Translate this page into:
Assessment of the Genetic Diversity of Apple (Malus × domestica Borkh.) Cultivars Grown in the Kashmir Valley using Microsatellite Markers
⁎Corresponding author. jahangirdar53@gmail.com (Jahangir Ahmad Dar)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The diverse germplasm of any crop species represents an important genetic resource for mining genes or alleles necessary to meet future nutritional and disease resistance needs. A total of 29 SSR markers were used to elucidate genetic diversity among nineteen apple cultivars for the first time in the Kashmir valley. Different parameters like polymorphic information content, resolving power and marker index were calculated. A total of 218 polymorphic fragments were obtained. A high level of genetic diversity was observed in these 19 cultivars with 218 polymorphic fragments, between three and 14 alleles per primer pair, averaging 7.51 alleles per SSR. Cultivars differentiated through mutations like Oregon Spur, Reeka Red and Siliver Spur were also used as experimental cultivars in the present study and had identical allelic compositions at all loci. The cluster dendrogram and principal component analysis partitioned the cultivars into two main clusters based on Jaccard’s similarity coefficient. These findings will have impact on apple breeding and conservation programs as the present sample of apple cultivars are commercially very important at national and international level. So their characterization at morphological, biochemical, cytological and molecular level will help the apple breeders to use these in apple breeding.
Keywords
Apple germplasm
Genetic diversity
Kashmir Valley
SSR markers
1 Introduction
Apple is an important fruit crop in Kashmir Valley and ranks first in production as well as export among all the fruits in the region (hortikashmir.gov.in). It is one of the four most important fruit crops after citrus, grapes and banana, and one of the commercially most important horticultural crops in the temperate parts of the world (O’Rourke, 2003). Apple varieties are grown throughout the world including Central and West Asia, India, Western provinces of China, Europe and parts of America and Africa (Juniper et al., 1999). In India, apple is mainly grown in Jammu and Kashmir (the leading area), Himachal Pradesh, Uttarkhand, Arunachal Pradesh and Nagaland.
The cultivated apple in Kashmir is comprised of different groups of cultivars such as Delicious, Ambri, and Trel etc. In each type one or few cultivars are only commercially successful e.g. Kashmiri Ambri, American Trel, and Red Delicious etc. The rest of the cultivars in each group are sold in the market under the trade name of well-known cultivars. The monoculture of a few cultivars like Red Delicious, Kullu Delicious, Golden Delicious, American etc. associated with other constraints in the state like Apple Scab, Alternaria, Powdery Mildew and lack of cold chain storage have resulted in loss of diversity and depletion of indigenous apple germplasm and a number of apple cultivars are at the brink of extinction (Bhat et al., 2011). It is therefore important to characterize cultivars of each group so that well known cultivars are clearly distinguished from less known and commercially unsuccessful cultivars. The new cultivars with better characteristics could be identified and promoted to commercial level. The objective of this work was to analyze the genetic diversity of 19 apple cultivars in Kashmir with special reference to Ambri and Delicious cultivars using molecular markers. The information generated will help unambiguously to identify cultivars from each other.
Different types of molecular markers like RAPD, SSR, ISSR, AFLP, RFLP etc. have been used to assess the genetic diversity in crop species. The choice of the technique depends upon the objective of the study, financial constraints, skills and available facilities (Kafkas et al., 2008; Pavlovic et al., 2012). Among the different types of molecular markers, microsatellites have proved to be more reliable for DNA fingerprinting due to co-dominant inheritance, high polymorphism, abundance (Fernandez et al., 2009), reproducibility and relative ease of analysis (Schlotterer, 2004). SSR markers have been used to identify and determine genetic diversity and relationships among Malus × domestica accessions (Gasi et al., 2010; Patzak et al., 2012). Fougat (1984) and Raina (1989) characterized apple germplasm of Kashmir valley on the basis of morphology and cytology whereas Najar (2007) evaluated some apple germplasm of Kashmir Valley by ISSR based molecular markers. In the present study, SSR markers were used for the first time to identify and assess genetic diversity of apple germplasm from the Kashmir Valley.
2 Materials and methods
A total of nineteen apple cultivars (Table 1) were selected for the present study on the basis of high commercial importance in the apple market. They are sold at very high price and are also exported outside of the state to India and consist of the ’Delicious’ (indicated by D) and the ‘Ambri’ (indicated by A) groups. These cultivars were identified in private orchards and at the Govt. horticultural Nurseries of Kashmir. A single tree of each cultivar was selected and labeled with an accession number for collection of leaf samples for DNA extraction.
Cultivar
Code
Latitude
Longitude
Accession No.
Collection Site
District
Red Delicious
D1
34°02′N
74°53ʹE
RED DEL ZOU
Zoura
Srinagar
Kullu Delicious
D2
33° 57′N
74° 30′E
KUL DEL HAR
Hardu suresh
Budgam
Shimla Delicious
D3
34° 15′N
74° 83′E
SHIDEL ZAK
Zakura
Srinagar
Golden Delicious
D4
34° 09′N
74° 33′E
GOL DEL ZAN
Zangam Pattan
Baramullah
Cross Delicious
D5
34°02ʹN
74°53ʹE
CRO DEL ZOU
Zoura
Srinagar
Molies Delicious
D6
34° 18′N
74° 83′E
MOL DEL HOD
Hodura
Gandarbal
Gole Delicious
D7
34°18ʹN
74° 86ʹE
GOL DEL WAD
Wadimohalla
Srinagar
Balgarian Delicious
D8
34° 18′N
74° 83′E
BALDEL BAK
Bakura
Ganderbal
Oregon Spur
D9
34° 09′N
74° 33′E
ORE SPU ZAN
Zangam pattan
Baramullah
Reeka Red
D10
33° 72′N
74° 82′E
REE RED DAS
Dashpora Shopian
Shopian
Siliver Spur
D11
34° 09′N
74° 33′E
SIL SPUZAN
Zangam Pattan
Baramullah
Kashmiri Ambri
A1
34°02ʹN
74°53ʹE
KAS AMB ZOU
Zoura
Srinagar
Lal Ambri
A2
34°02ʹN
74°53ʹE
LAL AMB ZOU
Zoura
Srinagar
Ambri Cross
A3
34°02ʹN
74°53ʹE
AMB CRO ZOU
Zoura
Srinagar
Balgarian Ambri
A4
33° 72′N
74° 82′E
BAL AMB SHO
Shopian
Shopian
Vilayati Ambri
A5
34° 09′N
74° 33′E
VIL AMB ZAN
Zangam pattan
Baramullah
Delicious Ambri
A6
33° 72′N
74° 82′E
DEL AMB SHO
Shopian
Shopian
Dudh Ambri
A7
34°02ʹN
74°53ʹE
DUD AMB ZOU
Zoura
Srinagar
High Density Ambri
A8
33° 72′N
74° 82′E
HIG AMB SHO
Shopian
Shopian
2.1 DNA extraction and purification
DNA was isolated from young leaf samples using the cetyl trimethyl ammonium bromide (CTAB) protocol of Doyle and Doyle (1990). The extracted DNA was treated with RNase to remove the RNA. The DNA quantity was estimated after separation in 0.7% agarose gel stained with ethidium bromide in the presence of different known concentrations of lambda (λ) DNA. The final concentration of all the DNA samples was adjusted to50 ng µl−1 for subsequent PCR.
2.2 SSR analysis
For SSR analysis, PCR reaction mixture was prepared in 200 µl tubes. Final concentrations of the reagents were as follows: 1x PCR buffer, 1.5 mM MgCl2, 200 µM of each dNTP, 0.5 µM of each primer, 1 unit of taq DNA polymerase 5 U/µl and ultrapure water to reach the final volume of 20 µl. The volume of DNA used as template was 1.5 µl. PCR program was set as follows- initial denaturation: 95 °C for 5 min; denaturation: 95 °C for 30 s; annealing: 55 °C for 30 s; elongation: 72 °C for 60 s; repetition: 35 cycles. The last step was a final extension of 72 °C for 10 min.
The fluorescently-labeled PCR products were mixed with 0.3 µl of Gene Scan-500 ROX size standard (Applied Biosystems) and 12 µl of Hi-Di Formamide (Applied Biosystems) and separated by capillary electrophoresis on an ABI PRISM 3100. The experiment was replicated at least three times to verify the reproducibility of markers. The amplified fragments were scored with GeneScan 3.7 and Genotyper 3.7 software (Applied Biosystems) as 1 for presence and 0 for the absence of allele.
2.3 Data analysis
The following parameters were considered for each assay unit as described by Zargar et al. (2016); Number of polymorphic alleles (NPA); Number of monomorphic alleles (NMA); Fraction of polymorphic loci (β) = NPA/(NPA + NMA); Effective multiplex ratio (EMR) = nβ, where n is the total number of bands and β is the fraction of polymorphic loci;
Polymorphic information content (PIC) = 2fi (1-fi), where fi is the frequency of present bands and 1-fi is the frequency of absence bands;
Marker index (MI) = PIC × EMR; Resolving power (RP) = ∑Ib, where Ib can be calculated by the formula as Ib = 1- [2 × (0.5-p)], where p is the frequency of individual band present.
The scored binary data generated from SSR with present alleles scored as 1 and absent alleles as 0 was used for the construction of dendrogram by Jaccard’s similarity coefficient using NTSYS- pc version 2.02e (Rohlf, 1998). The principal component analysis was also performed to differentiate the cultivars. (See Fig. 1)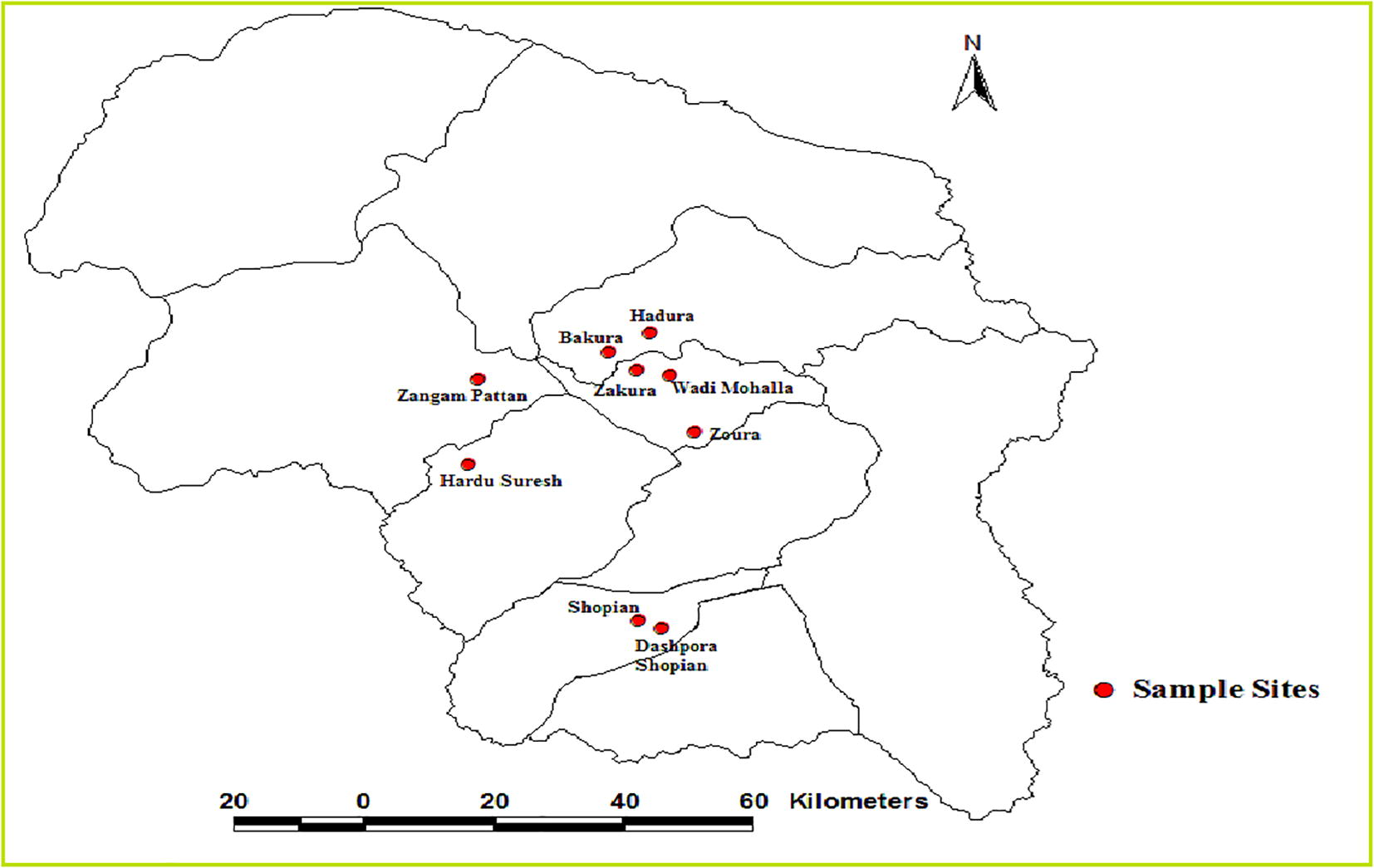
Map of Kashmir Valley showing collection sites of apple cultivars. Source: SOI Toposheet 1971.
3 Results
In the present study a highly informative set of 29 SSR primers (Table 2) was used to distinguish 19 apple cultivars from Kashmir valley. A total of 218 alleles were obtained by 29 SSR primers. The allele number for each primer varied from 3 (Hi06f09) to 14 (Hi08f12) with a mean number of 7.51 alleles per primer (Table 2). In general the size of the amplified DNA fragments scored ranged from 96 to 362 bp. The largest number of alleles was generated by Hi08f12 (14 alleles) followed by Hi05d10, CH03h06 and CH04f04 (13 alleles each). Primer pairs Hi06b06, CH03b01, CH04f03 and CH04f07 produced 10 alleles each in all the nineteen apple cultivars. On the other hand, the minimum number of alleles was amplified by Hi06f09 (3 alleles) followed by Hi08h03, Hi08a04, Hi11a01, Hi23d03 and CH04C03, each amplified 4 alleles in all the cultivars. In order to identify the most efficient primers that could distinguish all the cultivars either individually or in combination, three different indices like Polymorphic Information Content (PIC), Markers Index (MI) and Resolving Power (RP) were applied in the present study (Table 2). Allelic composition for each cultivar is presented in Table 3. NA: Number of alleles; NPA: Number of polymorphic alleles; PIC: Polymorphic information content; MI: Marker index; RP: Resolving power D1-Red Delicious, D2-Kullu Delicious, D3- Shimla Delicious, D4-Golden Delicious, D5-Cross Delicious, D6-Molies Delicious, D7-Gole Delicious, D8-Balgarian Delicious, D9-Oregon Spur, D10-Reeka Red, D11- Siliver Spur A1-Kashmiri Ambri, A2-Lal Ambri, A3-Ambri Cross, A4- Balgarian Ambri, A5-Vilayati Ambri, A6- Delicious Ambri, A7-Dudh Ambri, A8-High Density Ambri
Primer
Forward sequence (5′–3′)
Allele Range
NA
NPA
PIC
MI
RP
Hi05c06
F ATTGGAACTCTCCGTATTGTGC
143–183
5
5
0.45
2.25
2.319
R ATCAACAGTAGTGGTAGCCGGT
Hi05d10
F AATGGGTGGTTTGGGCTTA
147–362
13
13
0.31
4.03
5.263
R GTTTCTTTGGCTATTAGGCCTGC
Hi06f09
F AACCAAGGAACCCACATCAG
290–297
3
3
0.48
1.44
1.265
R GTTTCACTTACACACGCACACACG
GD147
F TCCCGCCATTTCTCTGC
158–172
8
8
0.32
2.56
3.269
R GTTTAAACCGCTGCTGCTGAAC
Hi08h03
F GCAATGGCGTTCTAGGATTC
150–172
4
4
0.37
1.48
1.897
R GGTGGTGAACCCTTAATTGG
Hi02a07
F TTGAAGCTAGCATTTGCCTGT
129–300
8
8
0.19
1.52
1.894
R TAGATTGCCCAAAGACTGGG
Hi01c06
F TTAGCCCGTATTTGGACCAG
144–163
5
5
0.40
2.00
3.241
R GTTTCACCTACACACACGCATGG
Hi06b06
F GGTGGGATTGTGGTTACTGG
171–283
10
10
0.27
2.70
3.473
R GTTTCATCGTCGGCAAGAACTAGAG
Hi02d11
F GCAATGTTGTGGGTGACAAG
210–275
6
6
0.33
1.95
3.157
R GTTTGCAGAATCAAAACCAAGCAAG
Hi08c05
F TCATATAGCCGACCCCACTTAG
173–265
9
9
0.34
3.06
4.315
R GTTTCACACTCCAAGATTGCATACG
Hi08a04
F TTGTCCTTCTGTGGTTGCAG
178–266
4
4
0.46
1.84
1.371
R GTTTGAAGGTAAGGGCATTGTGG
Hi08f12
F GGTTTGTAACCCGTCTCTCG
129–235
14
14
0.22
3.08
3.894
R GTTTCGTAGCTCTCTCCCGATACG
Hi08e06
F GCAATGGCGTTCTAGGATTC
150–184
5
5
0.36
1.80
3.055
R GTTTGGCTGCTTGGAGATGTG
Hi23b12
F TGAGCGCAATGACGTTTTAG
157–222
6
6
0.21
1.26
2.21
R GTTTCAGGCTTTCCCTTCAGTGTC
Hi11a01
F ACCGCCAAATGCTTTGTTAC
227–240
4
4
0.45
1.80
2.002
R GTTTCCTCCATTAAACTCCTCAGTG
AU223486-SSR
F TGACTCCATGGTTTCAGACG
222–228
5
5
0.36
1.80
2.424
R AGCAATTCCTCCTCCTCCTC
Hi23d02
F CCGGCATATCAAAGTCTTCC
174–234
4
4
0.42
1.68
2.423
R GTTTGATGGTCTGAGGCAATGGAG
CH03b01
F ACAAGGTAACGTACAACTCTCTC
158–234
10
10
0.29
2.90
3.684
R GTCACAAAACCGCCAGATG
U78948-SSR
F GATCGTCCGCCACCTTAAT
231–265
6
6
0.39
2.34
2.53
R AGGGTTTTCATCATGCACATT
CH03ho6
F TTGTCCCTTTTTACGTCTTTCC
163–191
13
13
0.26
3.38
4.210
R GTTATTGAGCAAGGCGGAGA
CH02e12
F CCAACTTTTTCTGCGGTAGTG
178–234
9
9
0.25
2.25
2.842
R TGGGACCCATATGGTTGAATAC
CH04C03
F TGCACACCAAACACAGGACT
212–246
4
4
0.42
1.68
2.423
R TATCAAACATTGGGGCACTG
CH04a06
F AGAAAATCTAAGAGCAGCAG
123–252
8
8
0.29
2.32
2.947
R TAAAACTCAAGTCGCCCGTC
CH04d11
F ATTAGGCAATACACAGCAC
110–163
8
8
0.26
2.08
2.441
R GCTGCTTTGCTTCTCACTCC
CH04d08
F AATTCCACATTCACGCATCT
131–159
8
8
0.32
2.56
3.473
R TTGAAAGACGGAAACGATCA
CH04F03
F CTTGCCCTAGCTTCAAATGC
177–207
10
10
0.30
3.00
2.894
R TCGATCCGGTTAGGTTTCTG
CH04e12
F CCTGAAATCTGCACAACTACCA
242–251
6
6
0.34
2.04
2.425
R GGTGGTGAAGAAGTAGACAGCC
CH04F07
F CAGATCATGAATGATTGAAA
96–202
10
10
0.22
2.20
2.631
R GAAAATCACACCCTCAAACCAT
CH04F04
F GTCGGTCACAACTCAGGACC
166–240
13
13
0.25
3.25
4.105
R CGACGTTCGATCTTCCTCTC
Average/primer
7.51
7.51
0.32
2.28
2.89
Primer
D1
D2
D3
D4
D5
D6
D7
D8
D9
D10
D11
Hi05c06
173,178
173,178
173,178
173,183
173,178
173,183
173
173,183
173,178
173,178
173
Hi05d10
260,270,357,362
268,270,338,362
268,270,338,362
229,339
246,268,338,362
339
246,260,357
229,339
268,338
268,270,338,362
268,270
Hi06f09
297
297
297
290,297
291,297
290,297
291
297
297
297
297
GD147
158
158,172
158,172
158,160
162,172
158,170
158,162
158,168
Nil
158
Nil
Hi08h03
172
172
172
171
172
171
172
171
172
172
172
Hi02a07
300
Nil
Nil
129,135
277
129,131,133
277
133
Nil
Nil
Nil
Hi01c06
145,163
145,163
145,163
160
145,163
144,156,160
163
160
145,163
145,163
145,163
Hi06b06
252,275
258,275
258,275
251
252,258
251,270
252
251
258,275
258,275
258,275
Hi02d11
215,275
265,275
265,275
214
215,275
210
215,261
214
265,275
265,275
265,275
Hi08c05
247,257
247,251,257
247,251,257
253,265
247,257
256
247,251
250,256
251,257
247,251,257
247,251,257
Hi08a04
263,266
263
263
263,266
263
263
263,266
263,266
263
263
Nil
Hi08f12
159
159,172
147,159,172
129
162,172
129,231
159,162
145
147,159,172
147,159,172
159,172
Hi08e06
151,172
151,172
151,172
150
151,172
150
151,172
150
151,172
151,172
151,172
Hi23b12
157
159
159
170
Nil
170
186
157,170
159
159
159
Hi11a01
231,234
234
234
231,234,240
234,240
234
231,234
231,240
234
234
234
AV223486-SSR
223,227
227
227
222,225
223,227
222
223
225
227
227
227
Hi23d02
231,234
234
234
174,180
234
174
231,234
174
234
234
234
CH03b01
158,172,180,198
158,198
158,198
179
180,198
179,181
180
177,183
158,198
158,198
158,198
U78948-SSR
231,234
234
234
262,265
234,263
262
231,234
262,265
234
234
234
CH03ho6
174,184
174,190
174,190
163,173,183
170,172,174
173,191
180,182,184
163,183
174,190
174,190
174,190
CH02e12
180,218
180
180
178
180,218
208,216
218
214
180
180
180
CH04C03
215
215
215
212
215
212
215
212,214
215
215
215
CH04a06
124
124
124
123,125
124,142
123,141
128,142
123
124
124
124
CH04d11
155
155
155
154
155
110,154
143,155,160
147
155
155
155
CH04d08
148
131,148
131,148
142,152
131,159
132,152
152
132,154
131,148
131,148
131,148
CH04F03
202
202,204
202,204
201,207
193,202
201,203
177,202
195,201
202,204
202,204
202,204
CH04e12
243
243
243
242,246
243,251
242
243,251
246,250
243
243
243
CH04F07
124
124,159
124
104,112
124
110,112
178,202
96
124
124
159
CH04F04
167,180,218
180,234
180
166,178
180,218
184,208
167,218,231,234
166
180,185
180,185
180,185,234
Primer
A1
A2
A3
A4
A5
A6
A7
A8
Hi05c06
173,178
173,183
145,178
143,178
173,178,183
173,183
173,183
183
Hi05d10
240,256,338
229,339
151,160,270,338
147,256,268,338
256,268,338
339
339
339
Hi06f09
291
297
291,297
297
291,297
290,297
290
297
GD147
162,166
158,168
166,172
158,168
162,166,172
166,172
162,166
158,168
Hi08h03
172
150
158
158
172
171
Nil
171
Hi02a07
277
129,133
Nil
263
298
135
131,135
133
Hi01c06
163
160
145,163
163
145,163
144,160
160
160
Hi06b06
252,283
251,276
171,272
171,275
252,275
251,274
251
251
Hi02d11
215
214
215,265
215,275
215
214,262
214
214
Hi08c05
247,251,257
250,256
173,251,257
173,282
251
250
256
250,256
Hi08a04
263,266
263,266
180,266
178
263,266
263,266
263,266
263,266
Hi08f12
162,166
145
180,176
182,168,176
166,172
145,235
145
145
Hi08e06
151,155,172
150
184,172
184,172
151,155,172
150,154
150,154
150
Hi23b12
159
157,170
222
222,172
159
157
170
170
Hi11a01
234
231,240
227
227
231,234
234
234
231,240
AV223486-SSR
223,227
225
Nil
228,227
227
225
222,225
225
Hi23d02
234
174
Nil
231
231,234
174,180
174,180
174
CH03b01
180
177,183
234,180
234,180
158,180,198
179,197
179
Nil
U78948-SSR
234
262,265
234
231,240
234
262,265
262,265
Nil
CH03ho6
172,182
163,183
174,182
164,184
172,174,182
173,181
171,183
163,183
CH02e12
216
214
180,216,234
208,210,216,218
180,216
178
214
204
CH04C03
215
212,214
246
246
215
212,214
212,214
212,214
CH04a06
142
123
251,128,142
252,126
124,142
123,141
141
123
CH04d11
143,155
147
163
163
143,155
154
154
147
CH04d08
159
132,152
148,159
133
148,159
148,158
158
132,154
CH04F03
177,193
195,201
177,204
197
177,202,204
177,203
177,191
195,201
CH04e12
243,251
246,250
243
243
243
242
242,250
246,250
CH04F07
124
96
124
197,202
159
96,122
96
96
CH04F04
216,234
166
180,185,216
167,216,231,240
180,234
178
214
166
3.1 Cultivar relationships based on SSR analysis
The UPGMA separated the apple cultivars into two main clusters (Fig. 2). Cluster I consisted of twelve cultivars while the remaining seven of the cultivars were found in cluster II. Both the clusters were divided into sub clusters. The ‘Red Delicious’ sub-group consisted of six cultivars: Red Delicious, Kullu Delicious, Shimla Delicious, Reeka Red, Oregon Spur and Siliver Spur. Kullu Delicious, Shimla Delicious had the same allele composition at all SSRs while ‘Reeka Red’ was closely related with difference at two of the 29 SSRs. Cross Delicious, Kashmiri Ambri and Vilayati Ambri also grouped together in a separate sub-cluster. In cluster II, two small sub-clusters were again formed. The Golden Delicious sub-cluster consisted of Golden Delicious, Molies Delicious, Delicious Ambri and Dudh Ambri whereas the remaining three cultivars, Balgarian Delicious, Lal Ambri and High Density Ambri formed the second sub-group within cluster II. The Jaccard’s similarity coefficient based on SSR data ranged from 0.05 to 0.93. (Fig. 2). The three cultivars: Oregon Spur (D9), Reeka Red (D10) and Siliver Spur (D11) which are said to be sports of Red Delicious were different from each other and grouped together with Kullu Delicious (D2) and Shimla Delicious (D3) in one sub cluster.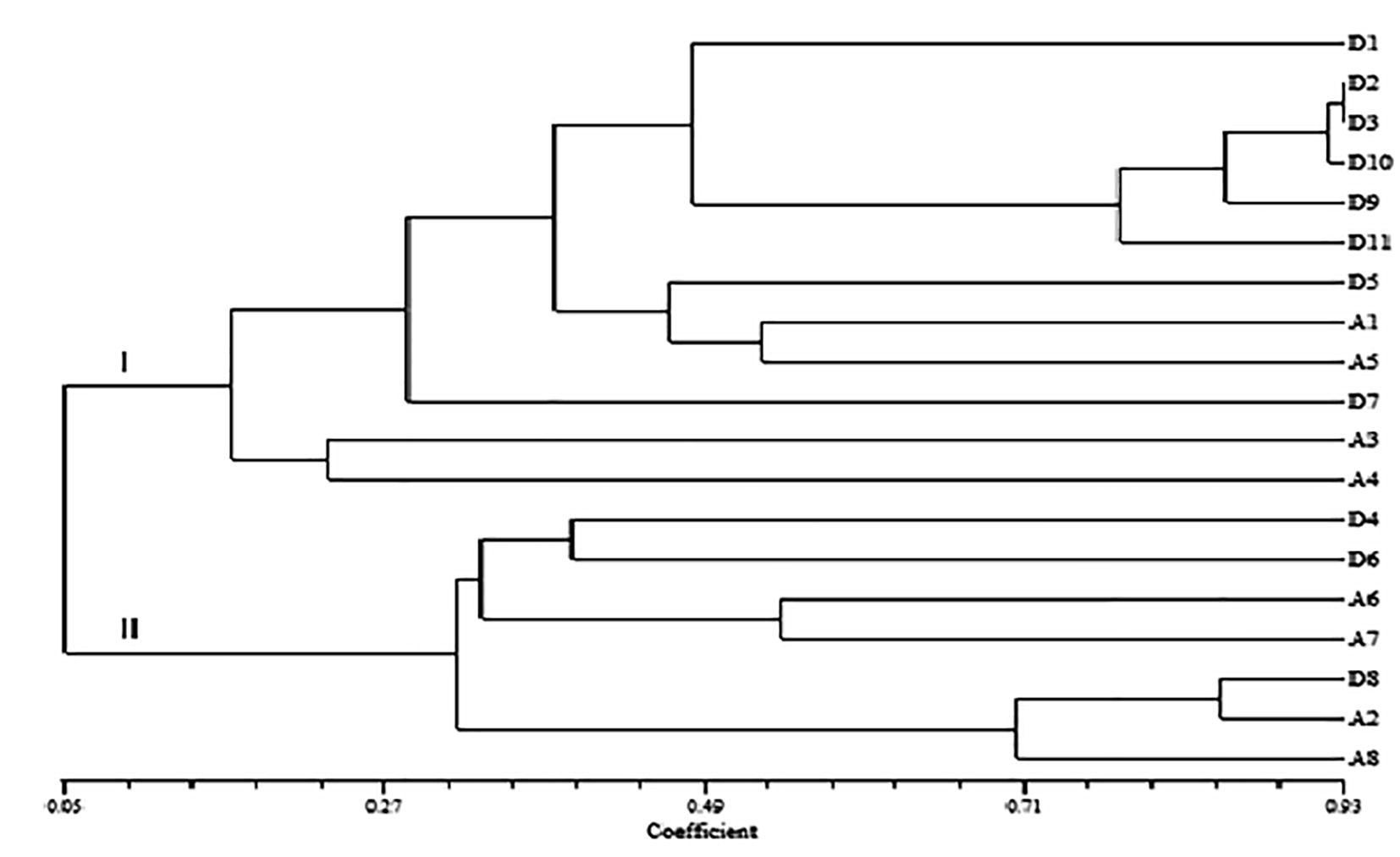
UPGMA cluster analysis based on Jaccard’s similarity coefficient. D1-Red Delicious, D2-Kullu Delicious, D3-Shimla Delicious, D4-Golden Delicious, D5-Cross Delicious, D6-Molies Delicious, D7-Gole Delicious, D8-Balgarian Delicious, D9-Oregon Spur, D10-Reeka Red, D11-Siliver Spur, A1-Kashmiri Ambri, A2-Lal Ambri, A3-Ambri Cross, A4-Balgarian Ambri, A5-Vilayati Ambri, A6-Delicious Ambri, A7-Dudh Ambri, A8-High Density Ambri.
The UPGMA cluster analysis revealed that some Ambri and Delicious cultivars form a separate subgroup. There are no possible reasons as the present study is just a preliminary survey in which only 19 cultivars and 29 SSR primers were used. The limited number of primers has generated little information. So the use of maximum number of primers to cover most of the linkage groups can provide more and more information. As such we can not say that the Ambri apple cultivars have developed from Delicious group due to some hybridizations events taking place in the orchards because there is no literature available regarding the origin of most of Ambri as well as Delicious cultivars. It may be possible that some Ambri cultivars would have been developed from Delicious by natural hybridisation events in the orchards.
PCA also supported the groups obtained with cluster analysis. Most of the Delicious cultivars grouped together along with few cultivars from the Ambri group. Five cultivars from the Delicious group namely Kullu Delicious (D2), Shimla Delicious (D3), Reeka Red (D10), Oregon Spur (D9) and Siliver Spur (D11) formed a separate group at one corner in PCA plot, thus indicating close similarity to each other. On the other hand, the second group consisted of seven cultivars which include Kashmiri Ambri (A1), Gole Delicious (D7), Ambri Cross (A3), Balgarian Ambri (A4), Vilayati Ambri (A5), Cross Delicious (D5) and Red Delicious (D1). The third group also was comprised of seven cultivars which includes Molies Delicious (D6), Golden Delicious (D4), Delicious Ambri (A6), Dudh Ambri (A7), High Density Ambri (A8), Lal Ambri (A2) and Balgarian Delicious (D8) (Fig. 3).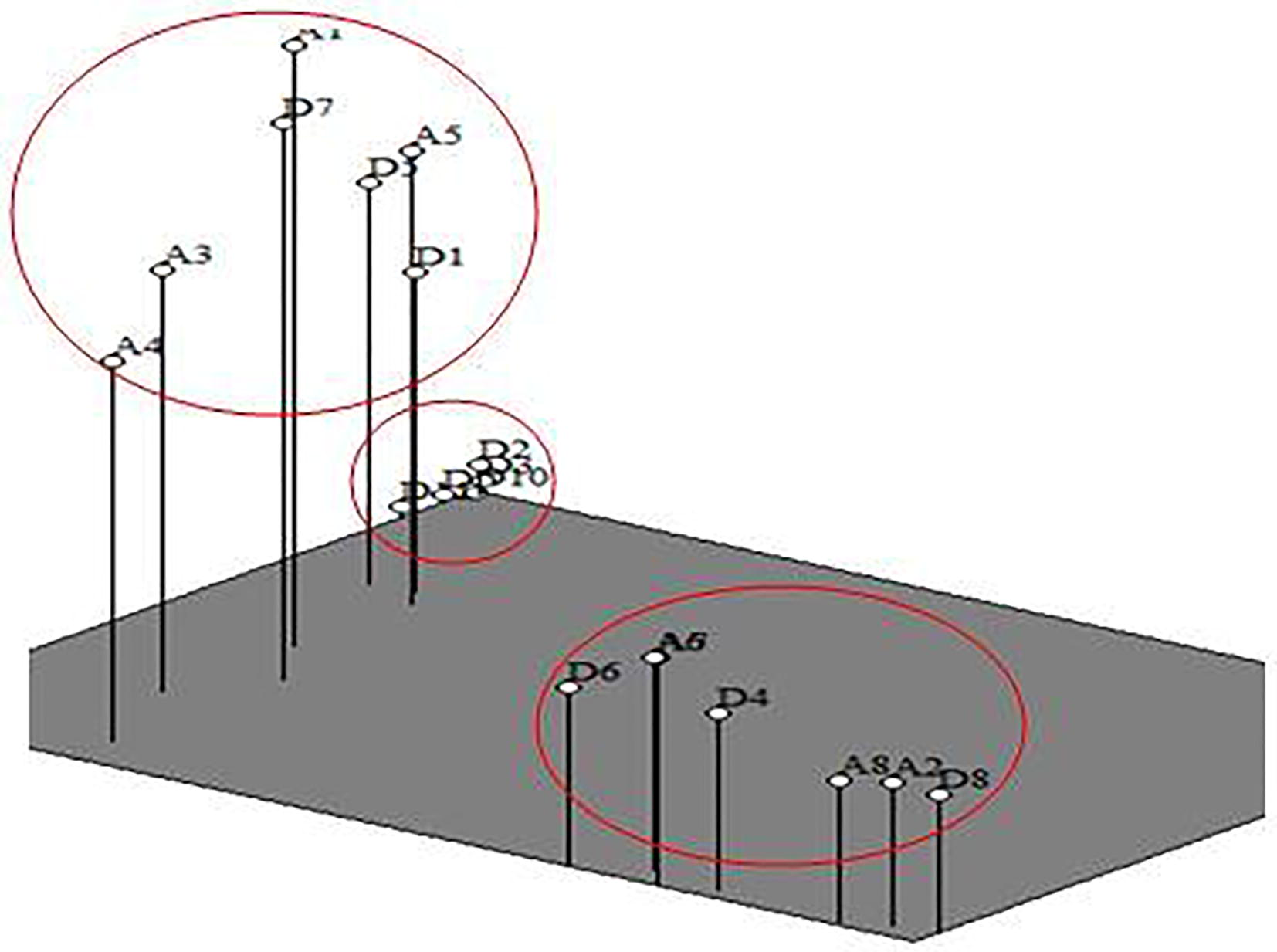
3D PCA plot of apple cultivars.
4 Discussion
Assessment of genetic diversity within a cultivated crop has important consequences in breeding and the conservation of genetic resources. Several molecular markers have been used widely for the analysis of genetic diversity and cultivar identification in large number of species. Molecular markers have succeeded in differentiating cultivars, classifying synonyms, identifying mislabeled cultivars, establishing genetic relationships and giving hints about the process of domestication (Anand, 2000; Wunsch and Hormaza, 2002). SSR markers are the preferred DNA markers for the analysis of genetic relationships and diversity within crop species due to their high polymorphism level, abundance, co-dominant inheritance (Fernandez et al., 2009), reproducibility and relative ease of analysis (Schlotterer, 2004). Hundreds of microsatellite markers have been developed in apple and some have been placed on genetic linkage maps (Liebhard et al., 2002; Silfverberg-Dilworth et al., 2006). Microsatellites have been also used as markers to predict important traits like resistance to apple scab (Vinatzer et al., 2004).
In the present investigation SSR data for 19 apple cultivars revealed a total of 218 polymorphic fragments with 29 primer pairs. The mean number of alleles per primer obtained was 7.51 which is similar to the results reported earlier by different groups (Wichmann et al., 2007; Pereira-Lorenzo et al., 2007). Gasi et al. (2010) selected ten genomic SSRs to assess genetic diversity in 39 cultivars of apple and reported that the average number of alleles per SSR is 10.4. Gao et al. (2007) analyzed 59 apple cultivars using 12 SSRs and detected an average of 14.7 alleles per primer. The higher average number of alleles per SSR primer may be attributed to multi allelic nature of SSR primers. The multi allelic SSRs produce more than two alleles even in diploid cultivars. Multi locus SSRs indicates how many alleles are present in the genome. There is nothing like triploid and tetraploid nature of these cultivars as the present samples were analysed based on cytology which proved all these cultivars diploid with 2n = 34.
Marker indices like PIC, MI, RP etc. are informative parameters to detect the levels of genetic diversity in an organism. In the current study, the primers with highest marker indices values will help in the screening of genetic polymorphism among apple cultivars. The respective values for each informative index have been reported in Table 2. It is anticipated that these primers would help apple researchers to pick up and conduct further downstream studies related to genetic amelioration.
Allelic compositions of most of the primer pairs have proved that Kullu Delicious and Shimla Delicious resemble the three sports (Oregon Spur, Reeka Red and Siliver Spur) investigated in the present study. By screening 29 SSR primers for their informativeness, the present study demonstrates that four primers Hi05d10, Hi08c05, CH03h06 and Ch04F04 have highest resolving power i.e. these detect enough base pair variation among nineteen apple cultivars to allow their distinction. Due to close interrelationships and narrow gene pool of the accessions in this study, additional markers/primers will be needed to fully characterize and distinguish a large set of cultivars. This study will enable us to identify a standard set of primers that can be used to distinguish the apple germplasm of our state.
5 Conclusion
The purpose of our study was to assess the genetic diversity of the apple germplasm of Kashmir Valley. SSR analysis based on 29 primer pairs have separated the cultivars of Delicious group and it was also found that Kullu Delicious and Shimla Delicious resemble in allelic composition with the sports like Oregon Spur, Reeka Red and Siliver Spur. All the observations made in this study will provide valuable evidence for decision making in choosing of markers for future work, characterization of germplasm, breeding and apple germplasm management.
Acknowledgements
The authors are highly thankful to Department of Biotechnology, Government of India for providing financial assistance as a part of the project entitled “Creating a Genomics Platform for Apple Research in India” vide no. DBT/PR/11040PBD/16/812/2008 dated on June 4, 2010. Dr. Sajad M. Zargar is highly acknowledged for assisting in statistics. We would like to thank to Director Horticulture, Kashmir Division for necessary permissions during field surveys. Thanks are also due to Mr. Manzoor Ahmad Bhat Pomology Expert for his unconditional help during field surveys.
Compliance with ethical standards.
Conflict of interest
All the authors declare that they have no conflict of interest.
References
- Anand, L., 2000. Molecular markers and their application in horticultural crops. In: Chadha, K.L., Ravindran, P.N., Sahijran (eds) Biotechnology in horticultural and plantation crops. Malhotra Publishing House, New Delhi.
- Effect of interstock on juvenility and tree size of ambri apple. Acta Hortic.. 2011;903:435-437.
- [Google Scholar]
- A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull.. 1990;19:11-15.
- [Google Scholar]
- Genetic diversity in Spanish and foreign almond germplasm assessed by molecular characterization with simple sequence repeats. J. Am. Soc. Horticultural Sci.. 2009;134:535-542.
- [Google Scholar]
- Fougat, R.S., 1984. Assessment of the germplasm of apple grown in Kashmir Valley. Ph.D. Thesis. University of Jammu, India.
- Analysis of genetic relationship for Malus germplasm resources by SSR markers. J. Fruit Sci.. 2007;24:129-134.
- [Google Scholar]
- Genetic assessment of apple germplasm in Bosnia and Herzegovina using microsatellite and morphologic markers. Scientia Horticulturae. 2010;126:164-171.
- [Google Scholar]
- http///hotrtikashmir.gov.in
- Molecular characterization of mulberry accessions in Turkey by AFLP markers. J. Am. Soc. Hortic. Sci.. 2008;4:593-597.
- [Google Scholar]
- Development and characterisation of 140 new microsatellites in apple (Malus × domestica Borkh.) Mol. Breed.. 2002;10:217-241.
- [Google Scholar]
- Najar, M.A., 2007. Molecular Characterization of Apple (Malus pumila Mill.) cultivars of Kashmir using DNA based markers. M. Phil. Dissertation, Dept. of Botany, University. Kashmir, Kashmir.
- World production, trade, consumption and economic outlook for apples. In: Ferree D.C., Warrington I.J., eds. Apples: botany, production, and uses. CAB international, UK: CABI publishing; 2003. p. :15-28.
- [Google Scholar]
- Genetic diversity of Czech apple cultivars inferred from microsatellite markers analysis. Hortic. Sci.. 2012;39(4):149-157.
- [Google Scholar]
- Characterization of onion genotypes by use of RAPD markers. Genetika. 2012;2:269-278.
- [Google Scholar]
- Evaluation of genetic identity and variation of local apple cultivars (Malus × domestica Borkh.) from Spain using microsatellite markers. Genet. Resour. Crop Evol.. 2007;54:405-420.
- [Google Scholar]
- Raina, R., 1989. Germplasm assessment of apple (Malus pumila Mill.) cultivars of Kashmir Valley. M. Phil. Thesis, University of Jammu, Jammu.
- Rohlf, M., 1998. NTSYS-pc: numerical taxonomy and multivariate analysis system. Version 2.2. Dept. of Ecology and Evolution. State University of New York.
- The evolution of molecular markers— just a matter of fashion? Nat. Rev. Genet.. 2004;5:63-69.
- [Google Scholar]
- Microsatellite markers spanning the apple (Malus × domestica Borkh.) genome. Tree Genet. Genomics. 2006;2:202-224.
- [Google Scholar]
- Isolation of two microsatellite markers from BAC clones of the Vf scab resistance region and molecular characterization of scab-resistant accessions in Malus germplasm. Plant Breed.. 2004;123:321-326.
- [Google Scholar]
- Molecular identification of old Hungarian apple varieties. Int. J. Hortic. Sci.. 2007;3:37-42.
- [Google Scholar]
- Cultivar identification and genetic fingerprinting of temperate fruit tree species using DNA markers. Euphytica. 2002;125:59-67.
- [Google Scholar]
- Unraveling the efficiency of RAPD and SSR markers in diversity analysis and population structure estimation in common bean. Saudi J. Biol. Sci.. 2016;23:139-149.
- [Google Scholar]







