Translate this page into:
Antimicrobial resistance profiles of Escherichia coli isolated from companion birds
⁎Corresponding author at: Department of Microbiology, Faculty of Veterinary Medicine, Istanbul University Cerrahpasa, TR-34320 Avcılar, Istanbul, Turkey belgis@istanbul.edu.tr (Belgi Diren Sigirci)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The increase of antimicrobial resistance in companion animals has already been reported worldwide; however, there is a deficiency of data and studies focusing on the resistance profiles of Escherichia coli isolates from Passeriformes and Psittaciformes which regarded to be among the most common and popular pet/companion bird species. For this reason, the current research was aimed to evaluate the presence of E. coli isolates from apparently healthy companion birds and their antimicrobial resistance profiles.
Two hundred sixty-five cloacal swab samples collected from apparently healthy companion birds (116 parakeets, 59 canaries, 56 parrots, 30 Indian nightingales, 3 finches, and 1 Golden finch) were examined by conventional bacteriological procedures for the identification of E. coli. Susceptibilities against 16 antimicrobials from 8 different classes and extended-spectrum beta-lactamase (ESBL) production was also determined. Moreover, all isolates were analysed by PCR assays for ESBL, Metallo-β-lactamases, serin-carbapenemase, AmpC beta-lactamase, plasmid-mediated quinolone resistance (PMQR), and aminoglycoside resistance genes.
E. coli were isolated from 37.7% of the samples. Majority of the isolates were found resistant to tetracycline (84%) followed by sulfamethoxazole/trimethoprim (46%), streptomycin (34%), and kanamycin (25%). Eleven parakeet and 2 parrot isolates were found resistant to all quinolone class antimicrobial agents, and three parakeet isolates showed resistance to all aminoglycosides. Additionally, 67% of the isolates exhibited multi-resistance, defined herein as 3 or more antimicrobial classes.
PMQR determinants (qnrB and qnrS) were determined in 3 E. coli isolates from a parrot and two parakeets. Furthermore, seven aminoglycoside resistance genes [aac(3)-IIa(aacC2), strA, strB, aadA (aadA1 or aadA2), aphA1, aphA2, and ant(2’’)-Ia(aadB)] were found in 3 parakeet isolates. Also, the most promising result of this study is that ESBL production was not determined phenotypically and genotypically.
Here, we present the first report that various aminoglycosides and quinolone resistance genes of E. coli isolates from parakeets, and a parrot was present in Turkey. In summary, the consequences of the current research emphasise that the companion birds may act as substantial reservoirs carrying antimicrobial resistance and estimate the potential risk for humans, it is critical to define their role as reservoirs.
Keywords
Antimicrobial resistance
Companion birds
Parakeet
Parrot
1 Introduction
The population of companion animals has been increased unquestionably in the whole world (Pomba et al., 2017). Pet birds which are the third most common animal as a companion after dogs and cats, are regarded as close friends of humans and play an important role in human life (Cong et al., 2014). The majority of caged birds are from two orders: Passeriformes including canaries, finches, and Psittaciformes including parrots, parakeets, and lovebirds (Boseret et al., 2013).
The emergence of antimicrobial resistance (AMR) is not a novel phenomenon, nor a surprising one. AMR has been regarded about One Health issue which associated not only human health but also animal health and the environment (Argudín et al., 2017). All over the world, the emergence and spread of AMR continue incessantly and unfortunately leaving devastating health and economic consequences. Household pets are recognised as significant risks for zoonotic diseases and considered as a source of multidrug-resistant (MDR) bacteria. Many authors emphasise that the drug-resistant bacteria or AMR genes may be transmitted from animals to man and vice versa, through contaminated food, the environment or via contact (Damborg et al., 2016, Argudín et al., 2017, Pompa, 2017). The close contact between household pets and humans creates many opportunities for transmission. Besides, the treatment of the domestic birds is carried out empirically, potentially promoting the AMR (Giacopello et al., 2015). E. coli, a member of the Enterobacteriaceae family, is the most prevalent commensal inhabitant of the gastrointestinal tracts of humans and animals. As a commensal, it lives in a mutually beneficial association with hosts and rarely causes disease. It is, however, also one of the most common human and animal pathogens as it is responsible for a broad spectrum of diseases. Also, it can be part of the commensal resident of microbiota and opportunistically colonise birds are considered reservoirs of drug-resistant bacteria and related genes (Machado et al., 2018). It has estimated that the flexibility and adaptability of E. coli to continuously changing environs provides a large number of resistance mechanisms and commensal E. coli can be considered as a source of AMR (Szmolka and Nagy, 2013).
The increase of AMR in companion animals has already been reported, but there is a lack of studies focusing on the effect of antimicrobial on the pathogens isolated from Passeriformes and Psittaciformes. The purpose of the current research was to detect the presence of E. coli in good condition Psittacines and Passeriformes, which become a common companion in the home and to evaluate their antimicrobial resistance profiles.
2 Material and methods
In total 265 cloacal swab samples from apparently healthy companion birds (116 parakeets, 59 canaries, 56 parrots, 30 Indian nightingales, 3 finches, and 1 Golden finch) were collected in a random manner, from Faculty of Veterinary Medicine clinics, breeding aviaries, and from different pet shops, all located in Istanbul. The protocols were approved by the Animal Care Committee of Istanbul University (Approval no: 2015/105).
All the swabs were cultured in buffered peptone water and incubated at 37 °C for 24 h. After incubation, a loopful of each culture was streaked onto MacConkey agar plates and incubated at 37 °C for 24 h. Strains initially were identified by conventional methods as E. coli (Krieg and Holt, 2005) and confirmed with API 20E strips.
Isolates were tested for antibiotic susceptibility by standard disk diffusion procedures to 16 different antimicrobials from 8 antimicrobial classes: amikacin (30 μg), amoxicillin-clavulanic acid (30 μg), ampicillin-sulbactam (20 μg), aztreonam (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), gentamicin (10 μg), kanamycin (30 μg), levofloxacin (5 μg), meropenem (10 μg), nalidixic acid (30 μg), norfloxacin (10 μg), ofloxacin (10 μg), streptomycin (10 μg), sulfamethoxazole/trimethoprim (1.25/23.75 µg) and tetracycline (30 μg). Extended-spectrum beta-lactamase (ESBL) production was analysed by the double disk diffusion test, and the results were based on CLSI breakpoints (CLSI, 2018). As quality controls, Escherichia coli ATCC 25,922 were tested in each run. Multiresistance was considered as resistance at least three different antimicrobial classes.
Multiplex PCR assays were used to screen all E. coli isolates for ESBL (OXA, CTX-M, TEM, SHV, GES, VEB, PER, KPC, VIM, and IMP), metallo-β-lactamases (GIM, NDM, SIM, and SPM), serin-carbapenemase (IMI, SME and NMC-A), AmpC beta-lactamase (ACC, ACT, DHA, CMY, FOX, LAT, MIR, and MOX) and PMQR genes (qnrA, qnrB, qnrS, qnrC, qnrD, qepA, and aac(6′)-Ib-cr) (Dallenne et al., 2010, Voets et al., 2011, Ciesielczuk et al., 2013). Moreover, PCR assays were performed to determine the aminoglycoside resistance genes (strA, strB, aph(3)-Ia, aac(3)-IIa, aac(3)-III, aac(3)-IV, aac(6′)-Ib, ant(3″)-Ia, aadA, aphA1, aphA2, ant(2′’)-Ia, armA, and armB) (Maynard et al., 2004, Saenz et al., 2004, Doi et al., 2007, Costa et al., 2008, Karczmarczyk et al., 2011, Zhang et al., 2014).
3 Results
Entirely 100 (37.7%) of the 265 cloacal swabs were positive for E. coli. 82/116 (70.6%) of the parakeets, 5/59 (8.4%) of the canaries, 8/56 (14.2%) of the parrots, 4/30 (13.3%) of the Indian nightingales, and 1/3 (33.3%) of the finches isolates were identified as E. coli (Table 1).
Sources
No. of isolates/samples (%)
Breeder no
Clinic no
Pet shop no
1
2
3
4
1
2
3
4
5
1
2
3
4
5
6
7
8
Species
Parakeets
9/13
13/15
11/17
–
3/4
–
6/9
11/12
3/6
6/9
–
9/10
9/14
0/5
–
2/2
–
82/116 (70.6%)
Canaries
–
–
0/10
0/5
2/4
0/1
–
–
0/4
0/5
3/5
0/9
0/10
–
0/6
–
–
5/59 (8.4%)
Parrots
–
2/2
0/10
0/5
–
0/1
0/1
0/2
–
2/11
–
3/10
1/7
0/5
–
–
0/2
8/56 (14.2%)
Indian Nightingales
–
1/5
1/6
0/3
–
–
–
1/3
–
0/8
–
–
–
–
–
1/5
–
4/30 (13.3%)
Finches
–
–
–
–
1/2
–
–
–
–
–
–
–
–
–
–
–
0/1
1/3 (33.3%)
Golden Finch
–
–
–
–
0/1
–
–
–
–
–
–
–
–
–
–
–
–
0/1
Total
9/13
16/22
12/43
0/13
6/11
0/2
6/10
12/17
3/10
8/33
3/5
12/29
10/31
0/10
0/6
3/7
0/3
100/265 (37.7%)
Table 1. Number of E. coli isolates/Number of samples among species and sources
It was determined that 11 of the isolates were susceptible to all tested antimicrobials. Other isolates presented different resistance rates to the tested antimicrobials. Majority of the isolates were found resistant to tetracycline (84%) followed by sulfamethoxazole/trimethoprim (46%), streptomycin (34%), and kanamycin (25%). The results are presented in Fig. 1.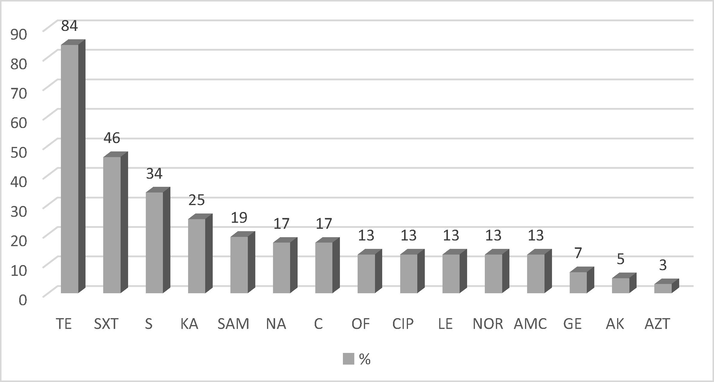
The rates of resistance decreasing in the order. TE: Tetracycline, SXT: Sulfamethoxazole/Trimethoprim, S: Streptomycin, KA: Kanamycin, SAM: Ampicillin-Sulbactam, C: Chloramphenicol, NA: Nalidixic Acid, OF: Ofloxacin, CIP: Ciprofloxacin, LE: Levofloxacin, NOR: Norfloxacin, AMC: Amoxicillin-Clavulanic Acid, GE: Gentamycin, AK: Amikacin, AZT: Aztreonam.
Thirteen isolates (11 parakeets and 2 parrots’ samples collected from the same breeder) were resistant to all quinolone class antimicrobial agents phenotypically. Also, 3 of the strains isolated from parakeets showed resistance to all aminoglycoside class of antimicrobial agents. In particular, MDR occurred in 67% of the isolates. MDR was observed in 57/116 (49.1%) of the parakeets, 3/59 (5%) of the canaries, 6/56 (10.7%) of the parrots, and 1/3 (33.3%) of the finches. Canaries’ MDR isolates were identified from a same pet shop.
Also, ESBL production was not determined phenotypically. None of the isolates was resistant to meropenem.
PCR analysis revealed that none of the susceptible isolates harboured the tested genes. However, 6 of the 67 (8.9%) MDR isolates harboured at least one of these genes (Table 1). PMQR determinants were found in 3 isolates: qnrB (1%) and qnrS (2%). Furthermore, seven aminoglycoside resistance genes were found in 3 resistant isolates: aac(3)- IIa(aacC2) (1%), strA (2%), strB (3%), aadA(aadA1 or aadA2) (1%), aphA1 (2%), aphA2 (1%),and ant(2′’)-Ia(aadB) (1%). ESBL production was not determined genotypically. The distribution of the genes among MDR E. coli isolates is presented in Table 2.
Source
Species
Aminoglycoside Resistance Genes
PMQR genes
aac(3)-IIa (aacC2)
strA
strB
aadA (aadA1 or aadA2)
aphA1
aphA2
ant(2′’)-Ia (aadB)
qnrB
qnrS
Breeder No 2
Parakeet 1
+
+
+
+
Parakeet 2
+
+
+
+
+
Parakeet 3
+
Parakeet 4
+
Parrot 1
.+
Pet shop No 8
Parakeet 5
+
+
4 Discussion
The complex hazard of AMR transmission from companion animals to man has not been fully established. Consequently, studies on AMR, performed in many countries of the world, unceasingly continue to provide more information. Unfortunately, data regarding AMR in companion birds are scarce. The current study is, to our knowledge, one of the scarce and comprehensive reviews on AMR that focuses explicitly on Passeriformes and Psittaciformes as companion birds.
In the current study, the obtained results demonstrated a 37.7% E. coli isolation rate in the Passeriformes and Psittaciformes. The isolation rates were 70.6% of the parakeets, 33.3% of the finches, 14.2% of the parrots, 13.3% of the Indian nightingales, and 8.4% of the canaries. In many studies focused on various bird species, E. coli isolation rates have been reported as 48% from the necropsy samples and cloacal swabs of psittacines living in captivity (Corrêa et al., 2013), 59.5% from the necropsy samples and cloacal swabs of canaries from domiciliary breeding locations (Horn et al., 2015), 46.5% from the cloacal swabs of psittacine birds maintained in the Wildlife Rehabilitation Center (de Souza Lopes et al., 2015), 10% from the cloacal swabs of captive cockatiels (De Pontes et al., 2018) and 36.1% from the cloacal swabs of free-living grey-breasted parakeets (Machado et al., 2018). The differences between rates could be based on multiple criteria, including geographical differences, sampling techniques, and detection procedures.
In the list of “Critically Important Antimicrobials for Human Medicine” of World Health Organization (WHO), chloramphenicol and tetracycline were stated as “highly important antimicrobials”. Erythromycin, streptomycin, ciprofloxacin, gentamycin and ampicillin were considered as “critically important antimicrobials” (World Health Organization, 2017). According to the “List of Antimicrobials of Veterinary Importance” of the World Organisation for Animal Health (OIE), tetracycline and ampicillin were considered as “very important” agents and erythromycin and streptomycin were regarded as extremely important (OIE, 2015). In this study, among the 100 E. coli isolates, 89 (89%) were resistant to at least one of the antimicrobials and the most frequently displayed resistances were determined to tetracycline (84%). Several authors have also observed a resistance rate to tetracycline; 41% from the cloacal swabs of captive cockatiels (De Pontes et al. (2018)), and 28.6% from the cloacal swabs of free-living grey-breasted parakeets (Machado et al. (2018)). Regarding previous reports in Turkey (65–100%), our results are consistent (Yılmaz and Dolar, 2017, Diren Sigirci et al., 2019). However, tetracycline resistance rate in Turkey is higher than the other countries. In our country, the wide uncontrolled use of antimicrobial compounds in human and veterinary practices, animal production, agriculture and industrial technology, and an increase in population mobility and the circulation of food might be the reason for this crucial problem. Factors responsible for the emergence and dissemination of resistant and MDR strains establish risk for human and animal health due to an increase in morbidity, mortality and the cost associated with the treatment of infections (Clemente et al., 2015).
Trimethoprim-sulfamethoxazole exhibits broad-spectrum activity through oral efficacy, and it is particularly proper used for therapy of birds. Diren Sigirci et al. (2019) reported that 38% of the isolates from cloacal swabs of the synanthropic birds were resistant to SXT. Relatively, the high resistance rate to sulphamethoxazole/trimethoprim (46%) was observed in this study.
Otherwise, 13% of the isolates obtained from the same breeder (11 parakeets and two parrots) were resistant to all analysed quinolone class antimicrobial agents. De Pontes et al. (2018) presented 41% resistance to quinolones from healthy captive cockatiels, and it is interpreted that this high percentage of resistance may be the result of empirical treatments without veterinary supervision. Also, the risk of contamination and spread should be taken into account in the places where the breeder is sold.
In the current study, three of the parakeet isolates sampled from the same breeder showed resistance to all aminoglycoside class of antimicrobial agents. Herein, if we consider rates severally, various resistances for other antimicrobials were recorded; streptomycin (34%), kanamycin (25%), gentamycin (7%), and amikacin (5%). In contrast to the present study, the higher resistance rates to streptomycin were also reported. Horn et al. (2015) determined that E. coli strains isolated from necropsy samples and cloacal swabs of canaries were resistant 4% to gentamycin and 40% to streptomycin. Therewithal, De Pontes et al. (2018) notified the high resistance rate to aminoglycosides (74%), and streptomycin (67%) from cockatiels kept in captivity.
A few studies were focused on the AMR of Gram-negative bacteria isolated from cage pet birds. Lopes et al. (2018) stated that ESBL-producing E. coli from faecal samples of psittacines had not been published yet. In this research, ESBL were not detected, which is the most promising result of this study. Contrary, the authors reported a 2.7% isolation rate for ESBL from cage birds, in Turkey (Yılmaz and Dolar, 2017). This promising result has to be confirmed by future studies and needs to be underlined.
MDR presence in companion animals may present a severe risk. Probably the most worrying result in the current study was the detection of 67% multiple drug resistance rates. MDR rate detected are quite higher than those described by 33.8% Hidasi et al. (2013), 55.7% Horn et al. (2015), and 59% de Pontes et al. (2018).
The majority of the studies focus mostly on phenotypic resistance profiles. Due to this deficiency, in the current study, PCR analysis was conducted for screening some of the AMR genes. As a result of the current data, it was revealed that the AMR genes were disseminated unequally among the isolates, besides some isolates harboured multiple genes.
PMQR is a significant phenomenon, and the most common PMQR genes found among Enterobacteriaceae isolates have been qnrS, qnrB and aac(6′)-Ib-cr (Araújo et al., 2017). In the current research, PMQR genes were found in 3 MDR isolates: qnrB (1%) and qnrS (2%). Whereas some researchers are already working on it, currently there is still little work focused on the cage birds. Alcalá et al. (2016) pointed out that it is also noteworthy the finding of PMQR genes in an only single isolate. Even though, a report concluded that PMQR were not detected from parrot (Clemente et al., 2015). This is the first paper of the qnrB gene presence from a parrot and qnrS from two parakeets, in Turkey.
Aminoglycosides are clinically important therapeutic agents commonly applied to treat infections particularly severe infections by Gram-negative bacteria, but they are less preferred at birds because of the toxicity disadvantage (Flammer, 2006). Unfortunately, a considerable resistance to aminoglycosides has been noticed, it is worth noting the detection of seven aminoglycoside resistance genes (aac(3)-IIa(aacC2), strA, strB, aadA(aadA1 or aadA2), aphA1, aphA2,and ant(2′’)-Ia(aadB)) in 3 MDR isolates from parakeets. Here, we demonstrated, for the first time in Turkey, the presence of aminoglycosides resistance in E. coli isolates from parakeets. It should be appreciated that direct contact with the owners of the cage birds may probably facilitate the possible transmission. It should be noted that resistant bacteria could also have been horizontally transmitted by the owners (Machado et al., 2018).
5 Conclusion
There is a deficiency of data concerning antimicrobial resistance in companion birds, more studies should be conducted, and more data should be collected in order to monitor future trends to build further knowledge. Nevertheless, infected/carrying/untreated birds could become a potential reservoir for humans, and they have consequences on public health. In an ideal situation, surveillance and early diagnosis should be performed in every imported bird bunch including animals captured from the wild. In both humans and animals, the use of antimicrobials caused an increase in the incidence of resistance in both pathogenic and endogenous bacteria, highlighting a serious health problem to human medicine. Information obtained from systematic surveillance studies is essential for monitoring changes in the antimicrobial resistance among pathogens, and appropriate antibiotic treatments.
In conclusion, the current results showed that resistance to various antimicrobials, unfortunately, and extensively, are available in companion birds, and this study present novel data that will support to fill knowledge. This outcome highlights the requirement to avoid the usage of unnecessary and inadequate antimicrobials, and to update continuous data by providing appropriate scientific support and to carry out multidisciplinary studies in the scope of “One health”.
6 Disclosure of funding
This research was supported by the Scientific Research Projects Coordination Unit of Istanbul University (Project number: 20265).
References
- Wild birds, frequent carriers of extended-spectrum β-lactamase (ESBL) producing Escherichia coli of CTX-M and SHV-12 types. Microb. Ecol.. 2016;72(4):861-869.
- [Google Scholar]
- High frequency of the combined presence of QRDR mutations and PMQR determinants in multidrug-resistant Klebsiella pneumoniae and Escherichia coli isolates from nosocomial and community-acquired infections. J. Med. Microbiol.. 2017;66(8):1144-1150.
- [Google Scholar]
- Bacteria from animals as a pool of antimicrobial resistance genes. Antibiotics.. 2017;6(2):12.
- [Google Scholar]
- Development and evaluation of a multiplex PCR for eight plasmid-mediated quinolone-resistance determinants. J. Med. Microbiol.. 2013;62:1823-1827.
- [CrossRef] [Google Scholar]
- Antimicrobial susceptibility and oxymino-β-lactam resistance mechanisms in Salmonella enterica and Escherichia coli isolates from different animal sources. Res. Microbiol.. 2015;166(7):574-583.
- [Google Scholar]
- Clinical and Laboratory Standards Institute Performance standards for antimicrobial susceptibility testing: twenty-fourth informational supplement. CLSI document M100S 28th Edition. Wayne, PA: CLSI, 2018.
- Detection of virulence factors in Escherichia coli and analysis of Salmonella spp. in psittacines. Pesquisa Veterinária Brasileira.. 2013;33(2):241-246.
- [Google Scholar]
- Prevalence of antimicrobial resistance and resistance genes in faecal Escherichia coli isolates recovered from healthy pets. Vet. Microbiol.. 2008;127:97-105.
- [Google Scholar]
- Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. J. Antimicrob. Chemoth.. 2010;65(3):490-495.
- [Google Scholar]
- Bacterial zoonoses transmitted by household pets: state-of-the-art and future perspectives for targeted research and policy actions. J. Comp. pathol.. 2016;155(1):27-40.
- [Google Scholar]
- Survey on pathogenic Escherichia coli and Salmonella spp. in captive cockatiels (Nymphicus hollandicus) Braz. J. Microbiol.. 2018;49:76-82.
- [Google Scholar]
- Prevalence and antimicrobial resistance profile of enterobacteria isolated from psittaciformes of illegal wildlife trade. Acta Sci. Vet.. 2015;43(1):1-9.
- [Google Scholar]
- Tetracycline Resistance of Enterobacteriaceae Isolated From Feces of Synanthropic Birds. J. Exot. Pet Med.. 2019;28:13-18.
- [Google Scholar]
- Community acquired extended-spectrum β-lactamase producers, United States. Emerg. Infect. Dis.. 2007;7:1121-1123.
- [Google Scholar]
- Antibiotic drug selection in companion birds. J. Exot. Pet Med.. 2006;15(3):166-176.
- [Google Scholar]
- Antibiotic-resistance patterns of Gram-negative bacterial isolates from breeder canaries (Serinus canaria domestica) with clinical disease. J. Exot. Pet Med.. 2015;24(1):84-91.
- [Google Scholar]
- Enterobacterial detection and escherichia coli antimicrobial resistance in parrots seized from the illegal wildlife trade. Journal of Zoo and Wildlife Medicine. 2013;44(1):1-7.
- [CrossRef] [Google Scholar]
- Identification and antimicrobial resistance of members from the Enterobacteriaceae family isolated from canaries (Serinus canaria) Pesqui. Vet. Bras.. 2015;35(6):552-556.
- [Google Scholar]
- Characterization of antimicrobial resistance in Salmonella enterica food and animal isolates from Colombia: identification of a qnrB19-mediated quinolone resistance marker in two novel serovars. FEMS Microbiol. Lett.. 2010;313:10-19.
- [Google Scholar]
- Facultatively anaerobic gram-negative rods. Family I. Enterobacteriaceae. Bergey's Manual of Systematic Bacteriology.“ Baltimore. Williams & Wilkins. 2005;1:408-420.
- [Google Scholar]
- Serogroup identification, phenotypic detection of hemolysis and extended spectrum beta-lactamases of Escherichia coli isolated from psittacine of illegal wildlife trade in Fortaleza, Brazil. Arq. Bras. Med. Vet. Zootec.. 2018;70(3):823-829. In this issue
- [CrossRef] [Google Scholar]
- Isolation and Antimicrobial Resistance Profiles of Enterobacteria from Nestling Grey-Breasted Parakeets (Pyrrhura Griseipectus) Rev. Bras. Ciênc. Avíc.. 2018;20(1):103-110.
- [Google Scholar]
- Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J. Clin. Microbiol.. 2004;42(12):5444-5452.
- [Google Scholar]
- OIE list of antimicrobial agents of veterinary importance. Paris. World Organisation for Animal Health. 2015. (http://www.oie.int/fileadmin/Home/eng/Our_scientific_expertise/docs/p (Accessed 10 January 2019)
- Public health risk of antimicrobial resistance transfer from companion animals. J. Antimic. Chemoth.. 2017;72(4):957-968.
- [Google Scholar]
- Mechanisms of resistance in multiple-antibiotic-resistant Escherichia coli strains of human, animal, and food origins. Antimicrob. Agents Chemother.. 2004;48(10):3996-4001.
- [Google Scholar]
- Multidrug resistant commensal Escherichia coli in animals and its impact for public health. Front. Microbiol.. 2013;4:258.
- [Google Scholar]
- A set of multiplex PCRs for genotypic detection of extended-spectrum β-lactamases, carbapenemases, plasmid-mediated AmpC β-lactamases and OXA β-lactamases. Int. J. Antimicrob. Agents.. 2011;37(4):356-359.
- [Google Scholar]
- Detection of extended-spectrum β-lactamases in Escherichia coli from cage birds. J. Exot. Pet Med.. 2017;26(1):13-18.
- [Google Scholar]
- Critically important antimicrobials for human medicine: ranking of antimicrobial agents for risk management of antimicrobial resistance due to nonhuman use. 2017. Accessed 10 January 2019
- [Google Scholar]
- A survey of the frequency of aminoglycoside antibiotic-resistant genotypes and phenotypes in Escherichia coli in broilers with septicaemia in Hebei. China. Br. Poult. Sci.. 2014;55(3):305-310.
- [Google Scholar]







