Translate this page into:
Association between Toll-like receptor 4 polymorphism and Acute Lymphoblastic Leukemia susceptibility in Saudi Arabian patients
⁎Corresponding author at: Zoology Department, College of Sciences, King Saud University, Post Office Box 2455, Riyadh, 11451, Saudi Arabia. Fadkhul2005@hotmail.com (Fadwa M. AlKhulaifi)
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University.
Abstract
Objective
Toll-like receptors (TLRs) play a critical role in initiating an immune response against infections. The present study aimed to investigate the relationship between Single Nucleotide Polymorphisms (SNP) and the expression of TLR4 gene with the risk and prognosis of acute lymphoblastic leukemia (ALL) in the Saudi population.
Methods
SNPs at rs4986790 (A/G) rs1927906 (C/T) and rs7856729 (G/T) were genotyped in 265 subjects, including150 patients and 115 community-matched healthy controls from a Saudi ethnic population. Using quantitative reverse transciptase PCR, the mRNA expression of TLR4 was compared between the two subgroup.
Results
Single nucleotide polymorphisms of the TLR4 in the 3’ UTR region and the fourth exon, were found to be associated with the risk of ALL. Individuals carrying the heterozygous genotype for the rs4986790 AG are significantly decreased ALL risk (OR: 0.313; 95% CI: 0.150–0.655; P = 0.002). Furthermore, the CT genotype of the rs1927906 was significantly associated with the protection against ALL (OR: 0.305; 95% CI: 0.167–0.557; P = 0.00007). Moreover, the mRNA expression of TLR4 was significantly higher in patients with ALL than in healthy individuals (P < 0.001).
Conclusion
This is the first study focusing on the TLR4 gene variants in leukemia patients in the Saudi population.These results suggested that TLR4 rs4986790 (AG) and rs1927906 (CT) polymorphisms may play an important protective role against ALL. In addition, the increased expression of the TLR4 gene in leukemic samples could serve as a novel potential prognostic factor for this receptor, especially in ALL cases.
Keywords
Acute lymphoblastic leukemia
Polymorphism
Gene expression
Toll like receptor
Saudi population

1 Introduction
Cancer is known to be a major public health issue. Between 1990 and 2018, Leukemia cases have shown a significant increase from 297,000 to 437,033 globally (Bawazir et al., 2019). The Gulf Cooperation Council (GCC) reports on cancers ranked leukemia as the 4th most common cancer among GCC nationals, it was also reported by the Saudi Cancer Registry that leukemia was the 5th commonly spread type of cancer within both genders in Saudi Arabia (GCCCP, 2011). It is worth mentioning that the total incidence of childhood leukemia in Saudi Arabia has jumped from 1.58/100,000 in 2001 to 2.35/100,000 in 2014 (Jastaniah et al., 2020).
Leukemias are a group of hematologic disorders associated with malignant neoplasm that arises from the hematopoietic origin. It’s characterized by uncontrolled proliferation and development of leukocytes (Sampaio et al., 2021). Leukemia can be classified as myeloid or lymphoid based on the expression of several antigens and markers and according to the predominant type of leukemic blast cells (Arber et al., 2016), if the affected cells are from granulocytes or monocytes lineage, leukemia will be known as myelogenous leukemia, and if they are from lymphocytes lineage, leukemia will be known as lymphoblastic leukemia (Shafique and Tehsin, 2018). Furthermore, leukemia can be more classified according to the stage of maturation arrest and the degree of cell differentiation into either acute (fast-growing) or chronic (slower- growing) with more mature and functional cells (Arber et al., 2016).
Acute Lymphoblastic Leukemias (ALL) is characterized by an uncontrolled spread of hematopoietic precursor cells of the lymphoid lineage within the bone marrow niche in most cases, and peripheral blood (Witkowski et al., 2019). Although, it has a bimodal age distribution, first early peak between 2 and 5 years of age, and a second steady increase at about age 50 years (Zhang et al., 2019).
Studies showed that multiple hematological malignancies and lymphoid precursor cells from different subtypes of acute leukemia express distinct and heterogeneous profiles of Toll-like receptors (TLRs) transcripts and could respond to stimulation by TLR agonists. Upraised TLR expression influence the bone marrow microenvironment in the very early fate decisions of the hematopoietic development and regulation (Dorantes-Acosta et al., 2013; Pandey et al., 2015).
Understanding the cellular and molecular foundation and genetic risk factors involved in leukemia etiology is essential. Several regulatory mechanisms exist that may lead to the onset of the disease and implicate in the makeup of the microenvironment of the tumor and the immune status in cancer. Consequently, uncontrolled activation of innate immunity and sustained cytokine production for a more extended period could serve as a platform for developing human cancers since TLRs and inflammasomes are the fundamental sensing molecules of innate immunity to induce specific pro-inflammatory responses (Keshavarz et al., 2021).
TLRs are a family of pattern-recognition receptors (PRRs) that identify and detect conserved sequences and structural motifs in a wide range of pathogens or PAMPs (pathogen-associated molecular patterns), along with damaged tissue or DAMPs (damage-associated molecular pattern) in addition to cancer debris, thus, provokes a host defense response (Ureña-Peralta et al., 2020). A candidate gene that might be associated with the risk of developing ALL is Toll-like receptor 4 (TLR4).
TLR4 is one of the major initiators of the innate immune response which promotes adaptive immunity. TLR4 is 1 of 11 known mammalian membrane-spanning receptors, acting as “bridging molecules” between innate and adaptive immunity and their genes have been reported to be polymorphic, especially due to their location in the ligand recognition area of the receptor. Recently, many research findings approved the functions and molecular mechanisms of TLRs in cancers (El-Omar et al., 2008; Farrugia and Baron, 2017).
Genetic variations caused by Single Nucleotide Polymorphisms (SNPs) in TLRs contribute to the alterations in the immune system, and those variations have been determined in different types of tumors, and their functionality goes far beyond the innate immune system, particularly the association of TLR4 polymorphism with cancer risk. Additionally, several studies examine the association between overall cancer risk or cancer-specific risk and SNPs of TLR4. Multiple SNPs of TLRs can affect the genetic susceptibility to hematologic malignancies, especially leukemia and non-Hodgkin lymphoma because TLRs are expressed on more primitive hematopoietic stem and progenitor cells (HSPCs). Considerable evidence has indicated that the deregulation of the innate immune system receptors may increase the risk for leukemia. Alterations in the expression of TLR4 were found in leukemic cells before and after chemotherapy induction (Ramzi et al., 2018; Paracatu and Schuettpelz, 2020). Therefore, TLR4 signaling is essential for tumor cell proliferation and immune response. Understanding such polymorphisms and their associated pathologies would provoke a novel therapeutic approach concentrating on halting the abnormal proliferative activity or developing some mediators that can override the signals to block abnormal hematopoietic differentiation. Accordingly, we thought that functional polymorphisms that occur on the TLR4 gene might have a significant role in the inflammation mechanism and might trigger cancerogenesis through disruption of inflammation.
The present study was conducted to explore the relationship between rs4986790 A/G, rs1927906 C/T and rs7856729 G/T variants of TLR4 polymorphisms with ALL risks in a Saudi population and to investigate new genetic biomarkers for ALL by analyzing the TLR4 polymorphisms at the fourth exon and 3’ UTR region.
2 Materials and methods
2.1 Ethical consideration
All procedures were in accordance with the Helsinki protocol and approved by the medical ethics committee in King Khalid University Hospital and the ethics committee of King Saud University, Riyadh, Saudi Arabia. (Ref. No. 20/0525/IRB).
2.2 Inclusion criteria for sample selection
Human whole blood samples of 265 individuals were collected for this study. The ALL samples comprised blood samples isolated from 150 Saudi patients diagnosed with Acute Lymphoblastic Leukemia and with no other known pathologies, hematological disorders, or previous cancer. In addition, 115 unrelated healthy individuals without any clinical signs of cancer or other diseases of both genders (female and male) served as controls.
The mean age of the study population was 22.45 ± 20.27 years for the patients with ALL and 18.68 ± 15.53 years for the healthy controls. No significant inter-group differences were recorded for age (P = 0.09).
2.3 DNA extraction
Two milliliters of blood samples were collected by venipuncture from each individual and then stored in ethylenediaminetetraacetic acid (EDTA)-containing tubes and stored at −20 °C before analysis.
Genomic DNA was extracted from blood samples using QIAamp DNA Mini Kit following the recommendation of the manufacturer. DNA purity and concentrations were determined by spectrophotometric measurement of absorbance at 260 and 280 nm (A260/A280 ratio) using Nanodrop ND-2000c spectrophotometer (Thermo Scientific, Wilmington, DE, USA).
2.4 RNA isolation and cDNA synthesis
Total RNA was isolated from 17 patients with newly diagnosed ALL and 17 matched healthy individuals using the QIAamp® RNA Blood Mini kit for total RNA purification from human whole blood (QIAGEN). RNA concentration was determined spectrophotometrically, the ratio of absorbances at 260 and 280 nm measured in a Nanodrop ND-2000c spectrophotometer (Thermo Scientific, Wilmington, DE, USA) was used as a parameter to quantify and evaluate the quality of the total RNA extracted. RNA was reversely transcribed into cDNA, following the manufacturer's instructions, the reverse transcription step was carried out using AMV Reverse Transcriptase (Promega, Madison, USA).
2.5 Single nucleotide polymorphisms (SNPs) selection, genotyping, and quality control
The tagging SNPs in TLR4 were selected from The International HapMap website (https://hapmap.ncbi.nlm.nih.gov) using genotype data. The SNPs with the frequency of minor alleles greater than 0.05 were selected to predict the possible function. TLR4 polymorphisms were determined by TaqMan allelic discrimination with a TaqMan SNP Genotyping Assay using Real-time PCR.
We genotyped three SNPs in the TLR4 (rs4986790 (Asp299Gly) A/G, assay ID: C__11722238_20), (rs1927906 C/T, assay ID: C__11722136_10), (rs7856729 G/T, assay ID: C__43308526_20). Furthermore, these SNPs are located in the fourth exon and 3’ UTR region . The SNPs positions are described in Table 1.
SNP
Chromosome
Cytogenetic location
Genotype
Genomic location
Consequence
rs4986790
9
9q33
AA
AG
GG117713024
fourth exon
rs1927906
TT
TC
CC117717837
3 Prime UTR
rs7856729
GG
GT
TT117719578
3 Prime UTR
TLR4 polymorphisms were genotyped using the allelic (VIC- and FAM-labeled) discrimination method according to the manufacturer’s instructions and as described previously by Livak (1999).
Briefly, forward and reverse primers were designed by the wild-type probe VIC® dye (linked to the 5́ end of the Allele 1 probe) and the variant probe 6FAM™ dye (linked to the 5́ end of the allele 2 probe) for the sequence of interest. Primers and TaqMan® Genotyping Master Mix were purchased from the assays-on-demand service of Applied Biosystems (Thermo Fisher Scientific, Applied Biosystems, USA).
DNA (10–20 ng) from each sample was added to 12.50 μl of TaqMan Genotyping Master Mix (2x) and 1.25 µl of SNP Genotyping Assay mix (2x). Amplification was performed in a final reaction volume of 25 μl.
The following amplification cycle conditions were used: 95 °C for 10 min as initial step, followed by 40 cycles of 95 °C for 15 s, and 40 cycles at 60 °C for 60 s. The PCR conditions were alike for all tested polymorphisms.
Analyses of amplification products and the allelic discrimination were achieved using ViiA™7, v.1.1. (Applied Biosystems, USA). A total of 5% of the selected samples were randomly selected for repeat analysis as quality control.
2.6 Quantitative of gene expression by Real-Time PCR
Quantitative real-time PCR was performed to determine the expression level of TLR4 mRNA in blood samples of ALL patients and controls.
Expression levels of TLR4 and the housekeeping gene Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (as internal control) genes were assessed using ViiA™7, v.1.1. (Applied Biosystems, USA). The reactions were performed in triplicate, each 20 µl reaction mixture contained 10 μl of SYBR Green PCR Master Mix, 5 μl forward and reverse primers and 2.5 + 2.5 of cDNA template + RNase-free water (Sequences of the primers were shown in Table 2).
Gene
Forward primer
Reverse primer
Product length
Annealing temperature
TLR4
CCGTTTTATCACGGAGGTGGT
CTGCCTAAATGCCTCAGGGG
135
60
GABDH
AATGGGCAGCCGTTAGGAAA
AAAAGCATCACCCGGAGGAG
133
60
Specific PCR conditions were as follows: 95 °C for 5 min followed by 45 cycles of 95 °C for 15 s, 60° C for 1 min, and 72 °C for 30 s, and 72 °C for 5 min.
Relative quantitation of TLR4 and GAPDH mRNA expression was calculated with the comparative threshold cycle method. The general process for analyzing the data from gene expression assays: The amplification plots were viewed, the baseline and threshold values were adjusted to determine the threshold cycles (CT) for the amplification curves and the relative expression of TLR4 was calculated using the comparative CT method (the 2^-ΔΔCt method) (Schmittgen and Livak 2008).
2.7 Statistical analysis
The Hardy-Weinberg equilibrium test was performed for each SNP to detect any deviation in the control samples. Allelic and genotypic frequencies of TLR4 SNPs were compared between patients with ALL and the control group to identify polymorphisms that could associate with the susceptibility to ALL by χ2 test, Odds ratios (OR) and 95% confidence intervals (CIs) were calculated to evaluate the disease risk. As well, P value < 0.05 was considered statistically significant.
3 Results
Three TLR4 SNPs (rs4986790 A/G, rs1927906 C/T, rs7856729 G/T) were genotyped and assessed for deviation from the Hardy–Weinberg equilibrium (HWE) (P > 0.05). A total of 265 were enrolled in this section, 150 patients with ALL and 115 unrelated individuals as healthy control. The strength of the association between these polymorphisms and risk of ALL cancer was measured by ORs with 95% CIs have shown in Tables 3–5, which summarizes allelic associations between the selected SNPs of the TLR4 in a patient with ALL and healthy individuals. Abbreviations; OR, odds ratio; CI, confidence interval; X2, Chi square; n, number of individuals; Boldfaced values indicate a significant difference at the P > 0.05 level. Abbreviations; OR, odds ratio; CI, confidence interval; X2, Chi square; n, number of individuals; Boldfaced values indicate a significant difference at the P > 0.05 level. Abbreviations; OR, odds ratio; CI, confidence interval; X2, Chi square; n, number of individuals; Boldfaced values indicate a significant difference at the P > 0.05 level.
Genotype
ALL patients n = 150
Controls n = 115
Control vs Patients
Count
%
Count
%
OR
CI
P-value
AA
138
92
90
78
Reference = 1
AG
12
8
25
21.7
0.313
0.150–0.655
9.520
0.002
GG
0
0
0
0
NA
NA
–
–
Allele
A
150
0.96
115
0.89
Ref.
G
0
0.04
0
0.11
0.342
0.168–0.696
9.46
0.0021
Genotype
ALL patients
n = 150
Controls
n = 115
Control vs Patients
Count
%
Count
%
OR
CI
P-value
TT
124
82
72
62.6
Reference = 1
CT
21
14
40
34.7
0.305
0.167–0.557
15.74
0.00007
CC
5
3
3
1.9
0.968
0.225–4.169
0.00
0.96490
CT+CC
26
17.3
43
37.4
0.351
0.199–0.619
13.60
0.00023
Allele
T
145
0.90
112
0.80
Ref.
C
5
0.1
3
0.2
0.461
0.282–0.754
9.80
0.00175
Genotype
ALL patients
n = 150
Controls
n = 115
Control vs Patients
Count
%
Count
%
OR
CI
P-value
GG
102
68
79
68.6
Reference = 1
GT
47
31
34
29.5
1.071
0.630–1.819
0.06
0.80073
TT
1
0.6
2
1.7
0.387
0.034–4.348
0.63
0.42566
GT+TT
48
32
36
31.1
1.033
0.612–1.741
0.01
0.90399
Allele
G
149
0.84
113
0.83
Ref.
T
1
0.16
2
0.17
0.986
0.621–1.568
0.00
0.95372
3.1 Association of TLR4 polymorphisms with ALL
3.1.1 TLR4 (rs4986790 A/G) polymorphism
In terms of the genotype and allele frequencies in rs4986790 A/G polymorphism, there was a remarkable difference between ALL patients and healthy controls, shown in Table 3. The homozygous state AA was detected in 92% of patients and 78% in healthy individuals. Our analysis revealed that subjects with the G allele were more likely to be normal compared with those bearing the A allele. Additionally, out of the three genotypes, only the AG, showed a significant association with a protective effect against ALL (OR: 0.313; 95% CI: 0.150–0.655; P = 0.002). It is worth noting that none of the investigated patients or healthy individuals in this study were a carrier of GG genotype.
3.1.2 TLR4 (rs1927906 C/T) polymorphism
Genotypes of rs1927906 in the wild type homozygous (TT), heterozygous (CT), and homozygous for the variant CC are shown in Table 4.
Our result indicated that the presence of the TLR4 rs1927906 C/T polymorphism in 3’ UTR region was associated with decreased cancer risk. Heterozygous CT was decreased significantly among patients with ALL (OR: 0.305; 95% CI: 0.167–0.557; P = 0.00007). Individuals with the CT genotype when considering the recessive model CC genotype in rs1927906 were significantly lower risk of developing ALL.
Furthermore, the recessive genotypes (CT + CC) were also significantly decreased in patient with ALL (OR: 0.351; 95% CI: 0.199–0.619; P = 0.00023). The majority of patients and controls were with homozygous variant genotype (TT). For the C allele, our analysis showed that the allele frequency is significantly decreased the risk of developing ALL, where the T allele frequency was higher in patients with ALL compared to C allele (OR: 0.461; 95% CI: 0.282–0.754; P = 0.00175). The results showed that C allele may be a protective factor against ALL.
3.1.3 TLR4 (rs7856729 G/T) polymorphism
Among the three SNPs in TLR4 that were analyzed, the allele and genotype frequencies of rs7856729 G/T polymorphism in the 3’ UTR region show no statistically significant difference between healthy individuals and patients with ALL. According to rs7856729 SNP, the P-value was > 0.05 for all genotypes and alleles. Table 5 illustrate the base pairs in the wild type homozygous (GG), heterozygous (GT), and homozygous for the variant (TT). Overall, the majority of patients and controls were with homozygous variant genotype (GG).
3.2 Association of TLR4 relative mRNA expression
To determine whether there was a link between TLR4 expression and ALL, we analyzed TLR4 mRNA expression level in 17 ALL patients and 17 matched healthy individuals using Quantitative RT- PCR. Analysis of the expression data revealed that TLR4 mRNA expression of ALL patients was higher than those of healthy donors. A significant statistical difference was detected between the patients compared to healthy individuals (fold change: 554.402, p value < 0.001) (Fig. 1).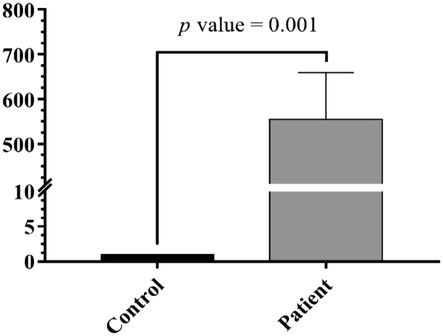
Higher Relative expression of TLR4 in the blood samples of ALL patients compared to healthy individuals (p < 0.001).
4 Discussion
TLRs play an important role in the innate immune system by regulating inflammatory reactions and activating adaptive responses to eliminate harmful pathogens. A large number of SNPs have been discovered in the genes of these receptors. TLR genes have polymorphic nature, so as a consequence, genetic variations in these genes have the ability to affect the pathogenesis of different diseases. Some associations between TLR4 polymorphisms and the susceptibility to infections, inflammation and risk of various cancers have been previously reported (Ding et al., 2017; El-Omar et al., 2008; Ferwerda et al., 2008).
This is the first study investigating whether polymorphisms of TLR4 are associated with ALL development among individuals in the Kingdom of Saudi Arabia. Thus, it would enable the researchers to improve the identification of early diagnostic tools of potential markers.
TLR4 acts in synergy with innate immune cells, and that may contribute to the inflammatory mechanism, which in a genetic perspective could enhance carcinogenesis in some conditions (Martínez-García et al., 2020) since TLR4 is expressed in the lymphocytes and participate in the regulation of B-cell differentiation and activation, consequently, genetic variation of TLR4 gene may contribute to the pathogenesis of leukemia (Rožková et al., 2010; Priyadarshini et al., 2013).
Several previous studies have suggested an influence of polymorphisms in TLR genes encoding factors related to the innate immune response on the etiology of some hematological malignancies subtypes; non-Hodgkin lymphoma, diffuse large cell lymphoma, Hodgkin's lymphoma, acute myeloid leukemia (AML) and chronic lymphocytic leukemia (CLL) and have been reported to be a risk factor (Forrest et al., 2006; Mollaki et al., 2009; Rybka et al., 2016; Ali et al., 2017).
In addition, various studies have been carried out to evaluate the correlation of TLR4 rs4986790 polymorphism with different types pf cancer, such as gastric cancer (Zhou et al., 2014), colorectal cancer (Moaaz et al., 2020), prostate cancer (Priyadarshini et al., 2013), and lung cancer (Kurt et al., 2016).
Conversely, some studies do not support the role of TLRs genes in cancer risk. For example, Rybka et al. (2016) did not observe any significant relationships between the nonsynonymous TLR4 polymorphisms rs4986790 in AML patients. A meta-analysis study for the association between rs4986790 polymorphism and cancer risk, did not find any strong association between this polymorphism and different types of cancer (Ding et al., 2017). Moreover, the TLR4 Asp299Gly variant was inversely associated with diffuse large cell lymphoma (Forrest et al., 2006) and positively associated with the risk of mucosa-associated lymphoid tissue lymphoma (Nieters et al., 2006), whereas the TLR2 -16933 T>A polymorphism was found to be associated with an increased risk of follicular lymphoma and a decreased risk of CLL (Hellmig et al., 2005).
This study showed that the genotypes AG of the rs4986790 SNP and CT and TT of the rs1927906 SNP are associated with decreased susceptibility to ALL in the Saudi population, thus may cause mild disease outcomes or can develop a balanced inflammatory response, while polymorphism of rs7856729 was found irrelevant to acute leukemia risk. The frequency of the wild allele (A) of the rs4986790 at TLR4 gene in healthy Saudis was 100%; similar to that reported in other populations such as Southern Han Chinese, Han Chinese in Bejing in China, and Japanese in Tokyo. Moreover, the minor allele (G) frequency of the same SNP in healthy Saudis was 0%; is similar to aforementioned populations but different to others including Qatar (0.051), Esan in Nigeria (0.0505) and British (0.0440) (1000 Genomes Project Consortium, 2015).
It has been reported that the substitution of adenine (A) to guanine (G) in the fourth exon in TLR4 rs4986790 may disrupt the extracellular domain of TLR4, consequently, this may reduce signaling via TLR4 which diminished production of inflammatory cytokines (Arbour et al., 2000).
Furthermore, studies have shown that wiled allele A is associated with enhanced activation of TLR4 in different inflammatory diseases due to increased levels of inflammatory cytokines like in type 2 diabetes and atherosclerosis (Steinhardt et al., 2010; Kolek et al., 2004).
However, the presence of the AA genotype in rs4986790 might increase the TLR4 activation and susceptibility to ALL since 92% of patients carry this genotype. So, further extensive studies with larger sample size and in other populations should be conducted to confirm this result.
A highly significant difference in TLR4 rs1927906 frequencies was found between the case and control groups. The total recessive, heterozygous genotype and C allele carriers were less susceptible to ALL. Contradictory to our results, concerning other types of cancer or diseases, TLR4 rs1927906 polymorphism is not associated with susceptibility to prostate cancer risk (Lindström et al., 2010) and type 2 diabetes (Kolz et al., 2008).
Generally, the T allele and TT genotype frequencies in this SNP in the healthy participant were 0.80 and 0.626 respectively. This is similar to the data from multiple populations such as in Finland (0.8333) and in Puerto Ricans (0.8317) However, the frequency of the Saudi T allele was higher than that in many populations such as Yoruba in Ibadan, Nigeria (0.4630), Mende in Sierra Leone (0.4941). Additionally, the minor allele (G) frequency of the same SNP in healthy Saudis was 0.2, this is similar to many populations such as: Indian Telugu from the UK (0.2206) and Bangladesh (0.2035) (1000 Genomes Project Consortium, 2015).
No association in our study was found between our TLR4 cohort and the candidate SNP for rs7856729 G/T and ALL. On the other hand, there have been very few publications based on this SNP ID. TLR4 rs7856729 and rs1554973 were significantly associated with IL-1α and IL-1β, and IL-10, which involved in the immune response, are correlated with cervical pro-inflammatory cytokine concentrations (Wang et al., 2021). We suggested that the Ethnic differences in genetic susceptibility genes could be a reason for these differences.
Over the recent years, many studies analyzed the role of TLR4 mRNA expression in hematological malignancies and different types of cancer. In our study, overexpression on TLR4 was observed (≥500-fold, P < 0.001) in patients with ALL. A possible effect of the studied rs4986790 and rs1927906 SNPs on the upregulation versus downregulation of TLR4 is not excluded. It has been suggested that TLRs high expression in patients might be associated with the induction of different immune mechanisms enabling malignant cells to survive and stimulating carcinogenesis (Rybka et al., 2015). The results of our study are in line with some previous studies concerning the role of TLR4 expression levels in different hematological malignancies and cancers (Ramzi et al., 2018; Bagratuni et al., 2019: Zhao et al., 2019).Therefore, our result may reflect that the high mRNA expressions of TLR4 may contribute to ineffective hematopoiesis and cell proliferation in ALL cases.
5 Conclusion
rs4986790 and rs1927906 polymorphism of TLR4 gene could modulate the susceptibility towards ALL disease in the local population of Saudi Arabia. A deeper understanding of the genetic variations of TLR4 will enable us to better identify biomarkers for early detection and prognosis and improve the decision‐making procedure of ALL treatments.
Acknowledgment
This project was supported by the Researchers Supporting Project number (RSP-2021/35), King Saud University, Riyadh, Saudi Arabia.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- A global reference for human genetic variation. Nature,. 2015;526(7571):p86.
- Lack of associations between TLR9 and MYD88 gene polymorphisms and risk of chronic lymphocytic leukemia. Asian Pacific J. Cancer Prev.: APJCP. 2017;18(12):3245.
- [Google Scholar]
- The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood, J. Am. Soc. Hematol.. 2016;127(20):2391-2405.
- [Google Scholar]
- TLR4 mutations are associated with endotoxin hyporesponsiveness in humans. Nat. Genet.. 2000;25(2):187-191.
- [Google Scholar]
- Toll-like receptor 4 activation promotes multiple myeloma cell growth and survival via suppression of the endoplasmic reticulum stress factor chop. Sci. Rep.. 2019;9(1):1-12.
- [Google Scholar]
- The burden of leukemia in the Kingdom of Saudi Arabia: 15 years period (1999–2013) Bmc Cancer. 2019;19(1):1-10.
- [Google Scholar]
- Comprehensive assessment of association between TLR4 gene polymorphisms and cancer risk: a systematic meta-analysis. Oncotarget. 2017;8(59):100593-100602.
- [Google Scholar]
- TLR stimulation of bone marrow lymphoid precursors from childhood acute leukemia modifies their differentiation potentials. Biomed Res. Int.. 2013;2013:1-13.
- [Google Scholar]
- Polymorphisms in Toll-like receptor genes and risk of cancer. Oncogene. 2008;27(2):244-252.
- [Google Scholar]
- The role of toll-like receptors in autoimmune diseases through failure of the self-recognition mechanism. Int. J. Inflamm.. 2017;2017:1-12.
- [Google Scholar]
- Functional consequences of toll-like receptor 4 polymorphisms. Mol. Med.. 2008;14(5):346-352.
- [Google Scholar]
- Polymorphisms in innate immunity genes and risk of non-Hodgkin lymphoma. Br. J. Haematol.. 2006;134(2):180-183.
- [Google Scholar]
- GCCCP: ten-year Cancer incidence among nationals of the GCC states 1998–2007. Report Riyadh: Gulf Centre for Cancer Control and Prevention; 2011.
- Association study of a functional Toll-like receptor 4 polymorphism with susceptibility to gastric mucosa-associated lymphoid tissue lymphoma. Leukemia Lymphoma. 2005;46(6):869-872.
- [Google Scholar]
- Incidence trends of childhood acute lymphoblastic leukemia in Saudi Arabia: increasing incidence or competing risks? Cancer Epidemiol.. 2020;67:101764.
- [Google Scholar]
- Toll-like receptors (TLRs) in cancer; with an extensive focus on TLR agonists and antagonists. IUBMB Life. 2021;73(1):10-25.
- [Google Scholar]
- Toll–like receptor 4 gene Asp299Gly polymorphism is associated with reductions in vascular inflammation, angiographic coronary artery disease, and clinical diabetes. Am. Heart J.. 2004;148(6):1034-1040.
- [Google Scholar]
- BMC medical genetics. 2008;9(1):1-12.
- Determination of the relationship between rs4986790 and rs4986791 variants of TLR4 gene and lung cancer. Inflammation. 2016;39(1):166-171.
- [Google Scholar]
- Cancer Epidemiology and Prevention Biomarkers. 2010;19(3):873-876.
- Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genetic Anal.: Biomol. Eng.. 1999;14(5–6):143-149.
- [Google Scholar]
- TLR2 and TLR4 surface and gene expression in white blood cells after fasting and oral glucose, lipid and protein challenges: influence of obesity and sex hormones. Biomolecules. 2020;10(1):111.
- [Google Scholar]
- Study of toll-like receptor 4 gene polymorphisms in colorectal cancer: correlation with clinicopathological features. Immunol. Invest.. 2020;49(5):571-584.
- [Google Scholar]
- Polymorphisms and haplotypes in TLR9 and MYD88 are associated with the development of Hodgkin's lymphoma: a candidate–gene association study. J. Hum. Genet.. 2009;54(11):655-659.
- [Google Scholar]
- Gene polymorphisms in Toll-like receptors, interleukin-10, and interleukin-10 receptor alpha and lymphoma risk. Genes Immun.. 2006;7(8):615-624.
- [Google Scholar]
- Pandey, S., Singh, S., Anang, V., Bhatt, A.N., Natarajan, K., Dwarakanath, B.S., 2015. Pattern recognition receptors in cancer progression and metastasis. Cancer Growth Metastasis, 8, pp. CGM–S24314.
- Contribution of aberrant toll like receptor signaling to the pathogenesis of myelodysplastic syndromes. Front. Immunol.. 2020;11:1236.
- [Google Scholar]
- Asp299Gly and Thr399Ile polymorphism of TLR-4 gene in patients with prostate cancer from North India. Indian J. Urol.: IJU: J. Urol. Soc. India. 2013;29(1):37.
- [Google Scholar]
- Association between TLR2 and TLR4 expression and response to induction therapy in acute myeloid leukemia patients. Int. J. Hematol.-Oncol. Stem Cell Res.. 2018;12(4):303.
- [Google Scholar]
- Toll-like receptors on B-CLL cells: expression and functional consequences of their stimulation. Int. J. Cancer. 2010;126(5):1132-1143.
- [Google Scholar]
- The expression of Toll-like receptors in patients with acute myeloid leukemia treated with induction chemotherapy. Leuk. Res.. 2015;39(3):318-322.
- [Google Scholar]
- Variations in genes involved in regulation of the nuclear factor–κB pathway and the risk of acute myeloid leukaemia. Int. J. Immunogenet.. 2016;43(2):101-106.
- [Google Scholar]
- Chronic myeloid leukemia-from the Philadelphia chromosome to specific target drugs: a literature review. World J. Clin. Oncol.. 2021;12(2):69-94.
- [Google Scholar]
- Analyzing real-time PCR data by the comparative CT method. Nat. Protoc.. 2008;3(6):1101-1108.
- [Google Scholar]
- Shafique, S. and Tehsin, S., 2018. Acute lymphoblastic leukemia detection and classification of its subtypes using pretrained deep convolutional neural networks. Technol. Cancer Res. Treatment, 17, p. 1533033818802789.
- A functional nonsynonymous toll-like receptor 4 gene polymorphism is associated with metabolic syndrome, surrogates of insulin resistance, and syndromes of lipid accumulation. Metabolism. 2010;59(5):711-717.
- [Google Scholar]
- Lack of TLR4 modifies the miRNAs profile and attenuates inflammatory signaling pathways. PLoS ONE. 2020;15(8):e0237066.
- [Google Scholar]
- The association of TLR2, TLR3, and TLR9 gene polymorphisms with susceptibility to Talaromycosis among Han Chinese AIDS patients in Guangdong. Front. Cell. Infect. Microbiol.. 2021;11:149.
- [Google Scholar]
- Investigation of NF-κB-94ins/del ATTG and CARD8 (rs2043211) gene polymorphism in acute lymphoblastic leukemia. Front. Endocrinol. 2019:501.
- [Google Scholar]
- TLR4 expression correlated with PD-L1 expression indicates a poor prognosis in patients with peripheral T-cell lymphomas. Cancer Manage. Res.. 2019;11:4743.
- [Google Scholar]
- Association between TLR4 (+ 896A/G and+ 1196C/T) polymorphisms and gastric cancer risk: an updated meta-analysis. PLoS ONE. 2014;9(10):e109605.
- [Google Scholar]






