleterious
rs121909741
Val [V]/Ile [I]
0.01
Intolerant
0.846
Possibly damaging
−0.978
Neutral
rs149460434
Thr [T]/Lys [K]
0
Intolerant
0.665
Possibly damaging
−3.176
Deleterious
rs149460434
Thr [T]/Met[M]
0.01
Intolerant
0.166
Benign
−0.836
Neutral
rs1276756236
Leu [L]/Ile [I]
0
Intolerant
0.967
Probably damaging
−1.897
Neutral
rs779591826
Ile [I]/Met [M]
0
Intolerant
0.973
Probably damaging
−2.473
Neutral
rs1262860274
Ile [I]/Thr [T]
0.01
Intolerant
0.102
Benign
−2.589
Deleterious
rs1186359171
Gly [G]/Ser [S]
0
Intolerant
0.982
Probably damaging
−5.869
Deleterious
rs1215469128
Leu [L]/Ser [S]
0
Intolerant
0.993
Probably damaging
−4.95
Deleterious
rs1347267249
Asn [N]/His [H]
0.01
Intolerant
0.413
Benign
−3.346
Deleterious
rs573292685
Asn [N]/Ser [S]
0.88
Tolerant
0.048
Benign
−1.171
Neutral
rs764799427
Asp [D]/Val [V]
0.08
Tolerant
0.027
Benign
−3.592
Deleterious
rs1380319602
Ile [I]/Asn [N]
0
Intolerant
0.863
Possibly damaging
−5.167
Deleterious
rs760641937
Gly [G]/Ala [A]
0.26
Tolerant
0.401
Benign
−3.395
Deleterious
rs1413841367
Leu [L]/Pro [P]
0
Intolerant
0.996
Probably damaging
−5.774
Deleterious
rs771075989
Val [V]/Met [M]
0.02
Intolerant
0.863
Possibly damaging
−1.251
Neutral
rs773581866
Arg [R]/Gly [G]
0
Intolerant
0.008
Benign
−0.341
Neutral
rs770126214
Leu [L]/Ile [I]
0.09
Tolerant
0.032
Benign
−1.069
Neutral
rs1374154306
Leu [L]/Gln [Q]
0
Intolerant
0.853
Possibly damaging
−4.116
Deleterious
rs748588515
Leu [L]/Phe [F]
0
Intolerant
0.914
Probably damaging
−3.922
Deleterious
rs757087261
Phe [F]/Ile [I]
0.08
Tolerant
0.219
Benign
−4.242
Deleterious
rs777020657
Phe [F]/Cys [C]
0.03
Intolerant
0.359
Benign
−6.281
Deleterious
rs777020657
F (Phe) > S (Ser)
0
Intolerant
0.936
Probably damaging
−6.607
Deleterious
rs769089021
Ser [S]/Asn [N]
0
Intolerant
0.999
Probably damaging
−2.946
Deleterious
rs780381836
Arg [R]/Gly [G]
0.01
Intolerant
0.548
Possibly damaging
−6.565
Deleterious
rs1480881050
Tyr [Y]/His [H]
0
Intolerant
0.903
Possibly damaging
−4.446
Deleterious
rs1250722271
Tyr [Y]/His [H]
0
Intolerant
0.928
Probably damaging
−3.256
Deleterious
rs1158317020
Tyr [Y]/Phe [F]
0.06
Tolerant
0.02
Benign
−1.172
Neutral
rs867996396
Ile [I]/Asn [N]
0
Intolerant
0.958
Probably damaging
−6.497
Deleterious
rs867996396
Ile [I] / Thr [T]
0
Intolerant
0.925
Probably damaging
−4.799
Deleterious
rs536261161
Lys [K]/Asn [N]
1
Tolerant
0.004
Benign
0.309
Neutral
rs745373269
Asp [D]/Asn [N]
1
Tolerant
0.003
Benign
0.872
Neutral
rs745373269
Asp [D]/His [H]
0.05
Tolerant
0.637
Possibly damaging
−1.072
Neutral
rs1367431424
Glu [E]/Ala [A]
0.2
Tolerant
0.155
Benign
−3.915
Deleterious
rs865881030
Glu [E]/Lys [K]
0
Intolerant
0.377
Benign
−3.096
Deleterious
rs778607566
Glu [E]/Gly [G]
0
Intolerant
0.944
Probably damaging
−5.575
Deleterious
rs1309254226
Glu [E]/Asp [D]
0.03
Intolerant
0.247
Benign
−2.277
Neutral
rs777225980
Ser [S]/Arg [R]
0.07
Tolerant
0.689
Possibly damaging
−2.569
Deleterious
rs76026576
Leu [L]/Phe [F]
0
Intolerant
0.998
Probably damaging
−3.757
Deleterious
rs1490504926
Asp [D]/Asn[N]
0.51
Tolerant
0.005
Benign
−0.126
Neutral
rs935009475
Asp [D]/Glu [E]
0.06
Tolerant
0.434
Benign
−3.438
Deleterious
rs774721090
DD (AspAsp) / D (Asp)
–
–
–
–
−6.952
Deleterious
rs140285191
Asp [D]/Asn[N]
0
Intolerant
0.516
Possibly damaging
−4.047
Deleterious
rs140285191
Asp [D]/Tyr [Y]
0
Intolerant
0.981
Probably damaging
−7.312
Deleterious
rs754932741
Met [M]/Ile [I]
0
Intolerant
0.17
Benign
−3.506
Deleterious
rs750579210
Arg [R]/Gly [G]
0
Intolerant
0.579
Possibly damaging
−4.146
Deleterious
rs1304842107
Lys [K]/Gln [Q]
0.07
Tolerant
0.166
Benign
−0.767
Neutral
rs765426962
Lys [K]/Thr [T]
0.13
Tolerant
0.065
Benign
−2.692
Deleterious
rs761756532
Lys [K]/Asn [N]
0.09
Tolerant
0.036
Benign
−1.633
Neutral
rs1329237779
Glu [E]/Lys [K]
1
Tolerant
0.03
Benign
−0.266
Neutral
rs1384542256
Glu [E]/Ala [A]
0.64
Tolerant
0.055
Benign
−3.333
Deleterious
rs776912318
Ser [S]/Arg [R]
1
Tolerant
0.009
Benign
−0.133
Neutral
rs776912318
Ser [S]/Cys [C]
0.02
Intolerant
0.031
Benign
−2.646
Deleterious
rs764161243
Lys [K]/Thr [T]
0.05
Tolerant
0.133
Benign
−4.412
Deleterious
rs780873643
Val [V]/Gly [G]
0
Intolerant
0.943
Probably damaging
−6.041
Deleterious
rs775691314
Ser [S]/Cys [C]
0
Intolerant
0.984
Probably damaging
−3.556
Deleterious
rs772619265
Ile [I]/Val [V]
0.4
Tolerant
0.02
Benign
−0.515
Neutral
rs760061096
Ile [I]/Lys [K]
0
Intolerant
0.824
Possibly damaging
−5.896
Deleterious
rs749789723
Leu [L]
–
–
–
–
–
–
rs1205719797
Leu [L]/His [H]
0
Intolerant
0.999
Probably damaging
−6.482
Deleterious
rs1364855365
Thr [T]/Ser [S]
0.58
Tolerant
0.065
Benign
−0.827
Neutral
rs368432491
Asn [N]/Ser [S]
1
Tolerant
0.006
Benign
1.283
Neutral
rs182778895
Ser [S]/Pro [P]
1
Tolerant
0.004
Benign
1.098
Neutral
rs777540740
Ser [S]/Cys [C]
0.06
Tolerant
0.031
Benign
−2.516
Deleterious
rs769325995
Tyr [Y]/His [H]
0.11
Tolerant
0.32
Benign
−4.3
Deleterious
rs374492763
Arg [R]/Gln [Q]
0.04
Intolerant
0.771
Possibly damaging
−3.702
Deleterious
rs374492763
Arg [R]/Leu [L]
0
Intolerant
0.906
Possibly damaging
−6.535
Deleterious
rs938526894
Leu [L]/Pro [P]
0
Intolerant
0.811
Possibly damaging
−4.885
Deleterious
rs1312418962
Met [M]/Thr [T]
0.03
Intolerant
0.411
Benign
−3.131
Deleterious
rs1421580115
Val [V]/Met [M]
0.04
Intolerant
0.007
Benign
0.763
Neutral
rs1324205444
Ala [A]/Thr [T]
0
Intolerant
0.842
Possibly damaging
−0.976
Neutral
rs1324205444
Ala [A]/Ser [S]
1
Tolerant
0.138
Benign
1.321
Neutral
rs780067980
Gly [G]/Arg [R]
0
Intolerant
1
Probably damaging
−7.439
Deleterious
rs369101584
Asn [N]/Ser [S]
0.01
Intolerant
0.919
Probably damaging
−4.649
Deleterious
rs757366672
Gly [G]/Cys [C]
0
Intolerant
0.507
Possibly damaging
−2.067
Neutral
rs767670296
Ile [I]/Asn [N]
0
Intolerant
0.99
Probably damaging
−6.223
Deleterious
rs1272816101
Ile [I]/Met [M]
0
Intolerant
0.985
Probably damaging
−2.411
Neutral
rs759952425
Tyr [Y]/His [H]
0
Intolerant
0.986
probaly damaging
−4.763
Deleterious
rs751917665
Thr [T]/Met [M]
0.03
Intolerant
0.075
Benign
−2.321
Neutral
rs1441375275
Ala [A]/Thr [T]
0
Intolerant
0.955
Probably damaging
−3.749
Deleterious
rs763345848
Gly [G]/Asp [D]
0
Intolerant
0.96
Probably damaging
−6.283
Deleterious
rs1461795294
Ser [S]/Gly [G]
0.47
Tolerant
0.013
Benign
−1.028
Neutral
rs773717998
Thr [T]/Asn [N]
0
Intolerant
0.986
Probably damaging
−4.736
Deleterious
rs1162318193
Ile [I]/Thr [T]
0
Intolerant
0.775
Possibly damaging
−4.7
Deleterious
rs1050103029
Val [V]/Ala [A]
1
Tolerant
0.071
Benign
0.25
Neutral
rs764683908
Gly [G]/Asp [D]
0
Intolerant
1
Probably damaging
−6.631
Deleterious
rs776435170
Ala [A]/Thr [T]
0.02
Intolerant
0.032
Benign
−0.943
Neutral
rs1236921754
Ala [A]/Val [V]
1
Tolerant
0.006
Benign
1.67
Neutral
rs746863503
Met [M]/Arg[R]
0
Intolerant
0.034
Benign
−1.288
Neutral
rs775407568
Met [M]/Ile [I]
0.06
Tolerant
0.001
Benign
−0.367
Neutral
rs771855037
Ala [A]/Thr [T]
0.01
Intolerant
0.102
Benign
−0.618
Neutral
rs140815551
Val [V]/Ile [I]
0.03
Intolerant
0.047
Benign
−0.733
Neutral
rs1348497054
Ser [S]/Cys [C]
0
Intolerant
0.929
Probably damaging
−4.52
Deleterious
rs1469035471
Val [V]/Leu [L]
1
Tolerant
0.105
Benign
0.367
Neutral
rs372845210
Leu [L]/Ile [I]
0.04
Intolerant
0.817
Possibly damaging
−1.156
Neutral
rs372845210
Leu [L]/Phe [F]
0
Intolerant
0.94
Probably damaging
−3.045
Deleterious
rs1380054283
Glu [E]/Gln [Q]
0
Intolerant
0.984
Probably damaging
−2.416
Neutral
rs999185720
Glu [E]/Asp [D]
0.31
Tolerant
0.162
Benign
−2.169
Neutral
rs745619267
Lys [K]/Asn [N]
0
Intolerant
0.821
Possibly damaging
−2.418
Neutral
rs76362149
Ala [A]/Ser [S]
0
Intolerant
0.457
Possibly damaging
−2.448
Neutral
rs781225543
Arg [R]/Gln [Q]
0
Intolerant
1
Probably damaging
−3.368
Deleterious
rs1321655963
Arg [R]/Cys [C]
0
Intolerant
0.998
Probably damaging
−6.914
Deleterious
rs755000812
Arg [R]/His [H]
0
Intolerant
0.996
Probably damaging
−4.126
Deleterious
rs1223071449
Phe [F]/Leu [L]
0.14
Tolerant
0.004
Benign
−2.507
Deleterious
rs1430684701
Leu [L]/Pro [P]
0
Intolerant
0.988
Probably damaging
−5.765
Deleterious
rs747262541
Ser [S]/Asn [N]
0.01
Intolerant
0.361
Benign
−1.303
Neutral
rs868182136
Gly [G]/Glu [E]
0
Intolerant
0.99
Probably damaging
−6.873
Deleterious
rs780255530
Met [M]/Val [V]
0
Intolerant
0.491
Possibly damaging
−3.484
Deleterious
rs758699271
Met [M]/Ile [I]
0
Intolerant
0.612
Possibly damaging
−3.451
Deleterious
rs946622803
Phe [F]/Leu [L]
0.49
Tolerant
0.006
Benign
0.13
Neutral
rs1381085405
Phe [F]/Tyr [Y]
0.23
Tolerant
0.652
Possibly damaging
−0.543
Neutral
rs1381085405
Phe [F]/Ser [S]
0.41
Tolerant
0.063
Benign
0.376
Neutral
rs750782646
Ile [I]/Asn [N]
0
Intolerant
0.811
Possibly damaging
−4.874
Deleterious
rs765728439
Met [M]/Thr [T]
0
Intolerant
0.403
Benign
−5.185
Deleterious
rs757805176
Gly [G]/Arg [R]
0
Intolerant
0.752
Possibly damaging
−2.96
Deleterious
rs757805176
Gly [G]/Arg [R]
0
Intolerant
0.752
Possibly damaging
−2.96
Deleterious
rs1199637811
Val [V]/Gly [G]
0.19
Tolerant
0.027
Benign
−2.318
Neutral
rs121909747
Leu [L]/Arg [R]
0
Intolerant
0.947
Probably damaging
−4.842
Deleterious
rs 121,909,747
Leu [L] / Pro [P]
0.01
Intolerant
0.618
Possibly damaging
−5.813
Deleterious
rs760200790
Leu [L]/Arg [R]
0.6
Tolerant
0.139
Benign
−1.45
Neutral
rs766191732
Phe [F]/Leu [L]
0.69
Tolerant
0.002
Benign
−1.523
Neutral
rs1464417991
Ser [S]/Pro [P]
1
Tolerant
0.005
Benign
−0.341
Neutral
rs762668792
Val [V]/Glu [E]
0
Intolerant
0.866
Possibly damaging
−5.082
Deleterious
rs1457657980
Met [M]/Val [V]
0.27
Tolerant
0.03
Benign
−0.744
Neutral
rs1161394690
M (Met) > MM(MetMet)
–
–
–
–
–
–
rs374702599
Ile [I]/Thr [T]
0.18
Tolerant
0.002
Benign
−0.852
Neutral
rs1419532672
Ile [I]/Val [V]
0.19
Tolerant
0.139
Benign
−0.555
Neutral
rs2229608
Ile [I]/Thr [T]
0.21
Tolerant
0.401
Benign
−3.639
Deleterious
rs760729620
Phe [F]/Ser [S]
0
Intolerant
0.861
Possibly damaging
−7.529
Deleterious
rs140791627
Ser [S]/Cys [C]
0.05
Tolerant
0.082
Benign
−2.035
Neutral
rs746136121
Phe [F]/Leu [L]
0.06
Tolerant
0.38
Benign
−3.987
Deleterious
rs1353890919
Phe [F]/Cys [C]
0
Intolerant
1
Probably damaging
−7.57
Deleterious
rs966424064
Ile [I]/Thr [T]
0
Intolerant
0.206
Benign
−4.159
Deleterious
rs779212294
Ile [I]/Met [M]
0.08
Tolerant
0.643
Possibly damaging
−2.032
Neutral
rs121909744
Pro [P]/Arg [R]
0
Intolerant
0.998
Probably damaging
−8.516
Deleterious
rs121909744
Pro [P]/Leu [L]
0
Intolerant
0.994
Probably damaging
−9.463
Deleterious
rs1309197020
Ile [I]/Met [M]
0
Intolerant
1
Probably damaging
−2.838
Deleterious
rs1309197020
I (Ile) / I (Ile)
1
Tolerant
–
–
0
Neutral
rs749661374
Met [M]/Val [V]
0.01
Intolerant
0.004
Benign
0.655
Neutral
rs778490867
Met [M]/Thr [T]
0
Intolerant
0.016
Benign
−2.558
Deleterious
rs28928874
Val [V]/Glu [E]
0
Intolerant
0.976
Probably damaging
−5.619
Deleterious
rs367980651
Arg [R]/Cys [C]
0
Intolerant
0.983
Probably damaging
−7.42
Deleterious
rs75144723
Arg [R]/His [H]
0
Intolerant
0.986
Probably damaging
−4.632
Deleterious
rs754405476
Ala [A]/Thr [T]
0
Intolerant
0.992
Probably damaging
−3.706
Deleterious
rs1379813904
Ala [A]/Glu [E]
0
Intolerant
0.987
Probably damaging
−4.592
Deleterious
rs751226875
Ile [I]/Thr [T]
0
Intolerant
0.678
Possibly damaging
−3.063
Deleterious
rs762675284
Ala [A]/Ser [S]
0.07
Tolerant
0.665
Possibly damaging
−1.839
Neutral
rs1262058831
Ala [A]/Val [V]
0
Intolerant
0.99
Probably damaging
−3.433
Deleterious
rs758246412
Ala [A]/Glu [E]
0.02
Intolerant
0.515
Possibly damaging
−2.432
Neutral
rs1203908311
Asn [N]/Ser [S]
0
Intolerant
0.992
Probably damaging
−4.589
Deleterious
rs765196886
Ile [I]/Leu [L]
0.59
Tolerant
0.009
Benign
−0.308
Neutral
rs761784655
Ile [I]/Thr [T]
0
Intolerant
0.213
Benign
−3.564
Deleterious
rs776395971
Val [V]/Ala [A]
0
Intolerant
0.895
Possibly damaging
−3.625
Deleterious
rs1238600269
Ala [A]/Thr [T]
0
Intolerant
0.219
Benign
−2.469
Neutral
rs1418589512
Cys [C]/Arg [R]
0.13
Tolerant
0.532
Possibly damaging
−4.842
Deleterious
rs759480075
Tyr [Y]/His [H]
0.15
Tolerant
0.976
Probably damaging
−3.381
Deleterious
rs771274850
Ile [I]/Thr [T]
0.01
Intolerant
0.653
Possibly damaging
−2.456
Neutral
rs749710583
Ala [A]/Glu [E]
0.75
Tolerant
0.025
Benign
−1.512
Neutral
rs749710583
Ala [A]/Val [V]
0.18
Tolerant
0.025
Benign
−2.403
Neutral
rs774542648
Gly [G]/Arg [R]
0
Intolerant
0.734
Possibly damaging
−7.25
Deleterious
rs1272353608
Pro [P]/His [H]
0
Intolerant
0.598
Possibly damaging
−5.957
Deleterious
rs1381049817
Tyr [Y]/Cys [C]
0
Intolerant
0.439
Benign
−8.015
Deleterious
rs1336322605
Phe [F]/Leu [L]
0.67
Tolerant
0.002
Benign
1.022
Neutral
rs140138702
Leu [L]/Val [V]
0.26
Tolerant
0.085
Benign
−0.015
Neutral
rs770197371
Leu [L]/Arg [R]
0
Intolerant
0.944
Probably damaging
−4.636
Deleterious
rs748401954
Phe [F]/Ser [S]
0
Intolerant
0.999
Probably damaging
−7.315
Deleterious
rs374342938
Gly [G]/Ala [A]
0.15
Tolerant
0.02
Benign
0.329
Neutral
rs769037887
Val [V]/Met [M]
0.01
Intolerant
0.216
Benign
0.077
Neutral
rs556023421
Phe [F]/Leu [L]
0
Intolerant
0.919
Probably damaging
−5.436
Deleterious
rs5397
Leu [L]/Val [V]
0.41
Tolerant
0.04
Benign
−0.284
Neutral
rs5398
Phe [F]/Leu [L]
0
Intolerant
0.424
Benign
−5.276
Deleterious
rs757137603
Thr [T]/Ile [I]
0.09
Tolerant
0.12
Benign
−4.225
Deleterious
rs1441249949
Phe [F]/ Ile [I]
0
Intolerant
0.616
Possibly damaging
−4.39
Deleterious
rs1195253424
Val [V]/Ile [I]
0
Intolerant
0.971
Probably damaging
−0.908
Neutral
rs753575081
Pro [P]/Thr [T]
0
Intolerant
0.994
Probably damaging
−7.274
Deleterious
rs777806589
Lys [K]/Arg [R]
0.38
Tolerant
0.601
Possibly damaging
−1.518
Neutral
rs766600474
Ser [S]/Thr [T]
1
Tolerant
0.104
Benign
1.006
Neutral
rs766600474
Ser [S]/Ala [A]
0
Intolerant
0.304
Benign
−1.65
Neutral
rs1379944645
Glu [E]/Lys [K]
0.04
Intolerant
0.279
Benign
−3.223
Deleterious
rs1353603250
Glu [E]/Gln [Q]
0.19
Tolerant
0.759
Possibly damaging
−2.031
Neutral
rs1446857276
Glu [E]/Asp [D]
0.31
Tolerant
0.405
Benign
−0.5
Neutral
rs1283734332
Ile [I]/Thr [T]
0
Intolerant
0.999
Probably damaging
−4.518
Deleterious
rs1445660295
Ala [A]/Val [V]
0.02
Intolerant
0.444
Benign
−3.184
Deleterious
rs201797691
Ala [A]/Thr [T]
0.06
Tolerant
0.566
Possibly damaging
−1.562
Neutral
rs201797691
Ala [A]/Pro [P]
0.03
Intolerant
0.979
Probably damaging
−2.518
Deleterious
rs200160167
Ala [A]/Glu [E]
0.05
Tolerant
0.302
Benign
−1.482
Neutral
rs200160167
Ala [A]/Val [V]
0.03
Intolerant
0.566
Possibly damaging
−2.596
Deleterious
rs762305192
Phe [F]/Leu [L]
0
Intolerant
0.866
Possibly damaging
−5.144
Deleterious
rs776826621
Lys [K]/Asn [N]
0.2
Tolerant
0.005
Benign
−1.549
Neutral
rs5399
Lys [K]/Asn [N]
0.41
Tolerant
0.004
Benign
−2.272
Neutral
rs1412427073
Gly [G]/Ser [S]
0.28
Tolerant
0.004
Benign
−1.628
Neutral
rs966895511
Ala [A]/Ser [S]
0.83
Tolerant
0.004
Benign
−0.449
Neutral
rs1199349184
Ala [A]/Val [V]
0.24
Tolerant
0.004
Benign
−1.611
Neutral
rs374630641
Arg [R]/Ser [S]
0.94
Tolerant
0.001
Benign
0.45
Neutral
rs771586150
Pro [P]/Gln [Q]
0.47
Tolerant
0.007
Benign
−1.564
Neutral
rs776597156
Lys [K]/Asn[N]
0.04
Intolerant
0.017
Benign
−1.772
Neutral
rs770462591
Ala [A]/Asp [D]
0.41
Tolerant
0.002
Benign
−1.217
Neutral
rs770462591
Ala [A]/Val [V]
0.16
Tolerant
0.007
Benign
−1.309
Neutral
rs141574520
Phe [F]/Tyr [Y]
1
Tolerant
0.003
Benign
0.369
Neutral
rs147959014
Gly [G]/Glu [E]
0.01
Intolerant
0.104
Benign
−2.153
Neutral
rs752687355
Glu [E]/Gly[G]
0.03
Intolerant
0.009
Benign
−2.391
Neutral
rs781215842
Thr [T]/Ser [S]
0.82
Tolerant
0.004
Benign
0.326
Neutral
rs758607933
Val[V]/Met[M]
0
Intolerant
0.004
Benign
−1.155
Neutral
3.5 Analysis of nsSNPS using PolyPhen
To forecast the effects of function and structure 293 nsSNPs lying in the coding region of GLUT2 protein, PolyPhen-2 server was used. It permits three types of prediction score which is based on a “Sensitivity” as well as “Specificity”. Three prediction types are given by PolyPhen benign, possibly damaging or probably damaging. Out of 293 nsSNPs, 165 were considered to be benign. 65 were predicted to be possibly damaging and 77 were considered to be probably damaging (Table 2).
3.6 Analysis of SNPs using PROVEAN
PROVEAN gave score to predict neutral and deleterious effect of 293 nsSNPs present in coding region of GLUT2 protein. Score equal to or below −2.5 was considered as deleterious and above −2.5, as neutral. Out of 293 nsSNPs, 162 were predicted to be neutral and 138 were considered to be deleterious. It also gave the results of inframe deletion and insertion. Out of 4 inframe deletion 2 (rs772999215 and rs774721090) were shown to be deleterious 3 SNPs were inframe insertion (rs1161394690), (rs1463507753), (rs749789723) and no results were found for them (Table 2).
3.7 Analysis of nsSNPS using SNPeffect
The SNPeffect server was used to predict the effect of molecular phenotype of nsSNPs found in coding region of GLUT2 protein. Effect of 293 nsSNPs on aggregation tendency, amyloid propensity and chaperon binding tendency was specified by dTANGO, dWALTZ and dLIMBO score. Two significant effects, intrinsic aggregation propensity and protein stability were shown. In a protein sequence, aggregation propensity was identified by dTANGO. Out of 293 nsSNPs, 61 were detected to decrease the aggregation propensity, 27 to increase the aggregation propensity while 211 had no effect on aggregation propensity. dWALTZ indicated the score of amyloid propensity. 23 nsSNPs were shown to increase the amyloid propensity, 16 decreased the amyloid propensity and 257 had no effect on the amyloid propensity. According to dLIMBO, the chaperon binding tendency was given. None of them had any effect on chaperon binding tendency (Table 3). Variations with dTANGO, dWALTZ and dLIMBO between −50 and 50 were supposed to have no effect on aggregation tendency, amyloid propensity and chaperone binding tendency, respectively.
dbSNP rs#cluster id
Proteinresidue
dTANGO
Aggregationtendency
dWALTZ
Amyloidpropensity
dLIMBO
Chaperone bindingtendency
rs759495940
Met [M]/Ile [I]
0
No effect
0
No effect
0
No effect
rs773205789
Thr [T]/Pro [P]
0
No effect
0
No effect
0
No effect
rs200073044
Asp [D]/His [H]
−101
Decrease
0.76
No effect
0
No effect
rs1372689853
Lys [K]/Glu [E]
−110.2
Decreases
−0.49
No effect
0
No effect
rs767313610
Val [V]/Ile [I]
−5.8
No effect
0
No effect
0
No effect
rs369700669
Thr [T]/Asn [N]
−109.6
Decreases
0.01
No effect
0
No effect
rs1481905618
Phe [F]/Leu [L]
−1.1
No effect
0.12
No effect
0
No effect
rs1247912820
Thr [T]/Asn [N]
−9.9
No effect
0.97
No effect
0
No effect
rs1247912820
Thr [T]/Ser [S]
−2.7
No effect
0.34
No effect
0
No effect
rs372441014
Val [V]/Leu [L]
−0.5
No effect
0.12
No effect
0
No effect
rs766364438
Ile [I]/Thr [T]
−19.8
No effect
0.96
No effect
0
No effect
rs867315854
Ala [A]/Thr [T]
−5.6
No effect
0.04
No effect
0
No effect
rs761992056
Gly [G]/Asp [D]
−66
Decreases
2.67
No effect
0
No effect
rs369781481
Phe [F]/Leu [L]
–22.4
No effect
0.12
No effect
0
No effect
rs1409436045
Phe [F]/Tyr [Y]
−8.7
No effect
0.21
No effect
0
No effect
rs1049223265
Ile [I]/Thr [T]
0
No effect
−0.09
No effect
0
No effect
rs1437312005
Gly [G]/Ser [S]
0
No effect
0.76
No effect
0
No effect
rs1437312005
Gly [G]/Arg [R]
0
No effect
−0.09
No effect
0
No effect
rs775531825
Asn [N]/Ser [S]
0.1
No effect
−0.08
No effect
0
No effect
rs143528640
Ala [A]/Thr [T]
0
No effect
0
No effect
0
No effect
rs746158263
Pro [P]/Thr [T]
4
No effect
−0.02
No effect
0
No effect
rs1158195535
Gln [Q]/Lys [K]
−4.3
No effect
−559.66
Decreases
0
No effect
rs1158195535
Gln [Q]/Glu [E]
11
No effect
−62.94
Decreases
0
No effect
rs1451394300
Val [V]/Ile [I]
−0.9
No effect
−116.97
Decreases
0
No effect
rs1477523180
Ile [I]/Leu [L]
−2.5
No effect
−3.89
No effect
0
No effect
rs1360464436
Ile [I]/Lys [K]
−4.3
No effect
−417.77
Decreases
0
No effect
rs1176350402
Ile [I]/Thr [T]
−4.3
No effect
−571.42
Decreases
0
No effect
rs758670698
Arg [R]/Ile [I]
110.7
Increases
−0.73
No effect
0
No effect
rs760618624
His [H]/Tyr [Y]
1.1
No effect
0
No effect
0
No effect
rs775791143
Val [V]/Ile [I]
0
No effect
0.01
No effect
0
No effect
rs149108283
Val [V]/Asp [D]
1.7
No effect
−0.72
No effect
0
No effect
rs149108283
Val [V]/Ala [A]
0
No effect
0.01
No effect
0
No effect
rs1159338702
Gly [G]/Ser [S]
0
No effect
0.02
No effect
0
No effect
rs561765982
Val [V]/Ile [I]
0
No effect
0
No effect
0
No effect
rs983907950
Pro [P]/Ser [S]
24.1
No effect
−0.27
No effect
0
No effect
rs1211075508
Leu [L]/Pro [P]
0
No effect
0.23
No effect
0
No effect
rs1311902495
Asp [D]/Asn [N]
−1.9
No effect
1.14
No effect
0
No effect
rs1311902495
Asp [D]/Tyr [Y]
−1.9
No effect
1.01
No effect
0
No effect
rs771477447
Arg [R]/Gly [G]
0.7
No effect
0.07
No effect
0
No effect
rs145210664
Arg [R]/Gln [Q]
0.6
No effect
−0.52
No effect
0
No effect
rs546539032
Lys [K]/Asn [N]
2.3
No effect
0.11
No effect
0
No effect
rs747555903
Val [V]/Ile [I]
−0.1
No effect
−58.27
Decreases
0
No effect
rs977284195
Ile [I]/Thr [T]
−1
No effect
0.02
No effect
0
No effect
rs780903829
Ile [I]/Met [M]
−1
No effect
0.01
No effect
0
No effect
rs1373290524
Ser [S]/Thr [T]
1
No effect
0
No effect
0
No effect
rs1310901426
Thr [T]/Ala [A]
−0.1
No effect
0.02
No effect
0
No effect
rs1354126805
Thr [T]/Arg [R]
1.6
No effect
−0.03
No effect
0
No effect
rs754585542
Asp [D]/Asn [N]
−1.9
No effect
0.76
No effect
0
No effect
rs1217666649
Asp [D]/Glu [E]
0
No effect
0.01
No effect
0
No effect
rs1391257598
Pro [P]/Thr [T]
0
No effect
0.33
No effect
0
No effect
rs7637863
Pro [P]/Leu [L]
6.5
No effect
7.11
No effect
0
No effect
rs779977931
Thr [T]/Lys [K]
0
No effect
−0.35
No effect
0
No effect
rs1182852354
Ile [I]/Val [V]
0
No effect
−0.44
No effect
0
No effect
rs750405382
Met [M]/Val [V]
3.5
No effect
0.26
No effect
0
No effect
rs1207297111
Asn [N]/Asp [D]
1.7
No effect
−0.72
No effect
0
No effect
rs1342979475
Asn [N]/Thr [T]
0
No effect
0
No effect
0
No effect
rs1274084408
Pro [P]/Ala [A]
0
No effect
0
No effect
0
No effect
rs778655073
Pro [P]/Leu [L]
0
No effect
0
No effect
0
No effect
rs549263048
Trp [W]/Arg [R]
0
No effect
0
No effect
0
No effect
rs531049536
Trp [W]/Cys [C]
0.1
No effect
−0.05
No effect
0
No effect
rs150851401
Glu [E]/Lys [K]
−1.3
No effect
0.86
No effect
0
No effect
rs766762468
Thr [T]/Ser [S]
−3.4
No effect
0.72
No effect
0
No effect
rs766762468
T (Thr) > I (Ile)
56.4
Increases
−14.66
No effect
0
No effect
rs763255363
Ala [A]/Ser [S]
−17.7
No effect
6.87
No effect
0
No effect
rs144715667
Ile [I]/Val [V]
12
No effect
−212.15
Decreases
0
No effect
rs1415169647
Met [M]/Arg [R]
−820.3
Decreases
227.06
Increases
0
No effect
rs1407375423
Trp [W]/Cys [C]
−662.7
Decreases
200.31
Increases
0
No effect
rs1800572
Val [V]/Ile [I]
−16
No effect
5.29
No effect
0
No effect
rs1800572
V (Val) > L (Leu)
−161.8
Decreases
45.81
No effect
0
No effect
rs770135219
Val [V]/Ala [A]
−382.9
Decreases
93.15
Increases
0
No effect
rs1399091893
Ala [A]/Val [V]
456.9
Increases
−154.77
Decreases
0
No effect
rs1332764085
Val [V]/Ile [I]
−12.4
No effect
3.22
No effect
0
No effect
rs5400
Thr [T]/Ile [I]
142.4
Increases
−3.5
No effect
0
No effect
rs377238940
Ala [A]/Pro [P]
−7.5
No effect
−7.19
No effect
0
No effect
rs768407637
Gly [G]/Val [V]
395.8
Increases
−4.57
No effect
0
No effect
rs753980727
Trp [W]/Cys [C]
−4.8
No effect
0
No effect
0
No effect
rs746632604
Gly [G]/Ala [A]
3.2
No effect
−0.03
No effect
0
No effect
rs772002572
Thr [T]/Ile [I]
3.6
No effect
−0.02
No effect
0
No effect
rs1476520648
Ile [I]/Asn [N]
6.4
No effect
0.04
No effect
0
No effect
rs760201098
Ala [A]/Asp [D]
179.3
Increases
−3.71
No effect
0
No effect
rs1267904495
Met [M]/Thr [T]
−39.4
No effect
0.97
No effect
0
No effect
rs1212980167
Met [M]/Ile [I]
212.1
Increases
−4.25
No effect
0
No effect
rs367856967
Val [V]/Ala [A]
−393
Decreases
11.97
No effect
0
No effect
rs970550665
Ala [A]/Gly [G]
−345
Decreases
−1.75
No effect
0
No effect
rs775283150
Ile [I]/Phe [F]
13.1
No effect
−3.23
No effect
0
No effect
rs913419413
Leu [L]/Arg [R]
−1039.9
Decreases
−4.86
No effect
0
No effect
rs771843187
Ser [S]/Pro [P]
−962.7
Decreases
49.25
No effect
0
No effect
rs144125084
Val [V]/Leu [L]
−119
Decreases
5.37
No effect
0
No effect
rs993833041
Val [V]/Ala [A]
−567.9
Decreases
28.42
No effect
0
No effect
rs1300072764
Gly [G]/Glu [E]
−327.2
Decreases
5.99
No effect
0
No effect
rs778548964
Leu [L]/Phe [F]
87.5
Increases
−2.88
No effect
0
No effect
rs1016384738
Met [M]/Lys [K]
−378.4
Decreases
11.06
No effect
0
No effect
rs770941010
Gly [G]/Arg [R]
−228.7
Decreases
6.26
No effect
0
No effect
rs777718289
Gly [G]/Glu [E]
3.5
No effect
−0.81
No effect
0
No effect
rs777718289
Gly [G]/Val [V]
0.6
No effect
0.19
No effect
0
No effect
rs372621339
Pro [P]/Arg [R]
−2.6
No effect
1.25
No effect
0
No effect
rs747025551
Ile [I]/Thr [T]
−254.4
Decreases
−380.89
Decreases
0
No effect
rs376064965
Leu [L]/Phe [F]
90.6
Increases
99.92
Increases
0
No effect
rs1188886679
Ile [I]/Leu [L]
−87.4
Decreases
−29.47
No effect
0
No effect
rs1188886679
Ile [I]/Val [V]
9.3
No effect
−467.67
Decreases
0
No effect
rs192720796
Ile [I]/Lys [K]
−292.7
Decreases
−470.84
Decreases
0
No effect
rs910976682
Ala [A]/Val [V]
190.3
Increases
−165.8
Decreases
0
No effect
rs750836049
Gly [G]/Arg [R]
−136.3
Decreases
37.18
No effect
0
No effect
rs1278964539
Arg [R]/Lys [K]
−0.1
No effect
−0.07
No effect
0
No effect
rs1231468128
Leu [L]/Gln [Q]
−8
No effect
−0.23
No effect
0
No effect
rs1445887606
Tyr [Y]/His [H]
−9.1
No effect
−6.28
No effect
0
No effect
rs1271546287
Ile [I]/Thr [T]
−10.1
No effect
−12.03
No effect
0
No effect
rs760095835
Gly [G]/Asp [D]
−2.3
No effect
−0.62
No effect
0
No effect
rs1335888503
Val [V]/Leu [L]
0
No effect
−0.02
No effect
0
No effect
rs1293130515
Met [M]/Thr [T]
0
No effect
−0.02
No effect
0
No effect
rs1019696977
Ile [I]/Val [V]
0
No effect
−5.05
No effect
0
No effect
rs144822218
Gly [G]/Ser [S]
0
No effect
43.87
No effect
0
No effect
rs759047405
Gly [G]/Asp [D]
1.7
No effect
−6.74
No effect
0
No effect
rs1441606652
Ala [A]/Val [V]
0
No effect
7.16
No effect
0
No effect
rs368626129
Ala [A]/Thr [T]
0
No effect
0.02
No effect
0
No effect
rs368626129
Ala [A]/Ser [S]
0
No effect
0.01
No effect
0
No effect
rs200213178
Ala [A]/Thr [T]
−0.1
No effect
0.01
No effect
0
No effect
rs200213178
Ala [A]/Pro [P]
−0.1
No effect
0
No effect
0
No effect
rs748052042
Leu [L]/Phe [F]
0.7
No effect
0
No effect
0
No effect
rs1412289847
Gly [G]/Ser [S]
0.1
No effect
0.02
No effect
0
No effect
rs776498787
Gly [G]/Asp [D]
0.4
No effect
−0.61
No effect
0
No effect
rs779065938
Leu [L]/Val [V]
36.5
No effect
146.08
Increases
0
No effect
rs1469335096
Ala [A]/Thr [T]
−20.5
No effect
4.53
No effect
0
No effect
rs771182536
Ile [I]/Asn [N]
−603.7
Decreases
96.63
Increases
0
No effect
rs771182536
Ile [I]/Thr [T]
−306.2
Decreases
63.05
Increases
0
No effect
rs121909741
Val [V]/Ile [I]
−2.4
No effect
−10.45
No effect
0
No effect
rs149460434
Thr [T]/Lys [K]
502
Increases
54.29
Increases
0
No effect
rs149460434
Thr [T]/Met [M]
14.7
No effect
−16.93
No effect
0
No effect
rs1276756236
Leu [L]/Ile [I]
16.9
No effect
−17.56
No effect
0
No effect
rs779591826
Ile [I]/Met [M]
−147
Decreases
82.45
Increases
0
No effect
rs1262860274
Ile [I]/Thr [T]
−148.5
Decreases
77.8
Increases
0
No effect
rs1186359171
Gly [G]/Ser [S]
12.2
No effect
−13.3
No effect
0
No effect
rs1215469128
Leu [L]/Ser [S]
0.7
No effect
−9.46
No effect
0
No effect
rs1347267249
Asn [N]/His [H]
1
No effect
−3.73
No effect
0
No effect
rs573292685
Asn [N]/Ser [S]
0.9
No effect
−5.01
No effect
0
No effect
rs764799427
Asp [D]/Val [V]
905.2
Increases
−37.64
No effect
0
No effect
rs1380319602
Ile [I]/Asn [N]
−54
Decreases
−78.29
Decreases
0
No effect
rs760641937
Gly [G]/Ala [A]
209.4
Increases
−34.8
No effect
0
No effect
rs1413841367
Leu [L]/Pro [P]
−45.1
No effect
8.09
No effect
0
No effect
rs771075989
Val [V]/Met [M]
−2.4
No effect
0.47
No effect
0
No effect
rs773581866
Arg [R]/Gly [G]
174.1
Increases
−49
No effect
0
No effect
rs770126214
Leu [L]/Ile [I]
60.4
Increases
−3.97
No effect
0
No effect
rs1374154306
Leu [L]/Gln [Q]
−491.6
Decreases
317.25
Increases
0
No effect
rs748588515
Leu [L]/Phe [F]
15.7
No effect
32.66
No effect
0
No effect
rs757087261
Phe [F]/Ile [I]
−4.5
No effect
−9.69
No effect
0
No effect
rs777020657
Phe [F]/Cys [C]
−264.3
Decreases
1.76
No effect
0
No effect
rs777020657
F (Phe) > S (Ser)
−295.4
Decreases
40.37
No effect
0
No effect
rs769089021
Ser [S]/Asn [N]
−0.1
No effect
0
No effect
0
No effect
rs780381836
Arg [R]/Gly [G]
4.2
No effect
147.36
Increases
0
No effect
rs1480881050
Tyr [Y]/His [H]
−1.6
No effect
−8.07
No effect
0
No effect
rs1250722271
Tyr [Y]/His [H]
−2.3
No effect
−8.13
No effect
0
No effect
rs1158317020
Tyr [Y]/Phe [F]
7.6
No effect
−6.38
No effect
0
No effect
rs867996396
Ile [I]/Asn [N]
−2.3
No effect
−8.12
No effect
0
No effect
rs867996396
I (Ile) > T (Thr)
−2.3
No effect
−8.19
No effect
0
No effect
rs536261161
Lys [K]/Asn [N]
62.3
Increases
385.31
Increases
0
No effect
rs745373269
Asp [D]/Asn [N]
−4.2
No effect
0.87
No effect
0
No effect
rs745373269
Asp [D]/His [H]
−4.2
No effect
1.41
No effect
0
No effect
rs1367431424
Glu [E]/Ala [A]
11.5
No effect
0.95
No effect
0
No effect
rs865881030
Glu [E]/Lys [K]
−1.8
No effect
1.2
No effect
0
No effect
rs778607566
Glu [E]/Gly [G]
4
No effect
1.31
No effect
0
No effect
rs1309254226
Glu [E]/Asp [D]
−0.1
No effect
0.45
No effect
0
No effect
rs777225980
Ser [S]/Arg [R]
−0.2
No effect
−0.6
No effect
0
No effect
rs76026576
Leu [L]/Phe [F]
0
No effect
0.07
No effect
0
No effect
rs1490504926
Asp [D]/Asn [N]
−1.9
No effect
0.74
No effect
0
No effect
rs935009475
Asp [D]/Glu [E]
0
No effect
0.01
No effect
0
No effect
rs140285191
Asp [D]/Asn [N]
−1.9
No effect
1.25
No effect
0
No effect
rs140285191
Asp [D]/Tyr [Y]
−1.9
No effect
2.54
No effect
0
No effect
rs754932741
Met [M]/Ile [I]
0
No effect
0.08
No effect
0
No effect
rs750579210
Arg [R]/Gly [G]
−0.2
No effect
0.03
No effect
0
No effect
rs1304842107
Lys [K]/Gln [Q]
−0.2
No effect
0.03
No effect
0
No effect
rs765426962
Lys [K]/Thr [T]
−0.2
No effect
0.03
No effect
0
No effect
rs761756532
Lys [K]/Asn [N]
−0.2
No effect
0.03
No effect
0
No effect
rs1329237779
Glu [E]/Lys [K]
−1.9
No effect
0.76
No effect
0
No effect
rs1384542256
Glu [E]/Ala [A]
−1.9
No effect
0.76
No effect
0
No effect
rs776912318
Ser [S]/Arg [R]
−1.3
No effect
−0.29
No effect
0
No effect
rs776912318
Ser [S]/Cys [C]
0.1
No effect
−0.04
No effect
0
No effect
rs764161243
Lys [K]/Thr [T]
123
Increases
−19.4
No effect
0
No effect
rs780873643
Val [V]/Gly [G]
−69.1
Decreases
4.31
No effect
0
No effect
rs775691314
Ser [S]/Cys [C]
30.7
No effect
−9.87
No effect
0
No effect
rs772619265
Ile [I]/Val [V]
10.1
No effect
97.69
Increases
0
No effect
rs760061096
Ile [I]/Lys [K]
−201.9
Decreases
−43.38
No effect
0
No effect
rs1205719797
Leu [L]/His [H]
−203
Decreases
−200.41
Decreases
0
No effect
rs1364855365
Thr [T]/Ser [S]
−54.4
Decreases
69.79
Increases
0
No effect
rs368432491
Asn [N]/Ser [S]
5.5
No effect
−2.26
No effect
0
No effect
rs182778895
Ser [S]/Pro [P]
−4.4
No effect
−0.59
No effect
0
No effect
rs777540740
Ser [S]/Cys [C]
0.9
No effect
0.46
No effect
0
No effect
rs769325995
Tyr [Y]/His [H]
−0.9
No effect
−0.61
No effect
0
No effect
rs374492763
Arg [R]/Gln [Q]
0.6
No effect
0.05
No effect
0
No effect
rs374492763
Arg [R]/Leu [L]
0.5
No effect
0.06
No effect
0
No effect
rs938526894
Leu [L]/Pro [P]
−548.4
Decreases
0.15
No effect
0
No effect
rs1312418962
Met [M]/Thr [T]
−36.1
No effect
0.01
No effect
0
No effect
rs1421580115
Val [V]/Met [M]
−30.3
No effect
0.05
No effect
0
No effect
rs1324205444
Ala [A]/Thr [T]
−4.9
No effect
−0.03
No effect
0
No effect
rs1324205444
Ala [A]/Ser [S]
−15
No effect
−0.04
No effect
0
No effect
rs780067980
Gly [G]/Arg [R]
−12.6
No effect
0.05
No effect
0
No effect
rs369101584
Asn [N]/Ser [S]
42.5
No effect
−0.2
No effect
0
No effect
rs757366672
Gly [G]/Cys [C]
33.1
No effect
−190.41
Decreases
0
No effect
rs767670296
Ile [I]/Asn [N]
−470.3
Decreases
−49.94
No effect
0
No effect
rs1272816101
Ile [I]/Met [M]
−365.7
Decreases
−30.37
No effect
0
No effect
rs759952425
Tyr [Y]/His [H]
−553.7
Decreases
−590.8
Decreases
0
No effect
rs751917665
Thr [T]/Met [M]
14.8
No effect
−0.05
No effect
0
No effect
rs1441375275
Ala [A]/Thr [T]
−2.3
No effect
0.02
No effect
0
No effect
rs763345848
Gly [G]/Asp [D]
−8.9
No effect
−0.68
No effect
0
No effect
rs1461795294
Ser [S]/Gly [G]
−0.1
No effect
0
No effect
0
No effect
rs773717998
Thr [T]/Asn [N]
−0.9
No effect
0.03
No effect
0
No effect
rs1162318193
Ile [I]/Thr [T]
−61.4
Decreases
0
No effect
0
No effect
rs1050103029
Val [V]/Ala [A]
−116.3
Decreases
0.03
No effect
0
No effect
rs764683908
Gly [G]/Asp [D]
−148.2
Decreases
2.22
No effect
0
No effect
rs776435170
Ala [A]/Thr [T]
−37.9
No effect
0.01
No effect
0
No effect
rs1236921754
Ala [A]/Val [V]
274.6
Increases
0
No effect
0
No effect
rs746863503
Met [M]/Arg [R]
−234.8
Decreases
3.51
No effect
0
No effect
rs775407568
Met [M]/Ile [I]
86.5
Increases
0.83
No effect
0
No effect
rs771855037
Ala [A]/Thr [T]
−1.1
No effect
0.01
No effect
0
No effect
rs140815551
Val [V]/Ile [I]
−0.1
No effect
0.01
No effect
0
No effect
rs1348497054
Ser [S]/Cys [C]
0.2
No effect
−0.05
No effect
0
No effect
rs1469035471
Val [V]/Leu [L]
−0.2
No effect
0.16
No effect
0
No effect
rs372845210
Leu [L]/Ile [I]
0.5
No effect
0.06
No effect
0
No effect
rs372845210
Leu [L]/Phe [F]
0.5
No effect
0.03
No effect
0
No effect
rs1380054283
Glu [E]/Gln [Q]
−16.6
No effect
0.77
No effect
0
No effect
rs999185720
Glu [E]/Asp [D]
−0.1
No effect
0
No effect
0
No effect
rs745619267
Lys [K]/Asn [N]
−10.2
No effect
0.03
No effect
0
No effect
rs76362149
Ala [A]/Ser [S]
−0.1
No effect
0
No effect
0
No effect
rs781225543
Arg [R]/Gln [Q]
1.3
No effect
−0.54
No effect
0
No effect
rs1321655963
Arg [R]/Cys [C]
10
No effect
−10.88
No effect
0
No effect
rs755000812
Arg [R]/His [H]
8.8
No effect
−10.45
No effect
0
No effect
rs1223071449
Phe [F]/Leu [L]
−5
No effect
−25.18
No effect
0
No effect
rs1430684701
Leu [L]/Pro [P]
−10.5
No effect
−27.2
No effect
0
No effect
rs747262541
Ser [S]/Asn [N]
−3.6
No effect
46.65
No effect
0
No effect
rs868182136
Gly [G]/Glu [E]
286.1
Increases
249.54
Increases
0
No effect
rs780255530
Met [M]/Val [V]
63.2
Increases
−24.52
No effect
0
No effect
rs758699271
Met [M]/Ile [I]
62.1
Increases
−24.31
No effect
0
No effect
rs946622803
Phe [F]/Leu [L]
−20.9
No effect
28.63
No effect
0
No effect
rs1381085405
Phe [F]/Tyr [Y]
−39.4
No effect
54.3
Increases
0
No effect
rs1381085405
Phe [F]/Ser [S]
−178.9
Decreases
280.03
Increases
0
No effect
rs750782646
Ile [I]/Asn [N]
−318.3
Decreases
268.28
Increases
0
No effect
rs765728439
Met [M]/Thr [T]
−3.8
No effect
14.75
No effect
0
No effect
rs757805176
Gly [G]/Arg [R]
−191
Decreases
26.9
No effect
0
No effect
rs757805176
Gly [G]/Arg [R]
−191
Decreases
26.9
No effect
0
No effect
rs1199637811
Val [V]/Gly [G]
−117.4
Decreases
11.9
No effect
0
No effect
rs121909747
Leu [L]/Arg [R]
−125.4
Decreases
11.4
No effect
0
No effect
rs 121,909,747
L (Leu) > P (Pro)
−133.3
Decreases
12.33
No effect
0
No effect
rs760200790
Leu [L]/Arg [R]
−62.3
Decreases
8.37
No effect
0
No effect
rs766191732
Phe [F]/Leu [L]
−25.2
No effect
0.37
No effect
0
No effect
rs1464417991
Ser [S]/Pro [P]
−94.9
Decreases
1.83
No effect
0
No effect
rs762668792
Val [V]/Glu [E]
−565.8
Decreases
46.57
No effect
0
No effect
rs1457657980
Met [M]/Val [V]
23.2
No effect
−2.74
No effect
0
No effect
rs374702599
Ile [I]/Thr [T]
−72.2
Decreases
43.39
No effect
0
No effect
rs1419532672
Ile [I]/Val [V]
−0.1
No effect
−2.52
No effect
0
No effect
rs2229608
Ile [I]/Thr [T]
5.3
No effect
−1.6
No effect
0
No effect
rs760729620
Phe [F]/Ser [S]
−14.9
No effect
−2.61
No effect
0
No effect
rs140791627
Ser [S]/Cys [C]
85.1
Increases
−1.9
No effect
0
No effect
rs746136121
Phe [F]/Leu [L]
−35.7
No effect
2.96
No effect
0
No effect
rs1353890919
Phe [F]/Cys [C]
4.5
No effect
5.85
No effect
0
No effect
rs966424064
Ile [I]/Thr [T]
0
No effect
0
No effect
0
No effect
rs779212294
Ile [I]/Met [M]
0
No effect
0
No effect
0
No effect
rs121909744
Pro [P]/Arg [R]
9.1
No effect
−0.44
No effect
0
No effect
rs121909744
Pro [P]/Leu [L]
−0.1
No effect
4.65
No effect
0
No effect
rs1309197020
Ile [I]/Met [M]
−0.2
No effect
−0.05
No effect
0
No effect
rs1309197020
I (Ile) > I (Ile)
0
No effect
0
No effect
0
No effect
rs749661374
Met [M]/Val [V]
230.5
Increases
−18.16
No effect
0
No effect
rs778490867
Met [M]/Thr [T]
−27.3
No effect
2.19
No effect
0
No effect
rs28928874
Val [V]/Glu [E]
−56.9
Decreases
–33.24
No effect
0
No effect
rs367980651
Arg [R]/Cys [C]
17.1
No effect
−2.91
No effect
0
No effect
rs75144723
Arg [R]/His [H]
16.7
No effect
−2.79
No effect
0
No effect
rs754405476
Ala [A]/Thr [T]
−15.2
No effect
−42.65
No effect
0
No effect
rs1379813904
Ala [A]/Glu [E]
−158.8
Decreases
−15.31
No effect
0
No effect
rs751226875
Ile [I]/Thr [T]
−147.2
Decreases
−53.16
Decreases
0
No effect
rs762675284
Ala [A]/Ser [S]
−118.2
Decreases
48.11
No effect
0
No effect
rs1262058831
Ala [A]/Val [V]
346.5
Increases
54.41
Increases
0
No effect
rs758246412
Ala [A]/Glu [E]
−113.4
Decreases
126.69
Increases
0
No effect
rs1203908311
Asn [N]/Ser [S]
90
Increases
−44.34
No effect
0
No effect
rs765196886
Ile [I]/Leu [L]
18.3
No effect
38.73
No effect
0
No effect
rs761784655
Ile [I]/Thr [T]
−138.3
Decreases
205.72
Increases
0
No effect
rs776395971
Val [V]/Ala [A]
−96.7
Decreases
204.13
Increases
0
No effect
rs1238600269
Ala [A]/Thr [T]
10.6
No effect
20.07
No effect
0
No effect
rs1418589512
Cys [C]/Arg [R]
−104.2
Decreases
339.43
Increases
0
No effect
rs759480075
Tyr [Y]/His [H]
−83.6
Decreases
−6.52
No effect
0
No effect
rs771274850
Ile [I]/Thr [T]
−80
Decreases
−6.87
No effect
0
No effect
rs749710583
Ala [A]/Glu [E]
−69.6
Decreases
−5.77
No effect
0
No effect
rs749710583
Ala [A]/Val [V]
142
Increases
−15.87
No effect
0
No effect
rs774542648
Gly [G]/Arg [R]
6
No effect
0
No effect
0
No effect
rs1272353608
Pro [P]/His [H]
−169.7
Decreases
−11.73
No effect
0
No effect
rs1381049817
Tyr [Y]/Cys [C]
12.4
No effect
−0.04
No effect
0
No effect
rs1336322605
Phe [F]/Leu [L]
−0.3
No effect
0
No effect
0
No effect
rs140138702
Leu [L]/Val [V]
0
No effect
0
No effect
0
No effect
rs770197371
Leu [L]/Arg [R]
−614.7
Decreases
0.4
No effect
0
No effect
rs748401954
Phe [F]/Ser [S]
−6.2
No effect
0.01
No effect
0
No effect
rs374342938
Gly [G]/Ala [A]
0
No effect
0
No effect
0
No effect
rs769037887
Val [V]/Met [M]
−0.3
No effect
0.04
No effect
0
No effect
rs556023421
Phe [F]/Leu [L]
−0.4
No effect
0.01
No effect
0
No effect
rs5397
Leu [L]/Val [V]
0.9
No effect
0
No effect
0
No effect
rs5398
Phe [F]/Leu [L]
−4.5
No effect
0.01
No effect
0
No effect
rs757137603
Thr [T]/Ile [I]
19.1
No effect
0
No effect
0
No effect
rs1441249949
Phe [F]/ Ile [I]
−2.8
No effect
0.01
No effect
0
No effect
rs1195253424
Val [V]/Ile [I]
0
No effect
0
No effect
0
No effect
rs753575081
Pro [P]/Thr [T]
0
No effect
0
No effect
0
No effect
rs777806589
Lys [K]/Arg [R]
0
No effect
0.07
No effect
0
No effect
rs766600474
Ser [S]/Thr [T]
0
No effect
27.03
No effect
0
No effect
rs766600474
Ser [S]/Ala [A]
0
No effect
151.16
Increases
0
No effect
rs1379944645
Glu [E]/Lys [K]
−1.9
No effect
−13.73
No effect
0
No effect
rs1353603250
Glu [E]/Gln [Q]
−1.9
No effect
−13.48
No effect
0
No effect
rs1446857276
Glu [E]/Asp [D]
0
No effect
−14.56
No effect
0
No effect
rs1283734332
Ile [I]/Thr [T]
0
No effect
−14.46
No effect
0
No effect
rs1445660295
Ala [A]/Val [V]
0
No effect
16.65
No effect
0
No effect
rs201797691
Ala [A]/Thr [T]
0
No effect
0.17
No effect
0
No effect
rs201797691
Ala [A]/Pro [P]
0
No effect
0.21
No effect
0
No effect
rs200160167
Ala [A]/Glu [E]
1.7
No effect
−0.33
No effect
0
No effect
rs200160167
Ala [A]/Val [V]
0
No effect
31.4
No effect
0
No effect
rs762305192
Phe [F]/Leu [L]
0
No effect
−2.56
No effect
0
No effect
rs776826621
Lys [K]/Asn [N]
−0.2
No effect
0.23
No effect
0
No effect
rs5399
Lys [K]/Asn [N]
−0.2
No effect
0.05
No effect
0
No effect
rs1412427073
Gly [G]/Ser [S]
0
No effect
0.01
No effect
0
No effect
rs966895511
Ala [A]/Ser [S]
0
No effect
0
No effect
0
No effect
rs1199349184
Ala [A]/Val [V]
0
No effect
0
No effect
0
No effect
rs374630641
Arg [R]/Ser [S]
−0.2
No effect
0.03
No effect
0
No effect
rs771586150
Pro [P]/Gln [Q]
0
No effect
0
No effect
0
No effect
rs776597156
Lys[K]/Asn[N]
−0.2
No effect
0.03
No effect
0
No effect
rs770462591
Ala [A]/Asp [D]
3
No effect
−0.72
No effect
0
No effect
rs770462591
Ala [A]/Val [V]
8.5
No effect
0
No effect
0
No effect
rs141574520
Phe [F]/Tyr [Y]
−2.7
No effect
0.06
No effect
0
No effect
rs147959014
Gly [G]/Glu [E]
2.8
No effect
−0.72
No effect
0
No effect
rs752687355
Glu[E]/Gly[G]
−4.4
No effect
0.76
No effect
0
No effect
rs781215842
Thr [T]/Ser [S]
0
No effect
0
No effect
0
No effect
rs758607933
Val[V]/Met[M]
0
No effect
0
No effect
0
No effect
4 Discussion
For most living cells, glucose is a vital energy source and an important substrate for biochemical reaction. There are two types of glucose transporters. GLUTs and SGLTs. SGLT family members consist of 580–718 amino acids and 60–80-kDa weight. GLUTs are proteins containing 12 membrane-spanning regions with intracellularl carboxyl and amino terminals. The sequence of GLUT proteins has been known to show 28–65% resemblance with GLUT1. The GLUTs symport glucose by facilitated diffusion mechanism across the plasma membrane (Navale and Paranjape, 2016). On the basis of phylogenetic relationship, 14 human GLUT proteins are grouped into three classes. Class I includes GLUT 1–4 and 14, in class II has GLUT 5,9,7, 11 and class III contains GLUT 6,13,8,12,10 (Manolescu et al., 2007). GLUT2 is a low affinity glucose transporter with affinity of Km of ∼ 17 mM, fructose (∼76 mM), galactose (∼92 mM) and mannose (∼125 mM) but high affinity glucosamine transporter (Km= ∼0.8 mM). The amino acid sequence of GLUT2 shows 81% resemblance among the human, rat and mouse.
In this study, a phylogenetic tree of 10 species of hominidae family including human was constructed using NCBI and Clustal Omega. Evolutionary tree shows that human GLUT2 is most closely related to that of Pongo abelli whereas is least related to GLUT2 of Bos Taurus among these ten members (Fig. 2). GLUT2 was predicted to have 12 transmembrane helices with intracellular amino and carboxyl termini in cytosol. It was also found to contain extracellular and intracellular loops which are located between the segments of 1st and 2nd transmembrane domains and between 6th and 7th transmembrane segments (Fig. 3)
The human genome consists of three billion base pairs. The SNPs neither cause disease nor indicate the sign of disease progression, but they can facilitate to establish the possibility that someone has acquired a specific disease. The SNPs build up approximately 90 of all genetic variations of human and take place every 100–300 bases on the genome (Noreen and Arshad, 2015). On a large scale genotyping and genome sequencing projects are producing huge data for the sequencing of diseased and healthy individuals. In human beings as well as in the model organisms the sequence variants are present in the genome of protein non-coding and coding regions. In non-coding regions, variants usually have no effect on gene expression and a regular function (Noreen et al., 2021). Association of molecular epidemiological studies is mainly concerned with non synonymous single nucleotide polymorphism (nsSNPs) that resides in exonic regions of gene (Savas et al., 2004; Zhu et al., 2004). Studying the structural as well as functional effect of nsSNPs on protein can facilitate in chosing nsSNPs which are functionally important. The activity and functioning of proteins are affected mainly by the coding variants in which truncation or frame shift, insertion or amino acids deletion or switching are included. There are so many variants, thus it is matter of great challenge to recognize important variants for disease. More than a dozen algorithms have been developed for resolving this issue. Some recent predicted algorithms are developed which utilize some score approaches that are alignment focused such as PolyPhen (Adzhubei et al., 2010), SIFT etc. In this study some sequences as well as structural homology focused algorithms were used to screen those SNPs which can play fundamental role in structural and functional changes in GLUT2.
The SIFT is a tool that was used to predict tolerant and intolerant amino acid substitution effect on protein function (Ng and Henikoff, 2001). It mainly considers homologous sequence alignment and physical amino acid properties to forecast possible influence of nsSNPs on protein. Out of 293 nsSNPs, 167 were forecasted to be damaging (Table 2). PolyPhen-2 (Adzhubei et al., 2010) was used to predict structural and functional influence of the nsSNPs lying in GLUT2. It considers homologous sequences and 3D structures of protein to predict the possible potential effect of SNPs on protein. Out of 293 nsSNPs, 165 were considered to be benign. 65 were predicted to be possibly damaging and 77 were considered to be probably damaging as shown in Table 2.
PROVEAN gave score to forecast neutral and deleterious effect of 293 nsSNPs present in coding region of GLUT2 protein. If the score was equal to or below −2.5 the results were deleterious and if the score was above −2.5, it was neutral. Out of 293 nsSNPs, 162 were considered to be neutral and 138 were predicted to be deleterious. It also gave the results of inframe deletion and insertion. Out of 4 inframe deletion 2 (rs772999215), (rs774721090) were found to be deleterious while others have shown no results. 3 SNPs were inframe insertion (rs1161394690), (rs1463507753), (rs749789723) and no results were found for them (Table 2).
The SNPeffect server was used to predict the effect of molecular phenotype of nsSNPs present in coding region of GLUT2. To find the effect of 293 nsSNPs on aggregation tendency, amyloid propensity and chaperon binding tendency, the dTANGO, dWALTZ and dLIMBO score was obtained. In a protein sequence, aggregation propensity was detected by dTANGO. Out of 293 nsSNPs, 61 were predicted to decrease and 27 to increase the aggregation propensity while 211 had not affect. Amyloid propensity was given by dWALTZ. 23 were expected to increase the amyloid propensity, 16 to decrease the amyloid propensity and 257 had not affect. According to dLIMBO, the chaperon binding tendency was given. None of the nsSNPs had any affecte on chaperon binding tendency (Table 3).
The recessive autosomal disease characterized by acquisition of hepatorenal glycogen, Fanconi nephropathy, galactose and improper carbohydrate utilization is FBS (Fanconi, 1949). Santer et al firstly recognized the underlying defect and GLUT2 as a possible site for the central defect, and three mutations in the GLUT2 gene have been investigated in this concern. At neucleotide 1405, the C /T transition was reported to result in production of truncated GLUT-2 protein leading to development to FBS. (Santer et al., 1997). Our rsults are in line with this study because same nsSNP is predicted to be intolerant, probably damaging and deleterious by SIFT, PolyPhen and PROVEAN respectively.
In human SLC2A2 several SNPs have been described: that are situated at the coding region rs7637863 Pro[P]/Leu[L] at position 68 and rs5400 Thr[T]/Ile[I] at 110 position. The connection of these SNPs with type 2 diabetes is contentious (Barroso et al., 2003). Since then, this “historical” GLUT2 SNP has been analyzed in several genetic studies, and incompatible conclusions were reached: the SNP Thr[T]/Ile[I] at 110 position was showed either related (Barroso et al., 2003; Burgdorf et al., 2011) or not related (Miller et al., 2001😉 with a risk of type 2 diabetes. Other study finds out that, rs5400 was related with the alteration from impaired glucose tolerance to hypercholesterolemia and type 2 diabetes (Laukkanen et al., 2005). During an OGTT performed on subjects stratified according to rs5400 genotype, no variation was shown in insulin secretion (Burgdorf et al., 2011), indicates that neither insulin content nor β cell mass are affected. (Laukkanen et al., 2005). In our study rs5400 was predicted to be tolerant, benign, neutral and responsible for increased aggregation tendency by SIFT, PolyPhen, PROVEAN and dTANGO respectively. Interestingly, this GLUT2 variant was related with high sugar consumption (Eny et al., 2008), indicating a GLUT2 arbitrate mechanism of glucose sensing that could modulate intake of food and sugar preference and consequently be associated with type 2 diabetes mellitus (Mueckler, 1994).
The rs7637863 Pro[P]/Leu[L] at position 68 GLUT2 SNP transported less sugar per unit of transporter, but this characteristic did not support any homeostatic disturbance, at any rate in the physiological situation. The outcome of this study showed that no affect occurs but only SIFT predicted it to be intolerant. Mostly the SLC2A2 gene mutations associated with FBS have been examined by DNA screening. The resulting GLUT2 mutants show transport function loss although biosynthesis of protein and targeting plasma membrane that is similar to wild-type protein for both of them. The syndrome of the patients having these mutations can consequently be recognized as lack of GLUT2 transport activity. Recently, compound heterozygous SLC2A2 mutations characterized by a deletion p.153–4delLI related with a missense mutation rs121909744 Pro[P]/Arg[R] at position 417 has been associated with mild FBS (mild glucosuria) (Grünert et al., 2012). Here, they showed that substitution of leucine Pro[P]/Leu[L] at position 417 diminished transport activity (Yang and Li, 2011). SIFT, PolyPhen and PROVEAN predicted (rs121909744) to have an influence on protein structure and function but SNPeffect showed no affect. Three algorithm (SIFT, PolyPhen and PROVEAN) predicted 101 SNPs to be damaging. Our results seem helpful to those epidemiologists who are involved in large-scale population-based studies. Moreover, on basis of our in silico predictions, actual role of nsSNPs can be assured by conducting demarcated in vitro and in vivo studies.
Acknowledgement
The authors would like to thank the Research Supporting Project number (RSP-2021/97) at King Saud University for funding this study, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7(4):248.
- [Google Scholar]
- Importance of maternal history of non-insulin dependent diabetic patients. Br. Med. J.. 1991;302(6786):1178-1180.
- [Google Scholar]
- Evidence from glut2-null mice that glucose is a critical physiological regulator of feeding. Diabetes. 2006;55(4):988-995.
- [Google Scholar]
- Exome sequencing as a tool for Mendelian disease gene discovery. Nat. Rev. Genet.. 2011;12(11):745-755.
- [Google Scholar]
- Candidate gene association study in type 2 diabetes indicates a role for genes involved in β-cell function as well as insulin action. PLoS Biol.. 2003;1(1):e20
- [Google Scholar]
- Glucose competence of the hepatoportal vein sensor requires the presence of an activated glucagon-like peptide-1 receptor. Diabetes. 2001;50(8):1720-1728.
- [Google Scholar]
- Positive emotional learning is regulated in the medial prefrontal cortex by GluN2B-containing NMDA receptors. Neuroscience. 2011;192:515-523.
- [Google Scholar]
- Milk synthetic response of the bovine mammary gland to an increase in the local concentration of arterial glucose. J. Dairy Sci.. 2002;85(3):494-503.
- [Google Scholar]
- Predicting the functional effect of amino acid substitutions and indels. PLoS ONE. 2012;7(10):e46688
- [Google Scholar]
- SNPeffect 4.0: on-line prediction of molecular and structural effects of protein-coding variants. Nucl. Acids Res.. 2012;40(D1):D935-D939.
- [Google Scholar]
- Deshpande, N., Addess, K.J., Bluhm, W.F., Merinoott, J.C., Townsend-Merino, W., ZHANG, Q., Knezevich, C., Xie, L., Chen, L., Feng, Z., Green, R.K., Flippen-Anderson, J.L., Westbrook, J., Berman, H.M. and Bourne, P.E., 2005. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucl. Acids Res., 33: D233-237.
- Eny, K. M., Wolever, T. M., Fontaine-Bisson, B., & El-Sohemy, A. (2008). Genetic variant in the glucose transporter type 2 is associated with higher intakes of sugars in two distinct populations. Physiological genomics, 33(3), 355-360.
- Chronic amino aciduria (amino acid diabetes or nephrotic-glucosuric dwarfism) in glycogenosis and cystine disease. Helv Pediat Acta.. 1949;4:359-396.
- [Google Scholar]
- Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat. Biotechnol.. 2004;22(10):1302-1306.
- [Google Scholar]
- Differential subcellular distribution of glucose transporters GLUT1–6 and GLUT9 in human cancer: ultrastructural localization of GLUT1 and GLUT5 in breast tumor tissues. J. Cell. Physiol.. 2006;207(3):614-627.
- [Google Scholar]
- Fanconi-Bickel syndrome: GLUT2 mutations associated with a mild phenotype. Mol. Genet. Metab.. 2012;105(3):433-437.
- [Google Scholar]
- Early diabetes and abnormal postnatal pancreatic islet development in mice lacking Glut-2. Nat. Genet.. 1997;17(3):327-330.
- [Google Scholar]
- Detection of human papillomavirus-16 DNA in archived clinical samples of breast and lung cancer patients from North Pakistan. J. Cancer Res. Clin. Oncol.. 2016;142(12):2497-2502.
- [Google Scholar]
- The extended GLUT-family of sugar/polyol transport facilitators: nomenclature, sequence characteristics, and potential function of its novel members. Mol. Membr. Biol.. 2001;18(4):247-256.
- [Google Scholar]
- Nomenclature of the GLUT/SLC2A family of sugar/polyol transport facilitators. Am. J. Physiol.-Endocrinol. Metabol.. 2002;282(4):E974-E976.
- [Google Scholar]
- Kellett, G. L., & Brot-Laroche, E. (2005). Apical GLUT2: a major pathway of intestinal sugar absorption. Diabetes, 54(10), 3056-3062.
- Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol.. 2001;305(3):567-580.
- [Google Scholar]
- Laukkanen, O., Lindström, J., Eriksson, J., Valle, T. T., Hämäläinen, H., Ilanne-Parikka, P., and Laakso, M. (2005). Polymorphisms in the SLC2A2 (GLUT2) gene are associated with the conversion from impaired glucose tolerance to type 2 diabetes: the Finnish Diabetes Prevention Study. Diabetes, 54(7), 2256-2260.
- Decreased glucose transporters correlate to abnormal hyperphosphorylation of tau in Alzheimer disease. FEBS Lett.. 2008;582(2):359-364.
- [Google Scholar]
- Facilitated hexose transporters: new perspectives on form and function. Physiology. 2007;22(4):234-240.
- [Google Scholar]
- Polymorphisms of GLUT2 and GLUT4 genes: use in evaluation of genetic susceptibility to NIDDM in blacks. Diabetes. 1990;39(12):1534-1542.
- [Google Scholar]
- Exploring the sequence determinants of amyloid structure using position-specific scoring matrices. Nat. Methods. 2010;7(3):237-242.
- [Google Scholar]
- Identification of glucokinase as an alloxan-sensitive glucose sensor of the pancreatic β-cell. Diabetes. 1986;35(10):1163-1173.
- [Google Scholar]
- Hypoglycemia in patients with type 2 diabetes mellitus. Arch. Intern. Med.. 2001;161(13):1653-1659.
- [Google Scholar]
- Glucose transporters: physiological and pathological roles. Biophys. Rev.. 2016;8(1):5-9.
- [Google Scholar]
- Ng, P. C., and Henikoff, S. (2001). Predicting deleterious amino acid substitutions. Genome Res., 11(5): 863-874.
- In Silico Analysis of SNPs in Coding Region of Human c-Myc Gene. Pakistan J. Zool.. 2015;47(5)
- [Google Scholar]
- Association of TLR1, TLR2, TLR4, TLR6, and TIRAP polymorphisms with disease susceptibility. Immunol. Res.. 2015;62(2):234-252.
- [Google Scholar]
- Knowledge and awareness about breast cancer and its early symptoms among medical and non-medical students of Southern Punjab, Pakistan. Asian Pacific J. Cancer Prevent. APJCP. 2015;16(3):979-984.
- [Google Scholar]
- Noreen, M., M. A. Shah, S. M. Mall, S. Choudhary, T. Hussain, I. Ahmed, S. F. Jalil & M. I. Raza (2012) TLR4 polymorphisms and disease susceptibility. Inflammation research : official journal of the European Histamine Research Society ... [et al.], 61, 177-88.
- Noreen, M., Muhammad Imran, Sher Zaman Safi, Muhammad Amjad Bashir, Sana Gul, Afrah Fahad Alkhuriji, Suliman Yousef Aloma, Hanan Mualla Alharbi, Muhammad Arshad, Protective role of TIRAP functional variant against development of coronary artery disease, Saudi Journal of Biological Sciences, 2021.
- Persson, B., and Argos, P. (1994). Prediction of transmembrane segments in proteins utilising multiple sequence alignments.
- Biological and chemical approaches to diseases of proteostasis deficiency. Annu. Rev. Biochem.. 2009;78(1):959-991.
- [Google Scholar]
- Topology prediction for helical transmembrane proteins at 86% accuracy–Topology prediction at 86% accuracy. Protein Sci.. 1996;5(8):1704-1718.
- [Google Scholar]
- Mutations in GLUT2, the gene for the liver-type glucose transporter, in patients with Fanconi-Bickel syndrome. Nat. Genet.. 1997;17(3):324.
- [Google Scholar]
- Fanconi-Bickel syndrome–the original patient and his natural history, historical steps leading to the primary defect, and a review of the literature. Eur. J. Pediatr.. 1998;157(10):783-797.
- [Google Scholar]
- Identifying functional genetic variants in DNA repair pathway using protein conservation analysis. Cancer Epidemiol. Biomark. Prev.. 2004;13:801-807.
- [Google Scholar]
- Glucose sensing in pancreatic β-cells: a model for the study of other glucose-regulated cells in gut, pancreas, and hypothalamus. Diabetes. 2001;50(1):1-11.
- [Google Scholar]
- Schymkowitz, J., Borg, J., Stricher, F., Nys, R., Rousseau, F. and Serrano, L., 2005. The FoldX web server: an online force field. Nucl. Acids Res., 33: W382-388.
- June). A hidden Markov model for predicting transmembrane helices in protein sequences. Ismb. 1998;6:175-182.
- [Google Scholar]
- Tennessen, J. A., Bigham, A. W., O’Connor, T. D., Fu, W., Kenny, E. E., Gravel, S., and NHLBI Exome Sequencing Project. (2012). Evolution and functional impact of rare coding variation from deep sequencing of human exomes. science, 337(6090), 64-69.
- Glucose transporters in the 21st Century. Am. J. Physiol.-Endocrinol. Metabol.. 2010;298(2):E141-E145.
- [Google Scholar]
- Cloning and functional expression in bacteria of a novel glucose transporter present in liver, intestine, kidney, and β-pancreatic islet cells. Cell. 1988;55(2):281-290.
- [Google Scholar]
- Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol.. 1998;283(2):489-506.
- [Google Scholar]
- Tusnady, G. E., and Simon, I. (2001). The HMMTOP transmembrane topology prediction server. Bioinformatics, 17(9), 849-850..
- Accurate prediction of DnaK-peptide binding via homology modelling and experimental data. PLoS Comput. Biol.. 2009;5(8):e1000475
- [Google Scholar]
- Intestinal absorption in health and disease—sugars. Best Pract. Res. Clin. Gastroenterol.. 2003;17(6):943-956.
- [Google Scholar]
- Renal Na(+)–glucose cotransporters. Am. J. Physiol. Renal Physiol.. 2001;280(1):F10-F18.
- [Google Scholar]
- miRDeep-P: a computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics. 2011;27(18):2614-2615.
- [Google Scholar]
- Zhu, Y., Spitz, M.R., Amos, C.I., Lin, J., Schabath, M.B. and WU, X., 2004. An evolutionary perspective on single-nucleotide polymorphism screening in molecular cancer epidemiology. Cancer Res., 64: 2251- 2257.
Appendix A
Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jksus.2021.101529.
Appendix A
Supplementary data
The following are the Supplementary data to this article:







