Translate this page into:
Prevalence, antibiogram, phenotypic and genotypic analysis of Clostridioides difficile toxigenic strains from stool samples
⁎Corresponding author.
-
Received: ,
Accepted: ,
This article was originally published by Elsevier and was migrated to Scientific Scholar after the change of Publisher.
Peer review under responsibility of King Saud University. Production and hosting by Elsevier.
Abstract
Clostridioides difficile (C. difficile) is a Gram-positive, spore-forming, toxin-producing, anaerobic bacterium that is a prominent cause of nosocomial antibiotic-associated diarrhea. Toxin-mediated C. difficile infections can cause a variety of diseases, from minor cases of diarrhea to life-threatening pseudomembranous colitis in mammals. This study aimed to detect C. difficile in patients admitted to King Saud Medical City (KSMC), to determine the phenotypic and genotypic types of the toxigenic C. difficile isolates, and to investigate the antimicrobial susceptibility patterns of these isolates. The study was conducted from January 2021 to December 2021 and involved 313 stool samples obtained from patients at KSMC and tested for the presence of C. difficile. Anaerobic incubation of each stool sample was carried out using C. difficile selective agar at 37 ± 2⁰C for 48–72 h to enable phenotypic characterization. For phenotypic detection and toxigenic isolate differentiation, a commercial kit, ImmunoCard Toxins A & B, was used. The E test and Brucella modified blood agar were used to determine antimicrobial susceptibility. Real-time polymerase chain reactions using GeneXpert were performed to detect the genotypes of toxigenic strains. Antimicrobial susceptibility testing revealed that all isolates were sensitive to vancomycin and metronidazole, while 89.3% and 82.1% were sensitive to moxifloxacin and tetracycline, respectively. Furthermore, the prevalence of toxigenic CDI was 9.3%.
Keywords
Clostridioides difficile
Toxigenic C. difficile
Antibiogram
CDI
Riyadh
1 Introduction
Infectious diarrhea brought on by antibiotic use is common in hospitals. Clostridium difficile (C. difficile), a Gram-positive opportunistic bacterium, is the most common cause. Colon damage and severe illness caused by C. difficile infection (CDI) are major public health concerns (Lanzoni-Mangutchi et al., 2022). C. difficile is a member of the Clostridioides genus and can be discovered in the digestive tracts of animals and humans. It is a rod-shaped bacterium, approximately 0.3 × 1.5 × 2 µm in size, that is anaerobic, spore-forming, motile, and contains a peritrichous flagellum (Willey, 2019). Due to the difficulty of isolating this bacteria in the laboratory, it was termed Clostridium difficile (Orrell and Melnyk, 2021). In 2016, Clostridium difficile officially changed to Clostridioides difficile (Lawson et al., 2016). Clostridioides difficile infection (CDI) is considered a major cause of nosocomial infection. CDI symptoms vary from mild watery diarrhea to pseudomembranous colitis (PMC) and toxic megacolon and, in severe cases, life-threatening colon perforation and sepsis (Huang et al., 2020; Lurienne et al., 2020). C. difficile is responsible for 20–30% of diarrhea acquired from using antibiotics, known as antibiotic-associated diarrhea (AAD), which is the causative agent of PMC (Riley and Kimura, 2018; Huang et al., 2020).
The development of endospores is the critical step in C. difficile’s spread from a contaminated hospital surface to a patient. Environmental factors, such as nutrition constraints, typically trigger the initiation of sporulation (Zhu, Sorg and Sun, 2018). More cases of CDI have been reported in the past year than MRSA infections (McDonald et al., 2018). In the United States, the CDC estimated that 30,000 people die from CDI each year, from a total of 450,000 cases. CDI is considered to represent an economic crisis, as the price of a CDI tablet in the United States has reached $4.8 billion (Sandhu and McBride, 2018).
In Saudi Arabia, limited studies have investigated CDI. One study documented the detection of the epidemic PCR ribotype 027 strain (Alzahrani and Aljohani, 2013), while another retrospective study detailed the epidemiology of CDI in the Eastern region, reported a 20% increase in CDI cases between 2001 and 2018 (Al-Tawfiq et al., 2020). One other study assessed the antimicrobial susceptibility patterns of isolated colonies of C. difficile, which is important for evaluating C. difficile antibiotic resistance (Hudhaiah and Elhadi, 2019). The purpose of the current study was to identify C. difficile in patients admitted KSMC, to determine the phenotypes and genotypes of toxigenic C. difficile isolates, and to investigate the antimicrobial susceptibility patterns of these isolates.
2 Materials and methods
2.1 Study setting
This prospective study was conducted at KSMC in Riyadh from January 2021 until December 2021. KSMC is a Ministry of Health tertiary hospital with a 1,400-bed capacity. In total, 313 patients were involved based on the eligibility criteria. To be included, patients had to be at least 18 years of age and have an anticipated hospital stay of>48 h. CDI has been defined as a symptomatic infection involving diarrhea (≥3 loose stools in 24 h) (CDC; NHSN, 2022) caused by toxigenic C. difficile. Stool samples are typically assessed for the presence of C. difficile toxins using molecular assay or Real time-PCR techniques. CDI was defined as a positive stool culture in a patient with diarrhea (McDonald et al., 2018). A case is considered HO-CDI if the CDI symptoms occur > 3 days after admission (CDC; NHSN, 2022).
2.2 Ethical considerations:
Ethical approval was obtained from the departmental research review committee of KSMC (H1RI-13-May21-01) prior to study onset.
2.3 Patient selection criteria
The inclusion criteria were patients who were at least 18 years old, male or female, who had an anticipated stay in the hospital longer than 48 h. All departments were involved, as were patients who presented with symptoms of diarrhea (watery or loose), abdominal pain, and fever. Patients younger than 18, without diarrhea, with negative samples for C. difficile, or who had recurrent or duplicated CDI were excluded.
2.4 Stool sample processing
Only unformed, liquid, and semi-liquid samples were processed. Stool was processed immediately or stored at − 20 °C until processed. Samples were inoculated with a brain–heart infusion (BHI) broth and placed in an 80 °C water bath for 10 min. All safety precautions were followed, including the use of personal protective equipment and processing the samples in a Biosafety class 2 cabinet (BSL-2). (Hudhaiah and Elhadi, 2019) All materials were treated prior to disposal.
2.5 C. Difficile culture and identification
The stool samples were cultured on C. difficile selective media containing cycloserine and cefoxitin and incubated for 48–72 h in an anaerobic environment using an anaerobic gas pack system (BD GasPak) (Hindi et al., 2020). The isolated colonies were identified then inculcated with a tryptone soya broth containing glycerol and then stored at − 80 °C. The characteristic phenotype of C. difficile is circular grey-white colonies with a raised center and irregular filamentous or opaque edges and a typical odor of a horse barn (Kouhsari et al., 2019).
2.6 C. Difficile toxin testing
Only the toxins generated by the toxigenic strains were detected after sample processing. The fast, qualitative, horizontal-flow EIA was used to detect toxins A and B in all samples (Meridian Bioscience Inc., Cincinnati, OH, USA). The commercial set consisted of a membrane mounted in a plastic frame with four openings (i.e., two for input and two for output reactions((Carroll and Mizusawa, 2020). Antibodies against both toxin A and toxin B were trapped in the membrane. Antibodies specific for toxins A and B that were fused to horseradish peroxidase comprised the enzyme conjugate reagent. The ImmunoCard Toxins A & B commercial kit included a test card, sample diluent, positive control, enzyme conjugate, wash buffer I, and substrate I (Fig. 1).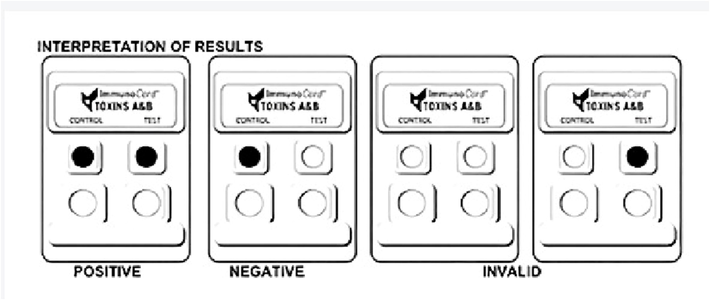
Interpretation of results.
2.6.1 Procedure for testing
The assay was performed according to the manufacturer’s instructions. First, the test tube was filled with 200 µL of sample diluent and then 3 drops of enzyme conjugate. The stool sample or control sample was mixed in for 10 s. The mixture was then incubated at 20–26 °C for 5 min to 24 h. The sample/control port was then filled with 150 µL and incubated at 20–26 °C for 5 min. Three drops of ImmunoCard wash buffer were dispensed until complete absorption was achieved, after which three drops of ImmunoCard substrate I were added. The test cards were then incubated for 5 min at 20–26 °C. There were several possible results: toxigenic C. difficile positive for toxins A and B, toxigenic C. difficile negative for toxins A and B, invalid, error; or no result.
2.7 Antimicrobial susceptibility testing
Antibiotic susceptibility testing (AST) was performed using Brucella modified blood agar using the lawn culture technique and E test. C. difficile colonies were isolated from the BHI agar medium, suspended to a density of 1.0 McFarland standard, and incubated anaerobically at 37 °C for 48 h (Kouhsari et al., 2019; Saber et al., 2020). C. difficile ATCC 700057 was used as the quality control strain for AST (Abuderman et al., 2018). The antimicrobial agents were moxifloxacin, vancomycin, metronidazole, and tetracycline.
2.8 Molecular analysis
GeneXpert for C. difficile is an rt-PCR assay (Alharbi et al., 2014) (Cepheid Inc., Sunnyvale, CA, USA) that detects the tcdB gene, cdtA gene, and the tcdC gene deletion at nucleotide 117 (Shah et al., 2020). The 18-bp deletion is located downstream of the mutated nucleotide at position 117. This test was performed on all samples. According to the manufacturer, this requires is a swab containing the organism to be cut inside the cartridge and placed inside the machine (Figure 2.3)(Shah et al., 2020). A stool sample was collected from the stool sample container via a swab and transferred to the sample reagent vial. The vial was vortexed for 10 s, following which the solution was pipetted into the “S” chamber of the cartridge. Finally, the cartridge was placed in the GeneXpert machine, and the C. difficile assay program was run. There were several potential results: toxigenic C. difficile positive, presumptive 027/NAP1/BI negative, toxigenic C. difficile positive, presumptive 027/NAP1/BI positive, toxigenic C. difficile negative, presumptive 027/NAP1/BI negative, invalid, error, or no result. The CT value, which reflects whether enough DNA amplification has occurred for a fluorescent signal to be detected, was recorded (Shah et al., 2020).
2.9 Statistical analysis
Statistical analysis was performed with numerical variables (Khan et al., 2019). Figures were created using SPSS software.
3 Results
3.1 Demographic information
This area presents the demographic information of the participants (n = 313), including the age and gender distributions (Table 1). In this study, 44.7% of participants were female, and 55.3% were males. Regarding age, 25.9% of participants were 30 or younger, 37.1% were aged between 31 and 50 years, and the rest were > 50 years of age.
Variables
Group
Frequency (n)
Percentage (%)
Sex
FEMALE
140
44.7
MALE
173
55.3
Age
≤30
81
25.9
31–50
116
37.1
>50
116
37.1
3.2 The distribution of CDI
Positive CDI cases were found in 9.3% of participants, while 90.7% were negative (Fig. 2).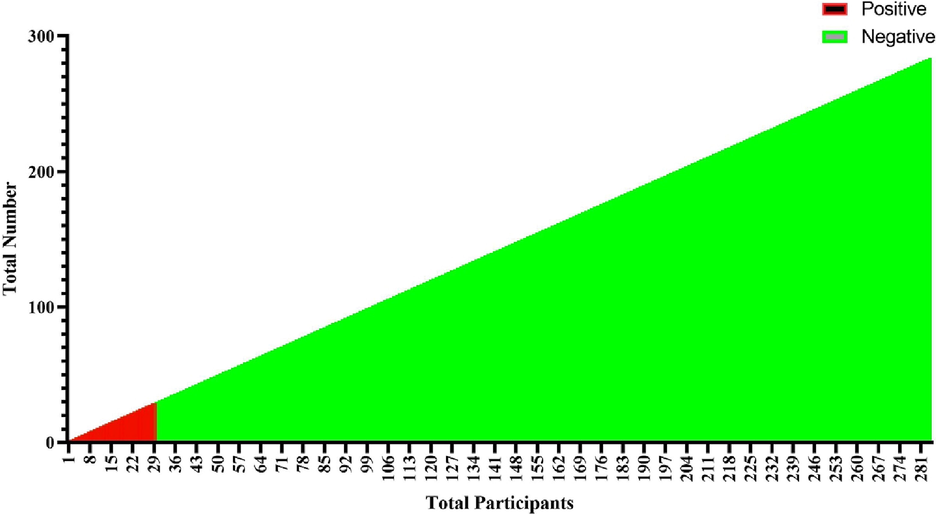
Distribution of CDI patients.
3.3 The distribution of CDI based on age
Participants were classified into 3 age groups: < 30 years, 31–50 years, and > 50 years. In the < 30 age group, only 1% were positive, and 24.9% were negative subjects. In the 31–50 age group, 2.9% were positive, and 34.2% were negative. In the > 50 group, 5.4% were positive, and 31.6% were negative. The numerical variables are shown in Fig. 3.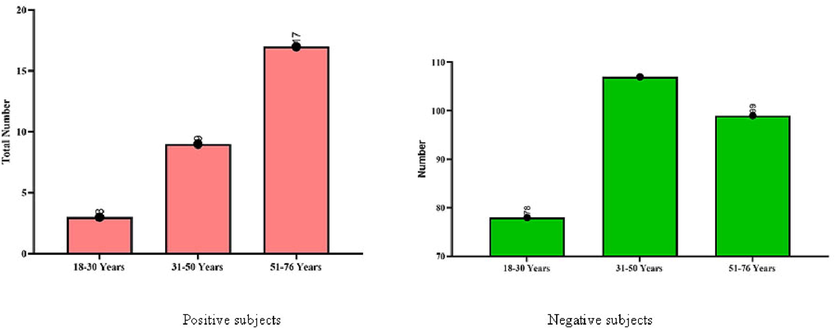
Distribution of CDI among positive and negative subjects with categorization of age.
3.4 The distribution of CDI based on gender
There was an insignificant relationship between gender and CDI (p = 0.4). Females accounted for 4.8% of subjects, and males accounted for 4.5%. A non-significant association was found between both groups (Fig. 4).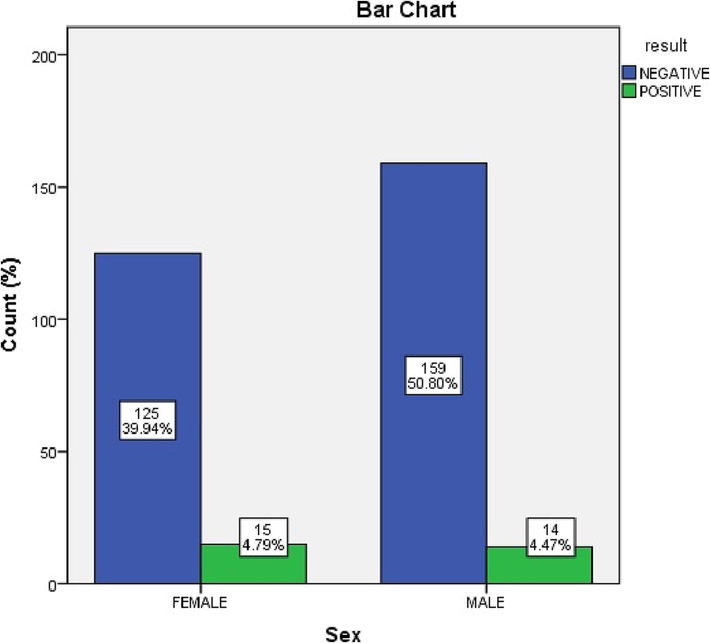
Bar Chart describes the frequencies between male and female subjects.
3.5 Detection of C. difficile
A molecular analysis study confirmed that 9.3% of samples were positive and 90.7% were negative subjects. This section presents the detection tests used to determine the presence of C. difficile and whether a strain was non-toxigenic or toxigenic. Fig. 5 demonstrates the frequencies of both positive and negative subjects.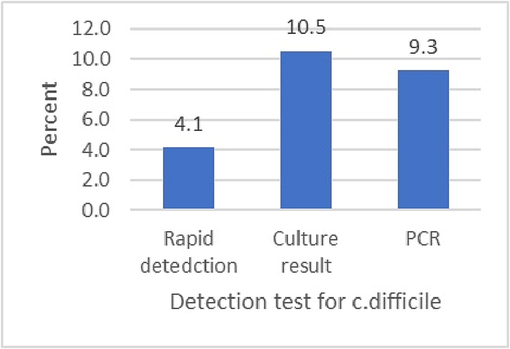
Detection of molecular analysis through positive and negative analysis.
3.6 Genetic profile of the toxigenic C. difficile
The gene profiles of all positive samples (n = 29) indicated that toxin B was detected in all positive samples, the binary toxin was detected in 13.8%, and no samples demonstrated tcdC. Fig. 6 shows the prevalence of the different toxins.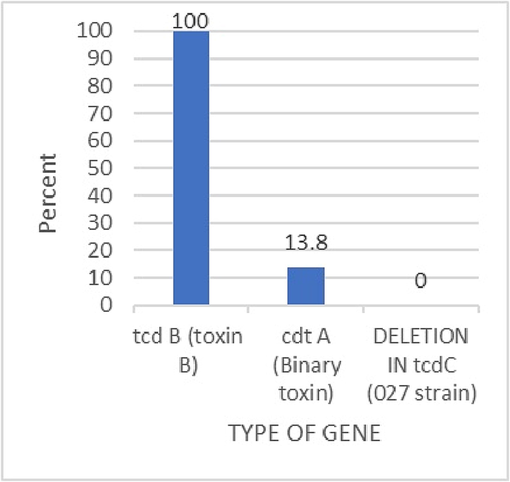
The gene profile of the toxigenic c. difficile were toxin B = 29, binary toxin = 4, and no detection for RT027.
3.7 Antimicrobial test:
All isolated samples (n = 28) were investigated for the antimicrobial agents: vancomycin, metronidazole, tetracycline, and moxifloxacin. The study results confirmed 100% of antibiotics such as vancomycin and metronidazole, 89.3% for tetracycline and 96.4% for moxifloxacin were present.
3.8 Correlation between age and gender wise distribution in CDI patients
The results in this Table 2 confirm that 29 positive patients have been confirmed by CDI tests. This study evidenced 51.7% of females and 48.3% of males. Female subjects (n = 15) were reported to be between 22 and 72 years old, while male subjects (n = 14) were found to be between 22 and 76 years old. However, the mean age in females was 46.9 ± 18.8 and 48.1 ± 15.8 in males. The overall mean age in the total gender (n = 29) was determined to be 48.3 ± 16.3 between the ages of 22 and 76 in 29 cases.
Gender
CDI positive levels (%)
Age levels
Mean age
Female (n =15)
15 (51.7%)
22–72 years
46.9 ± 18.8
Male (n = 14)
14 (48.3%)
22–76 years
48.1 ± 15.8
Total (n = 29)
29 (100%)
22–76 years
48.3 ± 16.3
4 Discussion
C. difficile is responsible for antibiotic-associated diarrhea in humans. Surveillance of C. difficile using phenotypic and genotypic approaches is a critical element in the strategy to understand and reduce the impact of C. difficile infections on global health systems. The main aim of this study was to determine the prevalence and genetic diversities of C. difficile - toxigenic strains from the stool sample in Saudi hospitals. Our participants number were (n = 313), including the gender and age distributions. 173 of them were male, and 140 were female. Most of the participants with positive C. difficile were adults (age>30).
This result is with consistent with the previous studies which suggested the increase in C. difficile risk infections with increasing age. C. difficile is a germ (bacteria) that causes life-threatening diarrhea. It is usually a side-effect of taking antibiotics. These infections mostly occur in: People 65 and older who take antibiotics and receive medical care. C. difficile risk factors include older age (65 and older) recent stay at a hospital or nursing home. a weakened immune system, such as people with HIV/AIDS, cancer, or organ transplant patients taking immunosuppressive drugs. C. difficile bacteria are commonly found in the environment, but most cases of C. difficile occur while you’re taking antibiotics or not long after you’ve C. difficile while on antibiotics and during the month after. That’s because antibiotics that fight bacterial infections by killing bad germs can also get rid of the good germs that protect the body against harmful infections, like CDI. If you take antibiotics for more than a week, you could be even more at risk.
There was an insignificant relationship between gender and CDI incidence in our data. Females accounted for 15 positive and 125 negative samples, while males accounted for 14 positive and 159 negative samples. Therefore, females accounted for 51.7% of positive samples, and males accounted for 48.3%.
This result was in contrast with the result explained by Mukil Natarajan., whose stated that (Natarajan et al., 2015),the Population rates of CDI are higher in women than in men (Rogers et al., 2013). Sex-specific differences in the gut microbiome have been shown to be mediated by hormone levels, and transference of intestinal bacterial communities can alter sex hormone levels in animal studies (Markle et al., 2013). Moreover, male castration attenuates these microbial differences suggesting that androgens may play a role (Yurkovetskiy et al., 2013). Human studies have also reported sex differences in intestinal microbiota (Mueller et al., 2006).
Concerning the prevalence Clostridium difficile in our research. C. difficile was detected in 29 samples; however, there was no detection of the ribotype 027 strain. Of the 313 included participants, 29 (9.3%) had CDI, 4 (1.6%) had non-toxigenic C. difficile, and 280 (89.1%) did not have CDI, of these, the ≤ 30 years group accounted for 10.34%, the 31–50 years group accounted for 31.03%, and the > 50 years group accounted for 58.61%, indicating a significant relationship between age and CDI.
When comparing our prevalence with the previous studies prevalence conducted in Saudi Arabia like Al-Madinah city, the prevalence of CDI was 21.7% (Sandokji et al., 2009) which was over double the prevalence in the current study, In Eastern Region was 18.7% (>80%), with ribotype 027 detected in three isolates (3.4%) (Hudhaiah and Elhadi, 2019), In Taif city, the prevalence was 13.6% (>3%), with ribotype 027 detected in four isolates (5.4%) (Saber et al., 2020). Worldwide the prevalence in Thailand, was 23.7%, with the ribotype 014/020 strain being predominant and no detection of the epidemic ribotype 027 strain (P. Putsathit et al., 2017). In Iran, was 28.6%, Furthermore, ribotype 027 has not been detected in Iran (Shoaei et al., 2019). In Zambia was 10% (Nehanda, Mulundu and Kelly, 2020). In the USA, the prevalence of the ribotype 027 is 22% (Giancola, Williams and Gentry, 2018). In France, the prevalence of CDI is 24.7%, with the ribotype 014/020/077 strain being the most dominant (23.6%) (Khanafer et al., 2018). We noticed that our prevalence was lower than the previous studies conducted locally or internationally. The improvement in strategies which help to prevent the spread of C. difficile in hospitals and other health care facilities which followed strict infection-control guidelines in the previous years might be related to decrease in the prevalence; like Avoid unnecessary use of antibiotics, Handwashing, Contact precautions, Thorough cleaning.
Epidemic and clinically important types of C. difficile are evolving and include several PCR ribotypes (Bauer et al., 2011). Therefore, sufficient diagnostic methods need to be continuously updated to comply with this bacterium's changing epidemiology. C. difficile toxins include toxin A, toxin B, and a binary toxin. Toxins A and B are encoded by the genes tcdA and tcdB, which are located on the pathogenicity island PaLoc, which also includes the negative and positive regulators tcdC and tcdR. The binary toxin is encoded by the genes cdtA and cdtB, which constitute another operon together with the positive regulator cdtR. A number of different genetic alterations in the tcdC gene have been observed. Most prominent are the in-frame deletion of 18, 39, or 54 bp and the truncating mutation at position 117 (1-bp deletion) or 184 (C → T transition).
When conducted the Gene profile of toxigenic C. difficile in our research, it appeared that the gene profiles of all positive (n = 29) samples were indicated the presence of toxin B. Four samples were positive for the binary toxin, and no samples were positive for tcdC deletion (i.e., the ribotype 027 strain). Other studies for the 5-plex PCR revealed four different toxin gene profiles: 36 tcdA+, tcdB+, cdtA+ / cdtB+; one tcdA+, tcdB–, cdtA+ / cdtB+; 98 tcdA+, tcdB+, cdtA– / cdtB–; and 24 non-toxigenic tcdA–, tcdB–, cdtA– / cdtB–. Deletion studies revealed that 26 strains contained a c. 700-bp deletion in tcdA, and 39 strains contained at least one possible inactivation feature in tcdC.(Persson, Torpdahl and Olsen, 2008).
The pathogenicity of C. difficile is mainly mediated by two exotoxins: toxin A (TcdA) and toxin B (TcdB). These toxins primarily disrupt the cytoskeletal structure and the tight junctions of target cells causing cell rounding and ultimately cell death. Detectable C. difficile toxemia is strongly associated with fulminant disease. However, besides the well-known intestinal damage, recent animal and in vitro studies have suggested a more far-reaching role for these toxins activity including cardiac, renal, and neurologic impairment. The creation of C. difficile strains with mutations in the genes encoding toxin A and B indicate that toxin B plays a major role in overall CDI pathogenesis. Novel insights, such as the role of a regulator protein (TcdE) on toxin production and binding interactions between albumin and C. difficile toxins, have recently been discovered and will be described.
All isolated samples (n = 29) were investigated for vancomycin, metronidazole, tetracycline, and moxifloxacin sensitivity. However, one positive sample failed to grow, bringing the total number of isolates investigated to 28. All isolates demonstrated vancomycin, and metronidazole sensitivity, 89.3% demonstrated tetracycline sensitivity, and 96.4% demonstrated moxifloxacin sensitivity.
Hudhaiah and Elhadi reported that the sensitivity to vancomycin, metronidazole, and moxifloxacin was 96.6%, 96.6%, and 97.7%, respectively (Hudhaiah and Elhadi, 2019). Saber et al. found that the sensitivity to vancomycin, metronidazole, moxifloxacin, and tetracycline was 100%, 100%, 48.6%, and 21.6%, respectively (Saber et al., 2020). The sensitivity to vancomycin, metronidazole, and moxifloxacin in Thailand is 100%, 100%, and 78.1%, respectively (Papanin Putsathit et al., 2017). In Iran, the sensitivity to vancomycin, metronidazole, and moxifloxacin being 100%, 100, and 61.1%, respectively. Furthermore, ribotype 027 has not been detected in Iran (Shoaei et al., 2019).
In Egypt, the sensitivity to vancomycin seen in 18 isolates (66.7%) and resistance in 1 (3.7%). Metronidazole sensitivity was observed in 13 isolates (48.2%) and resistance in 13 (48.2%). Moxifloxacin sensitivity was observed in 20 isolates (74.1%) and resistance in 5 (18.5%). Tetracycline sensitivity was observed in 7 isolates (25.9%) and resistance in 18 (66.7%) (Elgendy et al., 2020). Multiple studies on the antimicrobial resistance of C. difficile isolates from North America, Europe, and Asia in the last 15 years have demonstrated that the rates of moxifloxacin resistance of C. difficile isolates varied from 2% to 87%, and the rates of clindamycin resistance ranged from 15% to 97% (Tenover, Tickler and Persing, 2012). Almost 30% of ribotype 027 strains were resistant to multiple drugs, including clindamycin, moxifloxacin, and rifampin in North America, using the CLSI breakpoints for susceptibility testing of anaerobic bacteria (Tenover, Tickler and Persing, 2012). In a retrospective study of the antibiotic resistance pattern in the United States, approximately 98% of ribotype 027 strains were resistant to moxifloxacin; moreover, almost half of these isolates possessed high-level resistance based on the CLSI breakpoint (Wieczorkiewicz et al., 2016). Clostridium difficile strains of ribotype 078 (another hypervirulent genotype) isolated from humans and piglets in the Netherlands with active CDI showed resistance to ciprofloxacin, erythromycin, imipenem, and moxifloxacin according to the CLSI breakpoints (Keessen et al., 2013). Worldwide surveillance also indicated the emergence of Clostridium difficile strains resistant to multiple antibiotics in the past decade (Spigaglia, 2016).
The resistance of C. difficile to commonly used antibiotics for bacterial infections not only contributes to the occurrence/recurrence of CDI but also plays an important role in driving epidemiological changes and the emergence of new strain types (Spigaglia, 2016). Antibiotic resistance to C. difficile also leads to suboptimal clinical outcomes and may even lead to treatment failures of CDI. When uncommon antibiotics are chosen for the treatment of CDI, collateral damage to microbiota may occur and should not be ignored.
5 Conclusion
The prevalence of CDI detected in KSMC was 9.3%, lower than the prevalences reported in the Eastern region and Taif city of 23% and 13.6%, respectively. The antimicrobial susceptibility of the 28 strains revealed that 100% were sensitive to vancomycin and metronidazole, while 96.4% and 89.3% were sensitive to moxifloxacin and tetracycline, respectively. In the Eastern region and Taif city, 96.6% and 100% of samples were sensitive to vancomycin and metronidazole, respectively. Sensitivity to moxifloxacin was 97.7%, and 48.6%, respectively. In Taif city, tetracycline sensitivity was observed in 21.6% of samples. In the current study, most isolated strains were toxigenic (9.3%) . The ribotype 027 strain was not detected. While the prevalence and antibiogram of CDI differ between countries, a significant finding of the current study was that most isolated toxigenic strains were susceptible to most of the studied antimicrobials. Further studies are required to detect the ribotyping of the isolates. Limiting the prescription of broad-spectrum antibiotics is required to reduce the prevalence of CDI, particularly among older patients. Further studies in different regions of KSA are needed to compare the prevalence and findings of this study.
Funding
This work was completed without funding.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Molecular characterization of Clostridium difficile isolated from carriage and association of its pathogenicity to prevalent toxic genes. Microb. Pathog.. 2018;120:1-7.
- [CrossRef] [Google Scholar]
- A54T polymorphism in the fatty acid binding protein 2 studies in a Saudi population with type 2 diabetes mellitus. Lipids Health Dis.. 2014;13(1):1-6.
- [Google Scholar]
- Clostridioides (Clostridium) difficile-associated disease: Epidemiology among patients in a general hospital in Saudi Arabia. Am. J. Infect. Control. 2020;000:1-6.
- [CrossRef] [Google Scholar]
- Emergence of a highly resistant Clostridium dif- ficile strain (NAP/BI/027) in a tertiary care center in Saudi Arabia. Ann. Saudi Med.. 2013;33(2):198-199.
- [CrossRef] [Google Scholar]
- Clostridium difficile infection in Europe: a hospital-based survey. 2011;377:63-73.
- Laboratory Tests for the Diagnosis of Clostridium difficile. Clin. Colon Rectal Surg.. 2020;33(2):73-81.
- [CrossRef] [Google Scholar]
- CDC;NHSN 2022. ‘Multidrug-Resistant Organism & Clostridioides difficile Infection (MDRO / CDI) Module’, (January), pp. 1–52. Available at: https://www.cdc.gov/nhsn/pdfs/pscmanual/12pscmdro_cdadcurrent.pdf.
- Clinical and microbial characterization of toxigenic clostridium difficile isolated from antibiotic associated diarrhea in Egypt. Iranian Journal of Microbiology. 2020;12(4):296-304.
- [CrossRef] [Google Scholar]
- Prevalence of the Clostridium difficile BI/NAP1/027 strain across the United States Veterans Health Administration. Clinical Microbiology and Infection. Elsevier Ltd. 2018;24(8):877-881.
- [CrossRef] [Google Scholar]
- ‘The emergence of multidrug-resistant and hypervirulent Clostridium difficile clinical isolates’. Meta Gene. Elsevier. 2020;24(December 2019):100644
- [CrossRef] [Google Scholar]
- Successful treatment of pseudomembranous colitis with fecal microbiota transplantation – A case study on a patient rescued by extracorporeal cardiopulmonary resuscitation after cardiac arrest. Ann. Transplant.. 2020;25:1-5.
- [CrossRef] [Google Scholar]
- Prevalence and Genotypes of Nosocomial Clostridium difficile Infections in the Eastern Province of the Kingdom of Saudi Arabia: A Multi-Centre Prospective Study. J. Clin. Diagn. Res.. 2019;13(3):16-20.
- [CrossRef] [Google Scholar]
- Antimicrobial susceptibility profiles of human and piglet Clostridium difficile PCR-ribotype 078. Antimicrob. Resist. Infect. Control. 2013;2(1):1-6.
- [CrossRef] [Google Scholar]
- Genetic confirmation of T2DM meta-analysis variants studied in gestational diabetes mellitus in an Indian population. Diabetes Metab. Syndr.. 2019;13(1):688-694.
- [Google Scholar]
- Outcomes of Clostridium difficile-suspected diarrhea in a French university hospital To cite this version : HAL Id : hal-01950041 Outcomes of Clostridium difficile -suspected diarrhea in a French university hospital. Eur. J. Clin. Microbiol. Infect. Dis. 2018
- [Google Scholar]
- ‘The emergence of metronidazole and vancomycin reduced susceptibility in Clostridium difficile isolates in Iran’. Journal of Global Antimicrobial Resistance Taibah University. 2019;18:28-33.
- [CrossRef] [Google Scholar]
- Structure and assembly of the S-layer in C. difficile. Nat. Commun.. 2022;13(1):1-13.
- [CrossRef] [Google Scholar]
- Reclassification of Clostridium difficile as Clostridioides difficile (Hall and O’Toole 1935) Prévot 1938. Anaerobe. 2016;40:95-99.
- [CrossRef] [Google Scholar]
- ‘Perception of quality of life in people experiencing or having experienced a Clostridioides difficile infection: a US population survey’. Journal of Patient-Reported Outcomes Journal of Patient-Reported Outcomes. 2020;4(1):1-11.
- [CrossRef] [Google Scholar]
- ‘Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity’. Science (New York N.Y.). United States. 2013;339(6123):1084-1088.
- [CrossRef] [Google Scholar]
- Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA) Clin. Infect. Dis.. 2018;66(7):e1-e48.
- [CrossRef] [Google Scholar]
- ‘Differences in fecal microbiota in different European study populations in relation to age, gender, and country: a cross-sectional study’. Applied and environmental microbiology United States. 2006;72(2):1027-1033.
- [CrossRef] [Google Scholar]
- Natarajan, M. et al. 2015. ‘Gender Differences in Non-Toxigenic Clostridium difficile Colonization and Risk of Subsequent C. difficile Infection.’, Clinical research in infectious diseases, 2(2), pp. 1–14. Available at: http://www.ncbi.nlm.nih.gov/pubmed/28713874%0Ahttp://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=PMC5508598.
- Prevalence of Clostridium difficile and its toxins in hospital patients with diarrhoeal diseases in Lusaka, Zambia. Trans. R. Soc. Trop. Med. Hyg.. 2020;114(2):86-90.
- [CrossRef] [Google Scholar]
- Large Clostridial Toxins: Mechanisms and Roles in Disease. Microbiol. Mol. Biol. Rev.. 2021;85(3):1-30.
- [CrossRef] [Google Scholar]
- ‘New multiplex PCR method for the detection of Clostridium difficile toxin A (tcdA) and toxin B (tcdB) and the binary toxin (cdtA/cdtB) genes applied to a Danish strain collection’. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases England. 2008;14(11):1057-1064.
- [CrossRef] [Google Scholar]
- ‘Antimicrobial susceptibility of Clostridium difficile isolated in Thailand’. Antimicrobial Resistance and Infection Control Antimicrobial Resistance & Infection Control. 2017;6(1):1-6.
- [CrossRef] [Google Scholar]
- Putsathit, P. et al. 2017 ‘Prevalence and molecular epidemiology of Clostridium difficile infection in Thailand’, New Microbes and New Infections. Elsevier Ltd, 15, pp. 27–32. doi: 10.1016/j.nmni.2016.10.004.
- Riley, T. V. and Kimura, T. 2018. ‘The Epidemiology of Clostridium difficile Infection in Japan: A Systematic Review’, Infectious Diseases and Therapy. Springer Healthcare, 7(1), pp. 39–70. doi: 10.1007/s40121-018-0186-1.
- Depression, antidepressant medications, and risk of Clostridium difficile infection. BMC Med.. 2013;11(1)
- [CrossRef] [Google Scholar]
- ‘Prevalence, toxin gene profile, genotypes and antibiotic susceptibility of Clostridium difficile in a tertiary care hospital in Taif, Saudi Arabia’. Indian Journal of Medical Microbiology Indian Journal of Medical Microbiology. 2020;38(2):176-182.
- [CrossRef] [Google Scholar]
- Infectious Nosocomial Diarrhea in the Surgical Wards: Role of Parasites and Microbes Imply Stool Analysis. Journal of Taibah University Medical Sciences. 2009;4(1):73-81.
- [CrossRef] [Google Scholar]
- Evaluation of cycle threshold, toxin concentration, and clinical characteristics of clostridioides difficile infection in patients with discordant diagnostic test results. J. Clin. Microbiol.. 2020;58(5):1-11.
- [CrossRef] [Google Scholar]
- ‘Molecular epidemiology of Clostridium difficile infection in Iranian hospitals 11 Medical and Health Sciences 1117 Public Health and Health Services’. Antimicrobial Resistance and Infection Control Antimicrobial Resistance & Infection Control. 2019;8(1):6-13.
- [CrossRef] [Google Scholar]
- Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Therapeutic Advances in Infectious Disease. 2016;3(1):23-42.
- [CrossRef] [Google Scholar]
- Antimicrobial-resistant strains of Clostridium difficile from North America. Antimicrob. Agents Chemother.. 2012;56(6):2929-2932.
- [CrossRef] [Google Scholar]
- Fluoroquinolone and macrolide exposure predict Clostridium difficile infection with the highly fluoroquinolone- and macrolide-resistant epidemic C. difficile strain BI/NAP1/027. Antimicrob. Agents Chemother.. 2016;60(1):418-423.
- [CrossRef] [Google Scholar]
- Willey, J. M. 2019. Prescott’s Microbiology 11th Edition. 11th edn. Edited by McGraw-Hill Education, McGraw-Hill Education.
- ‘Gender bias in autoimmunity is influenced by microbiota’., Immunity. United States. 2013;39(2):400-412.
- [CrossRef] [Google Scholar]
- ‘Clostridioides difficile biology: Sporulation, germination, and corresponding therapies for C. difficile infection’. Front. Cell. Infect. Microbiol.. 2018;8(FEB):1-10.
- [CrossRef] [Google Scholar]







