Translate this page into:
16S rRNA gene identification and phylogenetic analysis of dhofar toad (Bufo dhufarensis) from riyadh province, saudi arabia
⁎Corresponding author. afrefaei@ksu.edu.sa (Abdulwahed Fahad Alrefaei)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
The Arabian Peninsula is home to many unique organisms due to its distinctive geographical location. Seven species of amphibians inhabit Saudi Arabia. One of them is the Dhofar toad (Bufo dhufarensis), a species of toad in the family Bufonidae found in Saudi Arabia, the United Arab Emirates, Yemen and Oman. This species lives near freshwater springs, pools, dams, rivers and rural gardens. By evaluating the genetic variety within evolutionary lineages using sequence analysis, we were able to determine the degree of genetic diversity of the Dhofar toad (Bufo dhufarensis). Additionally, we examine the probable connections between this species and other toads and frogs. We collected 27 samples from Bufo dhufarensis and prepared the samples for DNA extraction and PCR using suitable primers. The 16S ribosomal RNA (rRNA) gene sequences in toads from Riyadh Province were determined for the first time and deposited in a public gene data repository. Bufo dhufarensis, which was found in Riyadh Province, is very closely related to the toad Bufo dhufarensis, which was found in Oman, according to a phylogenetic tree based on the 16S rRNA sequence. This is the first report of the genetic diversity of Bufo dhufarensis in Riyadh Province based on the 16S rRNA gene.
Keywords
Bufo dhufarensis
16S ribosomal RNA
Phylogenetic tree
Genetic distance
Molecular analysis
1 Introduction
The Middle East region is distinguished by its geographical location situated between three regions – the Palearctic, Oriental and Afrotropic ecozones – which makes it diverse in its environment, topography and vitality. The Arabian Peninsula has a unique group of animal and plant species that have resulted from the dynamic geological history and changing climate of this region (Mittermeier et al., 1999, Metallinou et al., 2015, Myers et al., 2000).
Amphibians have a high degree of endemism and a restricted potential for growth due to their reliance on freshwater environments for reproduction and their intolerance to saltwater (Al-Obaid et al., 2017, Tinley, 1994). For these reasons, amphibians are an interesting system for investigating historical colonialism and biogeography in Riyadh Province, north Oman, east United Arab Emirates and southwest Saudi Arabia (Balletto, 1985, Frost et al., 2006, Gvoždík et al., 2010).
Amphibians offer unique opportunities for investigating evolutionary processes by virtue of their biological traits, such as morphological convergence, and their diversity of habitats, including fossorial, terrestrial and arboreal (Wells, 2010, Bossuyt and Milinkovitch, 2000).
Extracted DNA amplification by polymerase chain reaction (PCR) has been used for molecular analysis of amphibians, as it is an extremely sensitive and specific tool to identify taxon (Jayawardena et al., 2017, Al-Qahtani and Amer, 2019, Harris, 2001). The 16S ribosomal RNA (rRNA) gene is used almost exclusively in characterising phylogenetic trees of closely related or symbiotic organisms (Nakahara et al., 2004, Al-Qahtani and Amer, 2019, Metallinou et al., 2012). This gene has highly conserved and reliable molecular markers that have evolved slowly and are functionally preserved (EARDLY and Van Berkum, 2005). Pratihar et al. (2016) used 16S rRNA to examine the evolutionary connections among Duttaphrynus species, which is widely accepted as a suitable phylogenetic marker for resolving vertebrate relationships. Hedges et al. (1993) and Pratihar et al. (2016) both reported that a region of the mitochondrial 16S rRNA gene is useful for elucidating aspects of amphibian phylogeny, and it has become an essential molecular marker.
In this study, we collected Dhofar toad (Bufo dhufarensis) specimens from three localities in Riyadh Province and acquired DNA sequences from the 16S rRNA gene to examine the genetic variation of the Dhofar toad specimens and explore the relationships among this species and other toads and frogs.
2 Materials and methods
2.1 Sample collection
We collected tissue samples from 27 Dhofar toad (Bufo dhufarensis) individuals sampled from different sites (8 in Al-Kharj region, 10 in Al-Hareeq region, and 9 in Al-Aflaj region) in Riyadh Province, Saudi Arabia (Table 1). All individual samples were collected in September 2020, using the toe-clipping technique as described by Sambrook (1989). The tissue samples were stored in ethanol and sent to the King Saud University in Riyadh for analysis. For subsequent examination, the tissue samples were kept at −20 °C.
Sample No.
Species
Locality
GenBank Accession No.
A1
Dhofar toad Bufo dhufarensis
Al-Aflaj
MW242776
A2
Dhofar toad Bufo dhufarensis
Al-Aflaj
MW242775
A3
Dhofar toad Bufo dhufarensis
Al-Aflaj
MW242774
A4
Dhofar toad Bufo dhufarensis
Al-Aflaj
MW242773
A5
Dhofar toad Bufo dhufarensis
Al-Aflaj
MW242772
A6
Dhofar toad Bufo dhufarensis
Al-Aflaj
MW242771
A7
Dhofar toad Bufo dhufarensis
Al-Aflaj
MW242770
A9
Dhofar toad Bufo dhufarensis
Al-Aflaj
MW242769
A10
Dhofar toad Bufo dhufarensis
Al-Aflaj
MW242768
H11
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242767
H13
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242766
H14
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242765
H15
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242764
H16
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242763
H17
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242762
H18
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242761
H19
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242760
H20
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242759
H21
Dhofar toad Bufo dhufarensis
Al-Hareeq
MW242758
K28
Dhofar toad Bufo dhufarensis
Al-Kharj
MW242757
K29
Dhofar toad Bufo dhufarensis
Al-Kharj
MW242756
K30
Dhofar toad Bufo dhufarensis
Al-Kharj
MW242755
K31
Dhofar toad Bufo dhufarensis
Al-Kharj
MW242754
K32
Dhofar toad Bufo dhufarensis
Al-Kharj
MW242753
K33
Dhofar toad Bufo dhufarensis
Al-Kharj
MW242752
K34
Dhofar toad Bufo dhufarensis
Al-Kharj
MW242751
K35
Dhofar toad Bufo dhufarensis
Al-Kharj
MW242750
2.2 Genomic DNA extraction for PCR
Tissue samples were chopped into small pieces, and DNA was extracted from the tissue using DNAzol® (Invitrogen, UK), as directed by the manufacturer. Tissue samples were homogenised by a handheld glass/Teflon homogeniser. We transferred 40 mg of homogenous tissue to a 100 μl Eppendorf® tube, 500 μl of DNAzol was added to each sample, and pipet up and down gently to lyse the cells. The samples were then centrifuged at 9000 g for 5 min at 4 °C. We transferred the resultant supernatant to a new Eppendorf tube, added 500 μl of absolute ethanol to each tube, mixed for 10 s on a vibrating machine, and centrifuged at 9000 g for 8 min at 4 °C to precipitate DNA. We then removed the liquid and washed the DNA pellet with 200 µl of 75% ethanol. We centrifuged the samples again for 3 min and removed the liquid. The DNA pellets were left to air-dry for 20 min before we added 100 μl of nuclease-free water to resuspend the DNA. The extracted DNA samples were then stored at −20 °C. We measured DNA concentrations for each sample by spectrophotometry, based on an absorbance reading at 260 nm. On a 1% agarose gel stained with ethidium bromide, the isolated DNA was visually verified under UV light.
2.3 PCR amplification of the 16S rDNA
PCR was used to amplify the 16S rDNA with the primers 16S (5′-CGCCTGTTTATCAAAACAT-3′) and 16SSH (5′-CCGGTCTGAAC TCAGATCACG-3′), as described by Palumbi et al. (1991) with the following modifications: reactions were run with 8.5 µl Green Master Mix (2X; Thermo Fisher Scientific, Waltham, MA, USA), 3 µl forward and reverse primers (Macrogen, Seoul), 8.5 µl of distilled water, and 1 µl of DNA isolation to complete a 24 µl reaction mix. Each PCR was run with a negative control of nuclease-free water. We performed the PCR amplification using the following cycling conditions: 94 °C for 4 min, followed by 40 cycles of 94 °C for 1 min and 52 °C for 1 min, then 72 °C for 1 min, and a final extension at 72 °C for 5 min. The PCR amplification was visually verified under ultraviolet light by using a 1% agarose gel, stained with ethidium bromide. The PCR products were submitted for sequencing with both forward and reverse primers to Macrogen, Inc. (Seoul, Republic of Korea).
2.4 Constructing phylogenetic tree
We used the software program Molecular Evolutionary Genetics Analysis (MEGA), version 7 (Kumar et al., 2016), to examine the evolutionary connections between the sequences of Dhofar toad (Bufo dhufarensis) specimens in this study and constructed the phylogenetic tree. We used PCR to obtain the 16S rDNA in both directions from all samples in this study. The MEGA software aligned all sequence data on the 16S rRNA using the forward and reverse complements of the reverse primer. The gene fragments were aligned separately. All sequences obtained in this study were uploaded in GenBank under the accession number MW242750-MW242776, and other samples from different species used for phylogeny were retrieved from GenBank. We then compared the sequences obtained in our experimental dataset to reference ones extrapolated from GenBank (Fig. 1). The phylogenetic trees for the datasets acquired from GenBank and those found in this study were built separately, using NJ with genetic distance and the Tamura-Nei models, and were used to analyse the relationships between taxa by nucleotide sequence analysis (Kumar et al., 2016, Jeanmougin et al., 1998). The Felsenstein’s bootstrap method was used to calculate the associated taxa grouped in the bootstrap test (1000 replicates), which are shown next to the branches. The final dataset that contained PCR product from all samples was 460 base pairs. Estimates of Evolutionary Divergence between sequences were calculated as genetic distance matrix with MEGA X (version 10.2.6; Kumar et al. (2018)). Genetic distance heatmap were ploted using CLUSTVIS web tool (Metsalu and Vilo, 2015).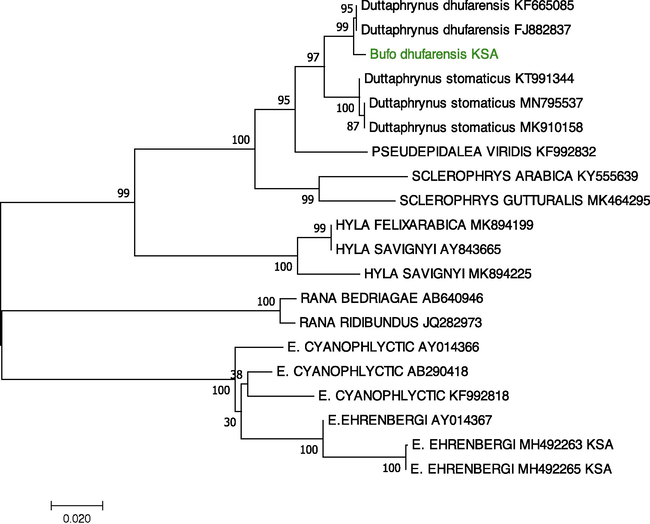
Phylogenetic tree created using the NJ method based on the 16S rRNA gene, indicating the relationships of Bufo dhufarensis to other toad and frog species. The NCBI GenBank accession numbers for all sequences are written after each species name. The sequences identified in this study are shown in green.
3 Results
We obtained sequence data of 16S rRNA for a total of 27 specimens of Bufo dhufarensis. We removed the ambiguous and gap-containing sites and found that all the samples collected in this study showed identical sequences. All these sequences are deposited in GenBank under the accession number MW242750-MW242776. We constructed an NJ phylogenetic tree with a 460-nucleotide sequence available from all samples (Fig. 1). The analyses of the NJ tree clearly clustered the species’ sequences in separate groups, and all grouped were supported by highly bootstrap values. Using GenBank-provided reference samples, we identified that all samples of Bufo dhufarensis in this study were in one major group. The NJ tree grouped Bufo dhufarensis with strong support as the sister taxon of the Oman population of Bufo dhufarensis (GenBank: KF665085 and FJ882837) (Liedtke et al., 2016, Jayawardena et al., 2017), with 99.35% identity. Moreover, genetic distance matrix showed a value of 0.007 of base substitutions per site between Bufo dhufarensis the Oman species Bufo dhufarensis (Table 2 and Fig. 2). Beyond this grouping, the relationships between the other derived species, such as Duttaphrynus stomaticus, formed a larger clade (Fig. 1). All of these sequences are novel and are being reported for the first time in this study.

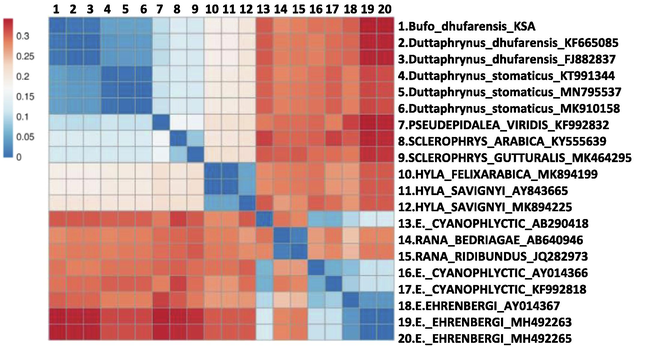
Genetic distance heatmap. The heatmap plot was constructed with genetic distance data (Table 2) using CLUSTVIS web tool (Metsalu and Vilo, 2015).
4 Discussion
Phylogenetic analysis uses molecular methods to classify and understand the relationships among closely related species in systematics and taxonomy based on studies of gene sequences. This method has become increasingly important in all fields of taxonomy because of the large amount of publicly available genetic sequence data (Yang and Rannala, 2010). Molecular phylogenetics is also used to classify closely related species sequences to recognize genes, regulatory elements and non-coding RNAs in newly sequenced genomes to explain modern and ancient individual genomes and refining phylogenetic trees. (Kellis et al., 2003, Pedersen et al., 2006).
The 16S rRNA genes have conserved and variable regions, where conserved areas reflect phylogenetic relationships among species (and are used as sites for PCR priming), are highly valuable tools for genomics and are widely used in phylogenic studies (Fouquet et al., 2007, Smith et al., 2008).
This paper provides the first overview analysis of the relationship of Bufo dhufarensis in Riyadh Province with other species obtained from GenBank using the 16S rRNA gene. All analyses of samples in this investigation were congruent, supported the sequences for the mitochondrial 16S rRNA gene. This tree also provides a preliminary theory regarding the relationship of Bufo dhufarensis with other species.
Based on the little available data from molecular studies in Bufo dhufarensis in the Arabian Peninsula, our results reveal that Bufo dhufarensis appeared in the phylogenetic tree as a sister group with the Bufo dhufarensis from Oman (GenBank: KF665085 and FJ882837) (Liedtke et al., 2016, Jayawardena et al., 2017), and both form one clade as shown in Fig. 1, with 99.35% identity. This is also supported by the low value of genetic distance in the estimation of evolutionary divergence (Table 2 and Fig. 2).
To explain such an expected relationship of Dhofar toad, we must understand the biogeographical distribution of the family Bufonidae. The toads of Saudi Arabia appear to have diversified from a monophyletic origin. The similarity of Bufo dhufarensis is due to their common origin, and any difference in the rRNA sequence of these two species might be because of their geographic separation and local environmental effects.
The results of the phylogenetic analyses clearly imply that all samples of Bufo dhufarensis we collected are grouped into one genetic lineage, which was supported by high bootstrap values. The tree analysis supports that Saudi Bufo dhufarensis and Oman Bufo dhufarensis are genetically closely related, and no significant differences are seen in their 16S rRNA sequences. These results clearly indicate that Dhofar toad in Saudi Arabia and Oman are the same species; the other group contained all the other species in different clusters, demonstrating that these results are robust.
According to our molecular study, there are only two Bufo dhufarensis variants in this species in Oman, which are shared with our samples of Bufo dhufarensis. However, the analysed populations have a strong population structure. The analysis of the 16S rRNA gene fragment shows incomplete lineage sorting, suggestive of a recent divergence event. The Bufo dhufarensis population from Riyadh Province is here considered to be the same species as Bufo dhufarensis, standing out as a distinct clade that we recognise as an evolutionarily important unit that has no haplotypes in common with any other population.
However, further phylogenetic and genetic diversity studies in the family Bufonidae and the use of comprehensive sampling of sequences from multiple mtDNA and nDNA genes could be used to accumulate enough phylogenetic information to resolve the diverged lineage. Hillis (1998) and Mayden et al. (2007) suggested that more sampling of either taxa or genes can result in increased accuracy of phylogenetic inference. There is a need to extend to studies with additional samples of B. dhufarensis are encouraged along with application of 16S rRNA from other provinces in Saudi Arabia, to get a better overview of the population structure.
Overall, our research provides a base for molecular genetics by 16S rRNA of Bufo dhufarensis in Riyadh Province, Saudi Arabia, and shows the potential for genomic data to help successful conservation management, both locally and internationally. Our findings also shows that both Saudi and Oman species need additional molecular research to determine their phylogenetic and systematic relationships.
Acknowledgements
We extend our appreciation to the Researchers Supporting Project number (RSP-2021/218), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- An overview of wetlands of Saudi Arabia: Values, threats, and perspectives. Ambio. 2017;46(1):98-108.
- [Google Scholar]
- First molecular identification of Euphlyctis ehrenbergii (Anura: Amphibia) inhabiting southwestern Saudi Arabia. The European Zoological Journal. 2019;86:173-179.
- [Google Scholar]
- Convergent adaptive radiations in Madagascan and Asian ranid frogs reveal covariation between larval and adult traits. Proceedings of the National Academy of Sciences. 2000;97:6585-6590.
- [Google Scholar]
- Use of population genetic structure to define species limits in the Rhizobiaceae. Symbiosis 2005
- [Google Scholar]
- Underestimation of species richness in Neotropical frogs revealed by mtDNA analyses. PLoS one. 2007;2:e1109
- [Google Scholar]
- The amphibian tree of life. Bulletin of the American Museum of natural History. 2006;2006:1-291.
- [Google Scholar]
- Slow worm, Anguis fragilis (Reptilia: Anguidae) as a species complex: genetic structure reveals deep divergences. Molecular Phylogenetics and Evolution. 2010;55:460-472.
- [Google Scholar]
- Reevaluation of 16S ribosomal RNA variation in Bufo (Anura: Amphibia) Molecular Phylogenetics and Evolution. 2001;19:326-329.
- [Google Scholar]
- Caecilian phylogeny and biogeography inferred from mitochondrial DNA sequences of the 12S rRNA and 16S rRNA genes (Amphibia: Gymnophiona) Herpetological Monographs 1993:64-76.
- [Google Scholar]
- Taxonomic sampling, phylogenetic accuracy, and investigator bias. Systematic Biology. 1998;47:3-8.
- [Google Scholar]
- Species boundaries, biogeography and evolutionarily significant units in dwarf toads: Duttaphrynus scaber and D. atukoralei (Bufonidae: Adenominae). Ceylon Journal of. Science. 2017;46
- [Google Scholar]
- Multiple sequence alignment with Clustal X. Trends in biochemical sciences. 1998;23:403-405.
- [Google Scholar]
- Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241.
- [Google Scholar]
- MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular biology and evolution. 2018;35:1547.
- [Google Scholar]
- MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular biology and evolution. 2016;33:1870-1874.
- [Google Scholar]
- No ecological opportunity signal on a continental scale? Diversification and life-history evolution of African true toads (Anura: Bufonidae) Evolution. 2016;70:1717-1733.
- [Google Scholar]
- Phylogenetic relationships of Danio within the order Cypriniformes: a framework for comparative and evolutionary studies of a model species. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution. 2007;308:642-654.
- [Google Scholar]
- METALLINOU, M., ARNOLD, E. N., CROCHET, P.-A., GENIEZ, P., BRITO, J. C., LYMBERAKIS, P., BAHA EL DIN, S., SINDACO, R., ROBINSON, M. & CARRANZA, S. 2012. Conquering the Sahara and Arabian deserts: systematics and biogeography of Stenodactylus geckos (Reptilia: Gekkonidae). BMC evolutionary biology, 12, 1-17.
- Species on the rocks: Systematics and biogeography of the rock-dwelling Ptyodactylus geckos (Squamata: Phyllodactylidae) in North Africa and Arabia. Molecular Phylogenetics and Evolution. 2015;85:208-220.
- [Google Scholar]
- ClustVis: a web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic acids research. 2015;43:W566-W570.
- [Google Scholar]
- Hotspots: Earth's biologically richest and most endangered terrestrial ecoregions. SA, Agrupación Sierra Madre, SC: CEMEX; 1999.
- Choricystis minor as a new symbiont of simultaneous two-species association with Paramecium bursaria and implications for its phylogeny. Symbiosis 2004
- [Google Scholar]
- The simple fool’s guide to PCR, version 2.0. Honolulu: University of Hawaii; 1991. p. :45.
- Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol. 2006;2:e33
- [Google Scholar]
- Phylogenetic relationships linking Duttaphrynus (Amphibia: Anura: Bufonidae) species based on 12S and 16S rDNA sequences. Mitochondrial DNA Part A. 2016;27:2881-2882.
- [Google Scholar]
- Molecular cloning: a laboratory manual. NY: Cold Spring Harbor; 1989.
- Extreme diversity of tropical parasitoid wasps exposed by iterative integration of natural history, DNA barcoding, morphology, and collections. Proceedings of the National Academy of Sciences. 2008;105:12359-12364.
- [Google Scholar]
- Survey of Saudi Arabian wetlands. Riyadh: IUCN/NCWCD Report; 1994.
- The ecology and behavior of amphibians. University of Chicago Press; 2010.
- Bayesian species delimitation using multilocus sequence data. Proceedings of the National Academy of Sciences. 2010;107:9264-9269.
- [Google Scholar]







